Abstract
The alternative sigma factor σB has an important role in the acquisition of stress resistance in many gram-positive bacteria, including the food-borne pathogen Bacillus cereus. Here, we describe the identification of the set of σB-regulated genes in B. cereus by DNA microarray analysis of the transcriptome upon a mild heat shock. Twenty-four genes could be identified as being σB dependent as witnessed by (i) significantly lower expression levels of these genes in mutants with a deletion of sigB and rsbY (which encode the alternative sigma factor σB and a crucial positive regulator of σB activity, respectively) than in the parental strain B. cereus ATCC 14579 and (ii) increased expression of these genes upon a heat shock. Newly identified σB-dependent genes in B. cereus include a histidine kinase and two genes that have predicted functions in spore germination. This study shows that the σB regulon of B. cereus is considerably smaller than that of other gram-positive bacteria. This appears to be in line with phylogenetic analyses where σB of the B. cereus group was placed close to the ancestral form of σB in gram-positive bacteria. The data described in this study and previous studies in which the complete σB regulon of the gram-positive bacteria Bacillus subtilis, Listeria monocytogenes, and Staphylococcus aureus were determined enabled a comparison of the sets of σB-regulated genes in the different gram-positive bacteria. This showed that only three genes (rsbV, rsbW, and sigB) are conserved in their σB dependency in all four bacteria, suggesting that the σB regulon of the different gram-positive bacteria has evolved to perform niche-specific functions.
The ability of bacteria to respond rapidly to changing environmental conditions is a prerequisite for survival in their habitats. This bacterial stress response is triggered by a change in the microorganism's growth conditions. Such a change triggers a cascade of events that will lead to the increased stress resistance of the bacterial cell, most often against not only the stress to which it was exposed but also other stresses, thereby ensuring its survival under a variety of conditions. A common strategy that bacteria use to counter stressful conditions is to activate a specific alternative sigma factor, which leads to the transcription of a set of genes (a so-called regulon), the products of which protect the cell against adverse conditions (17). In several gram-positive bacteria, the alternative sigma factor σB is the key sigma factor controlling the stress response (17, 26, 36).
In the gram-positive model organism Bacillus subtilis and the human pathogens Listeria monocytogenes and Staphylococcus aureus, numerous studies have shown that the activation of σB leads to an increased resistance to stress and that in pathogenic Listeria and Staphylococcus species, σB has a role in virulence (17, 36). In the bacteria that comprise the physiologically diverse Bacillus cereus group, the role of σB has also been studied. The most prominent members of the B. cereus group are B. anthracis (a mammalian pathogen that causes the disease anthrax), B. thuringiensis (an insect pathogen that is widely used as a biopesticide), and B. cereus, which is a common cause of food-borne infections (15). Recent genomics studies have revealed that these bacteria share a common core set of genes and that their phenotypic characteristics are determined largely by the presence or absence of large plasmids that may encode different virulence factors (30). Throughout the B. cereus group, the genes encoding σB and the regulators of its activity are conserved. In B. cereus, σB was shown to be activated under a number of stress conditions but most strongly upon a mild heat shock from 30°C to 42°C. Furthermore, the deletion of the sigB gene affected the heat-adaptive response of vegetative cells and some spore properties, most notably germination characteristics (4, 33). Also, in B. anthracis, σB was activated upon a heat shock, and the B. anthracis sigB deletion mutant was shown to have slightly lower virulence than the parental strain (8).
The regulation of σB activity in gram-positive bacteria occurs via a so-called partner-switching mechanism. Under nonstress conditions, σB is held in an inactive state by sequestration to an anti-sigma factor (RsbW). Under stress, an anti-sigma factor antagonist, RsbV, binds to RsbW, and this leads to the release of σB from its complex with RsbW. Subsequently, σB can bind to core RNA polymerase, leading to the transcription of σB-dependent genes. RsbW is not only the anti-sigma factor for σB, it also acts as a kinase on RsbV. In its phosphorylated form, RsbV is unable to bind to RsbW and activate σB. However, under stress conditions, the phosphorylated form of RsbV can be dephosphorylated by the action of a PP2C-type phosphatase, which then leads to the formation of the RsbV-RsbW complex and the activation of σB (reviewed in references 17, 26, and 36). In the B. cereus group, the PP2C phosphatase RsbY appears to be the only phosphatase responsible for the activation of σB under stress, as the deletion of the rsbY gene in B. cereus leads to an abolished σB response under several stress conditions, including a heat shock (34).
The identification of the complete σB regulons in B. subtilis (9, 24, 27), L. monocytogenes (16), and S. aureus (1, 22) by DNA microarray technology has revealed that a surprisingly low number of genes has an obvious role in the stress response of these organisms. Instead, σB-dependent specialized metabolic adaptations may contribute to stress resistance. Furthermore, in both L. monocytogenes and S. aureus, pathogenic traits seem to be governed by σB. These findings suggest that σB has evolved in response to specific niches that are populated by different bacteria.
Here, we describe the σB regulon of B. cereus, which was determined by comparing the transcriptome of the parental B. cereus strain ATCC 14579 and its sigB and rsbY deletion mutants upon a mild heat shock. It appears that the σB regulon of B. cereus is considerably smaller than those of other gram-positive bacteria, which may reflect the previously proposed close-to-ancestral state of σB in the B. cereus group (7). In other studies, the sets of σB-regulated genes in B. subtilis (9, 24, 27), L. monocytogenes (16), and S. aureus (1, 22) were determined by DNA microarray technology. This allowed us to compare the σB regulons of B. cereus, B. subtilis, L. monocytogenes, and S. aureus with each other to determine to what extent σB-regulated genes are conserved in the different bacteria and to assess how far the σB dependency of these genes is conserved.
MATERIALS AND METHODS
Strains, growth conditions, and RNA isolation.
B. cereus strain ATCC 14579 and its isogenic derivatives, the sigB deletion mutant FM1400 (33) and the rsbY deletion mutant FM1401 (34), were used in this study. Strains were cultured overnight in 100-ml Erlenmeyer flasks containing 20 ml brain heart infusion broth at 30°C with aeration by rotary shaking at 150 rpm. Subsequently, prewarmed brain heart infusion broth (20 ml in 100-ml Erlenmeyer flasks) was inoculated with 100 μl of a single culture grown overnight. These cultures were then incubated at 30°C as described above. One of the three cultures was used to measure the absorbance at 600 nm (A600). When the A600 of this culture reached 0.4 (mid-exponential growth phase), one of the other two cultures was transferred into a shaking water bath at 42°C and incubated for a further 10 min, which was followed by RNA isolation. We previously found that at 10 min after the start of the heat shock, the levels of σB protein reached maximum levels (34), which leads to high-level expression of σB-dependent genes at this time point. RNA was isolated immediately from the culture in the mid-exponential growth phase. RNA isolation was performed by transferring the cultures into a 50-ml Falcon tube, and the cultures were spun down at 13,000 × g for 20 s. After decanting the supernatant, the cell pellets were snap-frozen in liquid nitrogen. The time between removal from the incubator and freezing of the cell pellets was approximately 60 s. Within 20 min after freezing the cell pellets, RNAwiz (Ambion, Huntingdon, United Kingdom) was added to the pellets, and RNA was extracted as described previously (33). Residual chromosomal DNA was removed by treating samples with DNA-free (Ambion). Extracted RNA samples were stored in 70% ethanol-83 mM sodium acetate buffer (pH 5.2) at −80°C.
cDNA synthesis and microarray design and hybridization.
Before further analysis, the RNA samples were spun down at 13,000 rpm in a microtube centrifuge prechilled at 4°C. The RNA pellet was then washed with 70% ethanol (prechilled at −20°C) and resuspended in RNase-free water. Prior to cDNA synthesis, the quality of the extracted RNA was determined by analysis with an Agilent (Palo Alto, CA) 2100 Bioanalyzer according to the manufacturer's instructions. cDNA with amino-allyl-labeled dUTP (Ambion) was prepared in reverse transcription reactions using Superscript III (Invitrogen, Breda, The Netherlands). Cy3 and Cy5 labeling of the cDNAs was performed with the CyScribe Post-Labeling kit (GE Healthcare, Diegem, Belgium) as previously described (3). Labeled cDNAs were purified using the CyScribe GFX purification kit (GE Healthcare). Final cDNA yields and incorporation of the Cy labels were determined by Nanodrop analysis (Isogen Life Science, IJsselstein, The Netherlands). Custom-made B. cereus microarrays (produced by Agilent) (see below for details) were hybridized with 200 to 300 ng labeled cDNA. The microarray experiments for the comparison of the transcriptomes of the wild-type (WT) strain and the sigB and rsbY mutants were performed with three and two independent biological replicates, respectively. Microarray experiments (using two biological duplicates) were also performed to identify the set of genes that were differentially expressed in B. cereus ATCC 14579 after a heat shock from 30°C to 42°C for 10 min.
The microarrays used in this study were custom-made B. cereus ATCC 14579 microarrays using the 11K platform developed by Agilent Technologies. A total of 10,263 spots represented 5,058 B. cereus open reading frames, meaning that 96.6% of the predicted chromosomal B. cereus ATCC 14579 open reading frames (NCBI accession number NC_004722) are represented on the microarray. Ninety-five percent of the open reading frames for which probes could be designed were represented by two or three nonoverlapping probes on the array. The remaining 5% of the open reading frames were represented by a single oligonucleotide. After hybridization at 60°C for 17 h, the microarrays were washed according to the manufacturer's instructions with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.005% Triton X-102 at room temperature for 10 min and with prechilled (at 4°C) 0.1× SSC-0.005% Triton X-102 for 5 min. Slides were then scanned using an Agilent microarray scanner (G2565BA), and data were extracted from the scanned microarrays with Feature Extraction software, which includes a Lowess normalization step for the raw data.
Analysis of microarray data.
After removal of the data for the different controls printed on the microarray slides, the normalized data for each spot from the microarrays were analyzed for statistical significance using the Web-based VAMPIRE microarray suite (12, 13). A spot was found to be differentially expressed between two samples using the threshold of a false discovery rate smaller than 0.05. Subsequently, the data for the single spots were integrated to obtain expression ratios for an open reading frame. An open reading frame was found to be differentially expressed when all spots representing the open reading frame were significantly differentially expressed between samples. The average expression ratio was determined by calculating the average of the log values of the expression ratios over all experiments. This value (R) was then used to calculate the average expression ratio (10R). Finally, changes of 1.5-fold (for upregulated genes in the parental strain) and 0.67-fold (for downregulated genes in the parental strain) were also introduced as significance limits.
Validation of microarray data.
The expression ratios of six genes were determined using quantitative real-time PCR (qPCR) with the oligonucleotides listed in Table 1 on the RNA samples isolated from strains ATCC 14579 and FM1400. cDNA synthesis and qPCR were performed as described previously, with tufA as a reference gene (34). All qPCR reactions were performed in triplicate. Expression ratios between the strains were determined by REST-tool (25).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| tufAF | GCCCAGGTCACGCTGACTAT |
| tufAR | TCACGTGTTTGAGGCATTGG |
| BC0376F | GTAAACCGCCCTGGTGAAGA |
| BC0376R | GGAGCGCGTCCACTTACTTG |
| BC1008F | AAAGAGATTCACCCACTCATTGC |
| BC1008R | TTGCTCCTCAGTGTTACGGAAGT |
| BC1550F | CTACGGTCCGGCAATTGAAA |
| BC1550R | TCACCTTTGTAACTCTTTGTTGCATT |
| BC4641F | TGGTTGGAAATGGCATACACA |
| BC4641R | CGACAACTCCAGCAGCCATA |
| BC5149F | CCGCTCATACAATGGCAACA |
| BC5149R | CGGCTCCAGCGAAATCAAT |
| BC5391F | ACGTGGCAGTTCCCTTGGTA |
| BC5391R | GTTTTCCTGATTGCGGTTCA |
Prediction of σB-dependent promoter sites.
The prediction of σB-dependent promoter sites in B. cereus was performed by using the 500-bp upstream regions of the 26 genes identified in the σB regulon. Subsequently, the upstream regions were analyzed with AlignACE 3.1 (31) and DBTBS 4.1 (20) with a threshold of 5%. A total of 12 putative σB-dependent promoter sites were identified by these methods. These promoter sites were aligned with MUSCLE 3.6 (6), and a hidden Markov model (HMM) was constructed with the HMMER package (5). The HMM was used for a first screening for σB-dependent promoters in the genome sequence of B. cereus ATCC 14579 (14). Hits that were more than 300 nucleotides upstream of the start codon of an open reading frame or that had an E score higher than 1 × 10−3 were discarded. Two of the putative promoter sites identified in the first round of the HMMER search did not meet these standards. Therefore, an iteration of the alignment, the HMM, and the genome search were performed with the 10 remaining promoters. Predicted sites that were more than 300 nucleotides upstream of the start codon of an open reading frame or that had an E score higher than 1 × 10−3 were again discarded. The 10 σB-dependent promoter sites upstream of genes belonging to the σB regulon that were identified by this approach were aligned, and this alignment was visualized with WebLogo (2).
Comparison of the σB regulons of gram-positive bacteria.
To determine the conservation of σB-dependent genes in gram-positive bacteria, the set of genes that is directly regulated by σB was compiled for B. cereus (this study), B. subtilis (9, 24, 27), L. monocytogenes (16), and S. aureus (1). Direct regulation was defined as genes for which σB dependency was determined by experimental confirmation (for example, by promoter mapping through primer extension analysis) or by the prediction of a σB-dependent promoter (for example, by the application of an HMM to identify σB-dependent promoters) upstream of the gene. To obtain the set of genes directly regulated by σB in B. subtilis, three different studies in which the σB regulon was determined were compared, and only genes that were found to be σB dependent in at least two of the three studies were used in this comparative analysis. For the definition of the genes that were directly regulated by σB in S. aureus, data from the study described previously by Bischoff et al. (1) were used. Only genes directly regulated by σB and that had a homolog in S. aureus N315 (18) were included in the analysis. The more limited study of the σB regulon described previously by Pane-Farre et al. (22) is not further discussed here, but a comparison of the results from this σB regulon with those of the other bacteria is included in the Table S4 in the supplemental material and is in line with the data described previously by Bischoff et al. (1).
TIGR's Multi-Genome Homology Comparison tool (http://cmr.tigr.org/tigr-scripts/CMR/shared/MakeFrontPages.cgi?page=circular_display), applying a P value of ≤1.0 × 10−5 for significance, was used for comparing the set of σB-dependent genes of each organism.
Microarray data accession number.
Microarray data were submitted to the GEO database under accession number GSE6005.
RESULTS AND DISCUSSION
Validation of transcriptome data by qPCR.
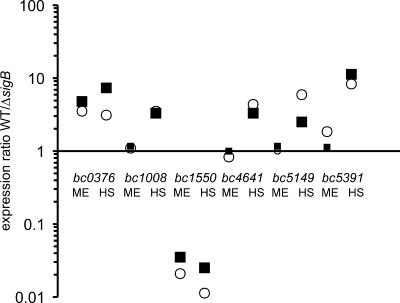
Because this is the first report of the use of B. cereus whole-genome microarrays, it was deemed essential to first validate the expression ratios generated by microarray analysis by qPCR. We used microarray data that were obtained in the comparison of the sigB mutant and its parental strain during mid-exponential growth at 30°C and upon a heat shock to 42°C for 10 min. We chose six genes with significantly different expression ratios under at least one growth condition, and the expression ratios found by microarray experiments exhibited a good correlation with the qPCR data (Fig. 1).
FIG. 1.
Validation of microarray results by qPCR. Three independent RNA samples were isolated from cultures of B. cereus strain ATCC 14579 (WT) and the sigB deletion mutant during mid-exponential growth at 30°C (ME) and after a heat shock to 42°C for 10 min (HS). The expression ratios between both strains of the indicated genes were measured using microarrays (closed squares) and qPCR (open circles). Small symbols indicate that no significant difference was measured for the expression levels of the target gene between the two strains.
Identification of σB-dependent genes of B. cereus upon mild heat shock.
To identify the set of σB-regulated genes in B. cereus, the transcriptomes of the sigB deletion mutant FM1400 and the rsbY deletion mutant FM1401 were compared with the transcriptome of the parental strain B. cereus ATCC 14579 after a heat shock to 42°C for 10 min. This mild heat shock strongly activates σB activity in the parental strain, while this response does not occur in both mutant strains (33, 34). Both the sigB and rsbY deletion mutants were used in these experiments to minimize the influence of mutant-specific, pleiotropic effects, which could give rise to false-positive results in the definition of the B. cereus σB regulon under this condition.
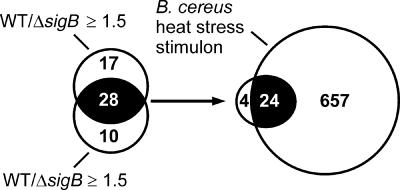
We observed that upon a heat shock to 42°C for 10 min, 45 genes were significantly upregulated in the parental strain in comparison with the sigB mutant (Fig. 2). Similarly, 38 genes were found to be upregulated after an identical heat shock in the WT strain compared to the rsbY mutant. Expression data for these experiments can be found in Table S1 in the supplemental material. A total of 28 genes from these two data sets were found to be overlapping, which suggests that these genes are candidates for being dependent on σB for their expression. No genes with expression ratios of ≤0.67 (WT/mutant) were found in either the sigB or the rsbY mutant, so neither σB nor σB-dependent proteins appear to have a role in the downregulation of genes in B. cereus upon a heat shock.
FIG. 2.
Comparison of differentially expressed genes between B. cereus ATCC 14579 (WT) and the sigB and rsbY deletion mutants. The numbers of genes that are differentially expressed between the parental strain (WT) and the rsbY and sigB mutants upon a heat shock to 42°C for 10 min are indicated in the circles. The black overlapping area indicates that the same genes were identified as being differentially expressed in both the sigB and the rsbY deletion mutants under the indicated conditions. Twenty-four genes were also upregulated upon a heat shock, and these genes form the σB regulon of B. cereus.
A further defining feature of a σB-dependent gene is that the gene exhibits upregulated expression upon stress. Therefore, we subsequently performed microarray experiments to define the set of genes that are upregulated upon a heat shock from 30°C to 42°C for 10 min in the parental strain B. cereus ATCC 14579 and compared these data sets (see Table S2 in the supplemental material) with the above-described set of putative σB-dependent genes (Fig. 2). In total, 681 genes were upregulated upon a heat shock from 30°C to 42°C for 10 min, which corresponds to 12.4% of the total number of genes in B. cereus. This substantial transcriptional reprogramming corresponds to data for the heat shock response in B. subtilis, where over 10% of the total number of genes was induced upon a heat shock (9). The genes that were upregulated upon a heat shock in B. cereus include dnaK, groEL, groES, and the different clp genes, which is in line with data from a previous proteome-based study of the heat shock response of B. cereus (23). Twenty-four of the 28 previously identified genes are also part of the B. cereus heat stress stimulon, and these genes were thus assigned to the σB regulon of B. cereus (Table 2). Indeed, the expression ratios between mid-exponentially-growing and heat-shocked B. cereus ATCC 14579 (Table 2) are practically identical to the expression ratios when the mutant and parental strains were compared with one another upon a heat shock (Table 2). This reflects both the inactivity of σB under exponential growth conditions and the absence of a σB response in the mutants. Four genes (bc1436, bc5148, bc5149, and bc5150), which exhibited higher levels of expression in B. cereus ATCC 14579 than in the sigB and rsbY mutants, were not assigned to the σB regulon, as they were not upregulated in their expression upon a heat shock in the parental strain.
TABLE 2.
Identification of σB-regulated genes in B. cereus ATCC 14579 upon mild heat shocka
| Gene | Expression ratio
|
Annotation | Aliasb | Experimentally defined and/or predicted promoter sequencec | E score by HMMER search | ||
|---|---|---|---|---|---|---|---|
| WT/ΔsigB | WT/ΔrsbY | 42°C/30°C | |||||
| bc0862 | 63.1 | 84.0 | 72.9 | Protease I | yfkM | In operon with bc0863 | |
| bc0863 | 72.1 | 31.5 | 34.0 | Catalase | katE | ATGTTTAC-13 bp-GGGTATC-N47-ATG | 4.7 × 10−5 |
| bc0995 | 1.9 | 2.0 | 2.2 | Hypothetical protein | None identified | ||
| bc0996 | 2.1 | 1.8 | 2.0 | Hypothetical protein | None identified | ||
| bc0998 | 123.6 | 86.0 | 70.9 | General stress protein | yflT | ATGTTTAA-14 bp-GTGTACT-N45-ATG | 6.4 × 10−5 |
| bc0999d | ND | ND | ND | Hypothetical protein | In operon with bc0998 | ||
| bc1000 | 98.1 | 42.4 | 36.3 | Hypothetical protein | In operon with bc0998 | ||
| bc1001 | 106.4 | 29.8 | 35.8 | Hypothetical protein | ATGTTTAA-13 bp-GTGTATG-N37-ATGf | 1.3 × 10−5 | |
| bc1002 | 42.6 | 31.7 | 38.3 | Anti-σB factor antagonist | rsbV | ATGTTTAA-13 bp-GGGTAAT-N33-ATG | 1.4 × 10−5 |
| bc1003 | 51.1 | 38.3 | 52.4 | Anti-σB factor | rsbW | In operon with bc1002 | |
| bc1004 | —e | 24.4 | 28.1 | RNA polymerase sigma factor σB | sigB | In operon with bc1002 | |
| bc1005 | 10.6 | 47.4 | 56.0 | Putative bacterioferritin | orf4 | In operon with bc1002 and under control of own promoter, ATGTTTAA-13 bp-GGGTACT-N20-ATG | 6.3 × 10−6 |
| bc1006 | 4.0 | —e | 5.1 | PP2C phosphatase, regulator of σB activity | rsbY | In operon with bc1002 and bc1005 | |
| bc1007 | 2.4 | 2.3 | 3.6 | Chemotaxis protein methyltransferase | In operon with bc1009 | ||
| bc1008 | 3.3 | 3.1 | 4.4 | Two-Component System Histidine Kinase | In operon with bc1009 | ||
| bc1009 | 188.5 | 110.2 | 87.5 | Hypothetical Protein | ATGTTTAA-13 bp-GGGTATG-N40-ATG | 1.1 × 10−5 | |
| bc1010 | 8.2 | 7.0 | 5.5 | Hypothetical Protein | ACGTTTAG-13 bp-GGGTATA-N285-ATG | 1.9 × 10−4 | |
| bc1011 | 33.9 | 25.8 | 23.7 | Hypothetical Protein | In operon with bc1010 | ||
| bc1012d | ND | ND | ND | Hypothetical Protein | ybjQ | In operon with bc1010 | |
| bc2108 | 2.9 | 2.8 | 3.8 | RNA polymerase ECF-type sigma factor | sigZ | None identified | |
| bc2638 | 4.7 | 4.4 | 4.3 | Spore germination protein LC | None identified | ||
| bc3130 | 20.7 | 29.3 | 10.3 | Hypothetical protein | In operon with bc3132 | ||
| bc3131 | 29.5 | 37.3 | 42.6 | Hypothetical protein | In operon with bc3132 | ||
| bc3132 | 7.6 | 2.0 | 2.0 | General stress protein 17M | AGGTTTAA-14 bp-GTGTATT-N48-ATG | 4.3 × 10−5 | |
| bc4641 | 3.3 | 3.9 | 3.8 | Hypothetical protein | ATGAATAA-13 bp-GGGTACG-N10-ATG | 9.3 × 10−5 | |
| bc5391 | 11.2 | 11.0 | 6.1 | Spore coat protein | gerQ | None identified | |
Genes were identified as being σB dependent by a significant difference in expression between the parental strain and both the sigB and rsbY deletion mutants upon a heat shock from 30°C to 42°C for 10 min. ND, not determined; ECF, extracytoplasmic function.
The aliases for the identified genes were based on the gene names of homologs in B. subtilis and/or B. anthracis.
Experimentally confirmed promoter sites (33, 35) are indicated in boldface type. Other promoters were identified by an HMM as outlined in Materials and Methods.
These genes were not represented on the microarray but were previously identified as being σB dependent (35) and are therefore included in this table.
These genes were disrupted by an erythromycin resistance cassette. Therefore, the expression data for these genes were omitted.
A second σB-dependent promoter site was predicted in the HMMER search upstream of this gene. The E score for this predicted site is higher (3.0 × 10−5) (see Table S3 in the supplemental material) than that for the experimentally confirmed promoter indicated in the table.
Function of σB-dependent genes in B. cereus.
The set of σB-dependent genes, which were identified by microarray analysis, includes genes that were previously identified in a study using a combined proteomics- and in vitro transcription-based approach (35), but a number of additional genes can now also be assigned to the σB regulon of B. cereus (Table 2). Note that two genes (bc0999 and bc1012) that were not represented on the DNA microarray are included in Table 2, as they were previously experimentally proven to be σB dependent (35). Τhe newly identified genes include bc1007, which encodes a predicted CheR-type methyltransferase, and bc1008, a histidine kinase fused to a Chase3-like N-terminal domain (37) and a C-terminal CheY-type response regulator domain, which could be involved in a complex regulatory route in the B. cereus group. A large proportion of genes in the B. cereus σB regulon appears to have no known function. However, three of these genes have homologs in other bacteria, where they have a role in the stress response. These are bc0998, which has a homolog in B. subtilis (yflT), where it has a role in resistance to ethanol and hyperosmotic conditions (10); bc0999, which is homologous to csbD in B. subtilis and which has a role in hyperosmotic and cold stress (10); and bc1000, which is homologous to the GlsB protein of Enterococcus faecalis, where it has a role in resistance to bile (32). It is interesting that in the region delimited by bc0995 and bc1012 (a 12.4-kb fragment), σB is involved in the expression of 16 of the 18 open reading frames in this region. This region also includes the genes encoding all known regulators of the σB activity of B. cereus. Possibly, the clustering of both regulators and important σB-dependent genes can be advantageous, or it may be a remainder of an early event in the evolution of σB in gram-positive bacteria.
Another newly identified member of the σB regulon of B. cereus is bc2108, which encodes the extracytoplasmic function-type sigma factor σZ. This apparent regulation of the transcription of bc2108 by σB may very well be indirect, as no candidate σB promoter site could be identified upstream of bc2108. Two other genes (bc2638 and bc5391), which also appear to be indirectly dependent on σB for their expression, have predicted roles in the processes of germination and sporulation. The bc2638 gene is predicted to encode the C subunit of a germination receptor. These receptors are important for sensing nutrients and triggering the germination of the bacterial spore (21). The bc2638 gene is not part of one of the seven germination receptor operons of B. cereus (11), and conceivably, the encoded protein can act together with A and B subunits of other germination receptor complexes. The bc5391 gene also has a proposed role in germination. The B. subtilis homolog of this gene (named gerQ or ywdL) is essential for the presence of the cortex-lytic enzyme CwlJ in the spore coat (29). In B. subtilis, GerQ is cross-linked into high-molecular-mass forms by a transglutaminase. This may be important for building a spore coat that contributes to the resistance of the spore to environmental insults (28). Consequently, the germination defect and decreased heat resistance of the spores of the sigB deletion mutant of B. cereus (4) may be explained by the lower expression levels of bc2638 and bc5391 during sporulation.
Prediction of σB-dependent promoters.
A HMMER search was performed using the genes of the σB regulon of B. cereus to identify σB-dependent promoters in the B. cereus ATCC 14579 genome (14). The HMMER search identified promoters regulating 21 of the 26 genes belonging to the σB regulon of B. cereus, suggesting that these genes are under the direct control of σB (Table 2). Promoters were identified in front of operons coding for 15 open reading frames of the island that are part of the σB regulon and in front of the bc0863-bc0862 and bc3132-bc3130 operons. Seven out of the 10 predicted promoter sites were experimentally defined previously (33, 35).
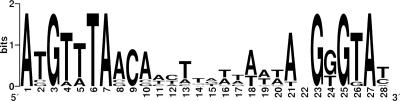
By aligning the predicted and experimentally verified σB-dependent promoter sites in B. cereus, a promoter consensus sequence for σB in B. cereus could be generated (Fig. 3). When the genome of B. cereus ATCC 14579 was searched with this consensus sequence, we found that the promoters of genes identified in this study were all found to have a low E score and therefore a significant fit to the HMM (see Table S3 in the supplemental material). Indeed, the 10 promoters with the lowest E scores (correlating with optimal fit to the HMM) were all identified in this study. Generally, the promoters with the best fit to the HMM have the highest change values (n-fold) in the microarray analysis, suggesting that differences between expression levels of the genes of the σB regulon are determined primarily by the recognition of the promoter sequence by σB. The other promoters are in all likelihood weakly (if at all) dependent on σB for their expression and/or may be expressed under other growth or stress conditions than those used in this study. Even though we cannot completely rule out that under other stress conditions, more σB-dependent genes may be identified, the fact that we identified the 10 promoters with the best fit to the HMM by microarray analysis implies that if additional σB-dependent genes can be identified under other conditions, their dependency on σB will probably be weak. Additionally, it is noteworthy that in our previous proteomics-based study, only a relatively small number of σB-dependent genes could be identified, even though we used an inducible overexpression system in B. cereus to obtain artificially high levels of σB and concomitant strong activation of σB-dependent genes (35). Both findings imply that the σB regulon of B. cereus is not much larger than approximately 25 to 30 genes.
FIG. 3.
Consensus sequence of the σB-dependent promoter sites in B. cereus. The promoter sequences indicated in Table 2 were visualized with WebLogo (2). The height of the nucleotide is indicative of its frequency at that position in the promoter sites. Variable spacing between the −35 and −10 binding sites for σB is signified by the absence of a nucleotide at position 22.
Evolutionary implications of the small σB regulon of B. cereus.
When the findings of this study are compared to the results for other gram-positive bacteria in which transcriptome profiling was used to determine the set of σB-regulated genes, it is striking that the σB regulon of B. cereus appears to be considerably smaller than those of other gram-positive bacteria. In L. monocytogenes, 54 genes were upregulated in a σB-dependent fashion upon stress exposure. However, the total number of σB-dependent genes in L. monocytogenes may be higher, as a microarray with limited genome coverage was used in that study (16). In both S. aureus and B. subtilis, the number of genes in the σB regulon is considerably larger and appears to be around 100 (1, 9, 22, 24, 26).
Possibly, the relatively small size of the σB regulon of B. cereus is a reflection of the way that σB has evolved throughout gram-positive bacteria. The similarities between the operon structures for sigB and sigF (encoding σF, the sigma factor that is active in the prespore) have been used to suggest that both operons have evolved from a common ancestor. Phylogenetic analysis indicates that the sigB operon of the B. cereus group most closely resembles the ancestral operon structure, whereas those of other gram-positive bacteria seem to have evolved further (7). Our finding of the relatively small size of the σB regulon of B. cereus seems to fit this hypothesis. While σB of B. cereus has a role in the development of stress resistance and is activated upon a number of different stress conditions (33), it does not seem to be very important for the regulation of metabolic pathways or the expression of virulence determinants as in the gram-positive pathogens L. monocytogenes and S. aureus (17). Possibly, the evolution of σB has progressed in a modular fashion in which functional groups of genes were added to the σB regulon in the different gram-positive bacteria based on selective pressures encountered in the different environments. In B. cereus, regulatory pathways other than σB may have evolved to regulate metabolic processes and the expression of virulence determinants, leaving B. cereus σB solely with the role of countering acute stresses and with a minor role in the process of sporulation.
Conservation of σB-dependent genes and σB-dependent gene expression in gram-positive bacteria.
In this study, the σB regulon of B. cereus was determined by DNA microarray analysis. This allowed us to compare the set of σB-regulated genes in B. cereus with the previously published σB regulons of B. subtilis, L. monocytogenes, and S. aureus, which encompasses the phylogenetic diversity of gram-positive bacteria that have a bona fide sigB gene (7). We performed this analysis to determine to what extent σB-regulated genes are conserved among gram-positive bacteria and to assess how far the σB dependency of these genes is conserved (Fig. 4).
FIG. 4.
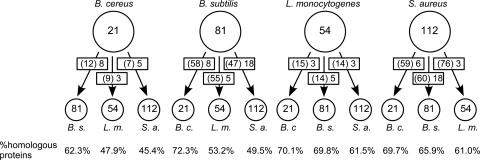
Conservation of genes from the σB regulon in B. cereus, B. subtilis, L. monocytogenes, and S. aureus. The number of genes that are directly regulated by σB in a given organism is shown in the large circle. The conservation of σB-dependent genes from this organism in three other gram-positive bacteria (B. c., B. cereus; B. s., B. subtilis; L. m., L. monocytogenes; S. a., S. aureus) was determined with TIGR's Multi-Genome Homology Comparison tool (http://cmr.tigr.org/tigr-scripts/CMR/shared/MakeFrontPages.cgi?page=circular_display) with a cutoff for significance of a P value of ≤1.0 × 10−5. The numbers of conserved genes of the σB regulon are indicated in the arrows toward the small circles. The numbers in parentheses are the numbers of genes from the σB regulon of the query organism that have a homolog in the target organism. The second number in the arrow indicates the number of genes from this group, which are dependent on σB for their expression in both organisms. For comparative purposes, the conservation of all protein-coding genes between the genomes is indicated at the bottom of the figure.
This comparison showed that genes from the σB regulon are generally conserved among the four gram-positive bacteria to the same extent as all the genes of the genome. For example, of the 81 σB-dependent genes of B. subtilis, 58 genes (71.6%) have a homolog in B. cereus. When all 4,100 genes of B. subtilis are considered, 72.3% have a homolog in B. cereus. It should be noted, however, that σB-dependent genes that are strongly induced upon stress in B. subtilis, such as ctc, gsiB, and gspA, have no homologs in B. cereus. This already suggests that σB-dependent genes that are important for the stress response in one organism are not necessarily conserved in other organisms, even when these organisms are relatively closely related, like B. subtilis and B. cereus. It is also noteworthy that the genes of the σB regulon of L. monocytogenes are relatively poorly conserved in other bacteria. The peculiar lifestyle of L. monocytogenes as a gastrointestinal intracellular pathogen may have led to the divergent evolution of the σB regulon in this organism.
The most important conclusion that can be drawn from Fig. 4 is that even though σB-dependent genes from one organism are generally well conserved in other organisms, their σB dependency generally is not. This is clearly illustrated by comparing the σB regulons of the phylogenetically relatively closely related bacteria B. subtilis and B. cereus. Of the 81 σB-dependent genes of B. subtilis, 58 have a homolog in B. cereus. However, only eight of these σB-dependent B. subtilis genes are also σB dependent in B. cereus. The complete data set for the genes that have conserved their σB-dependent expression in the compared bacteria discussed here can be found in Table S4 in the supplemental material.
Only three genes are conserved in their σB dependency in all bacteria discussed here. These are the sigB gene itself and the genes encoding RsbV and RsbW, the anti-anti-sigma factor and anti-sigma factor of σB, respectively. The bc0999 gene from B. cereus also has σB-dependent homologs in B. subtilis (ywmG or csbD) and S. aureus (sa0772). It is also present in L. monocytogenes (lmo2518), but the σB dependency of this gene has not been described, possibly because this gene was not represented on the microarray used for the determination of the L. monocytogenes σB regulon (16). However, a putative σB-dependent promoter upstream of lmo2518 can be identified (data not shown), which strongly suggests that lmo2518 is dependent on σB for its expression. In that case, it would be the fourth gene that is σB dependent in all four different gram-positive bacteria, and this would be an indication that csbD-like genes may have an important role in the σB-mediated stress response. Interestingly, csbD-like genes are widely distributed in eubacteria, strongly suggesting that they perform an important, but hitherto-unrecognized, function in eubacteria.
The poorly conserved control of stress response genes by σB in the different gram-positive bacteria is in line with data reported in a recent article by Lozada-Chávez et al. (19). That study predicted that bacterial regulatory networks are extremely flexible during evolution and that transcription factors appear to be primarily responsible for the plasticity of transcriptional regulatory networks. The evolution of σB and its regulon in gram-positive bacteria appears to be an experimentally determined proof of this prediction. Even though σB's main function appears to be to control the stress response in different organisms, it appears that it does so by using a unique strategy in each gram-positive bacterium through controlling the expression of different output genes.
Supplementary Material
Acknowledgments
We thank Jos Boekhorst and Mark W. de Been for their assistance in setting up tools for the analysis of the B. cereus microarrays.
Footnotes
Published ahead of print on 6 April 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.den Hengst, C. D., S. A. F. T. van Hijum, J. M. W. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. [DOI] [PubMed] [Google Scholar]
- 4.de Vries, Y. P., L. M. Hornstra, R. D. Atmadja, W. van Schaik, W. M. de Vos, and T. Abee. 2005. Deletion of sigB in Bacillus cereus affects spore properties. FEMS Microbiol. Lett. 252:169-173. [DOI] [PubMed] [Google Scholar]
- 5.Durbin, R., S. R. Eddy, A. Krogh, and G. Mitchison. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, United Kingdom.
- 6.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira, A., M. Gray, M. Wiedmann, and K. J. Boor. 2003. Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other gram-positive bacteria. Curr. Microbiol. 48:39-46. [DOI] [PubMed] [Google Scholar]
- 8.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoper, D., U. Volker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornstra, L. M., Y. P. de Vries, M. H. J. Wells-Bennik, W. M. de Vos, and T. Abee. 2006. Characterization of germination receptors of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 72:44-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiao, A., T. Ideker, J. M. Olefsky, and S. Subramaniam. 2005. VAMPIRE microarray suite: a Web-based platform for the interpretation of gene expression data. Nucleic Acids Res. 33:W627-W632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiao, A., D. S. Worrall, J. M. Olefsky, and S. Subramaniam. 2004. Variance-modeled posterior inference of microarray data: detecting gene-expression changes in 3T3-L1 adipocytes. Bioinformatics 20:3108-3127. [DOI] [PubMed] [Google Scholar]
- 14.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, E. S. Dusko, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 16.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda, M., et al. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Lozada-Chávez, I., S. C. Janga, and J. Collado-Vides. 2006. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 34:3434-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makita, Y., M. Nakao, N. Ogasawara, and K. Nakai. 2004. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 32:D75-D77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pane-Farre, J., B. Jonas, K. Forstner, S. Engelmann, and M. Hecker. 2006. The σB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296:237-258. [DOI] [PubMed] [Google Scholar]
- 23.Periago, P. M., W. van Schaik, T. Abee, and J. A. Wouters. 2002. Identification of proteins involved in the heat stress response of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 68:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 27.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 28.Ragkousi, K., and P. Setlow. 2004. Transglutaminase-mediated cross-linking of GerQ in the coats of Bacillus subtilis spores. J. Bacteriol. 186:5567-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ragkousi, K., P. Eichenberger, C. van Ooij, and P. Setlow. 2003. Identification of a new gene essential for spore germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 185:2315-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 31.Roth, F. P., J. D. Hughes, P. W. Estep, and G. M. Church. 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16:939-945. [DOI] [PubMed] [Google Scholar]
- 32.Teng, F., E. C. Nannini, and B. E. Murray. 2005. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J. Infect. Dis. 191:472-480. [DOI] [PubMed] [Google Scholar]
- 33.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. De Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Schaik, W., M. H. Tempelaars, M. H. Zwietering, W. M. de Vos, and T. Abee. 2005. Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. J. Bacteriol. 187:5846-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Schaik, W., M. H. Zwietering, W. M. de Vos, and T. Abee. 2004. Identification of σB-dependent genes in Bacillus cereus by proteome and in vitro transcription analysis. J. Bacteriol. 186:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Schaik, W., and T. Abee. 2005. The role of σB in the stress response of gram-positive bacteria—targets for food preservation and safety. Curr. Opin. Biotechnol. 16:218-224. [DOI] [PubMed] [Google Scholar]
- 37.Zhulin, I. B., A. N. Nikolskaya, and M. Y. Galperin. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in Bacteria and Archaea. J. Bacteriol. 185:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.