Abstract
While coagulase-negative staphylococci (CoNS), with their ability to form a thick, multilayered biofilm on foreign bodies, have been identified as the major cause of implant-associated infections, no data are available about biofilm formation by staphylococcal small-colony variants (SCVs). In the past years, a number of device-associated infections due to staphylococcal SCVs were described, among them, several pacemaker infections due to SCVs of CoNS auxotrophic to hemin. To test the characteristics of SCVs of CoNS, in particular, to study the ability of SCVs to form a biofilm on foreign bodies, we generated a stable mutant in electron transport by interrupting one of the hemin biosynthetic genes, hemB, in Staphylococcus epidermidis. In fact, this mutant displayed a stable SCV phenotype with tiny colonies showing strong adhesion to the agar surface. When the incubation time was extended to 48 h or a higher inoculum concentration was used, the mutant produced biofilm amounts on polystyrene similar to those produced by the parent strain. When grown under planktonic conditions, the mutant formed markedly larger cell clusters than the parental strain which were completely disintegrated by the specific β-1,6-hexosaminidase dispersin B but were resistant to trypsin treatment. In a dot blot assay, the mutant expressed larger amounts of polysaccharide intercellular adhesin (PIA) than the parent strain. In conclusion, interrupting a hemin biosynthetic gene in S. epidermidis resulted in an SCV phenotype. Markedly larger cell clusters and the ability of the hemB mutant to form a biofilm are related to the augmented expression of PIA.
Since the recovery of small-colony variants (SCVs) from clinical specimens >90 years ago, a number of studies have supported their pathogenic role. SCVs have been associated with long-lasting and recurrent infections, and it was suggested that this property was linked to the ability of SCVs to survive intracellularly, thereby being protected from the host immune system and the action of antibiotics (42). This subpopulation is characterized by reduced growth on routine culture media, with colonies that are <1/10 of the size of those of the normal morphotype.
These growth-deficient variants are formed by a wide range of gram-positive and gram-negative bacteria, such as Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa (32). The prevalence of these variants might be much higher than suspected because slow growth on standard culture media and colony polymorphisms frequently lead to insufficient or false identification in routine diagnostic laboratories (3). Biochemical differentiation is hampered by many phenotypic changes documented in SCVs, which can best be viewed as consequences of alterations in multiple metabolic and energy-dependent pathways (43). In addition, enhanced resistance to various antibiotics, particularly aminoglycosides, complicates antibiotic therapy and medical management (2, 32). While the recovery of hemin or menadione auxotrophic SCVs suggested a link between electron transport-defective strains and persistent infections, a site-directed S. aureus hemB mutant provided the first genetic evidence of this association (42). This mutant showed typical characteristics of clinical SCVs, such as slow growth, decreased pigment formation, low coagulase activity, and resistance to aminoglycosides, and was also able to persist intracellularly (2, 42).
Coagulase-negative staphylococci (CoNS), in particular Staphylococcus epidermidis, are the pathogens most commonly isolated from device-associated infections (44). Clinical experience with such infections shows that host defense mechanisms, as well as antibacterial chemotherapy, are often unable to cure these infections, which is very similar to clinical experience with infections due to SCVs. The persistent course of device-associated infections has been linked, in part, to the ability of the pathogens to establish adherent, multilayered biofilms on polymeric surfaces (44). Of particular interest, a number of device-associated infections due to staphylococcal SCVs were also described in the past years (1, 36, 37, 41, 45), among them several pacemaker-related infections due to SCVs of CoNS (36, 45). These SCVs, in part only identified as staphylococcal species by sequence analysis of a portion of the 16S rRNA gene (45), were recovered from blood cultures obtained over several weeks or months. Analysis for auxotrophism revealed hemin dependencies for isolated SCVs (36, 37, 45). Based upon these observations, it is reasonable to hypothesize that both biofilm formation and the SCV phenotype contributed to the recurrence and persistence of those infections. However, molecular aspects of coagulase-negative staphylococcal SCVs, as well as their biofilm-forming potential, have not been studied so far. Therefore, in order to test whether a defect in the hemB gene of S. epidermidis results in an SCV phenotype and to test the characteristics of SCVs of CoNS, in particular, to study the ability of SCVs to adhere to and accumulate on foreign bodies, we generated a stable electron transport mutant by interrupting the hemB gene of S. epidermidis. This mutant displayed the typical characteristics of clinical SCVs and was able to form a biofilm.
MATERIALS AND METHODS
DNA manipulations and transformation.
DNA manipulations, plasmid DNA isolation, and transformation of E. coli were performed by standard procedures. S. carnosus was transformed with plasmid DNA by protoplast transformation (11, 12), and S. epidermidis was transformed by electroporation (26). For DNA preparations from S. epidermidis, the cell wall was digested with 10 U lysostaphin/ml (Ambicin L, recombinant; Applied Micro Inc.). The subsequent steps in plasmid preparation are those of the alkaline lysis method established for E. coli. Chromosomal DNA from S. epidermidis was prepared as described by Marmur (30). DNA fragments were isolated with the Montage DNA gel extraction kit (Millipore, Bedford, MA). Selection for resistance to antibiotics in E. coli was performed with ampicillin (100 μg/ml), erythromycin (200 μg/ml), and chloramphenicol (10 μg/ml). E. coli strains were grown in LB medium, whereas S. epidermidis was routinely cultured in Trypticase soy broth (TSB) or agar (TSA).
Construction of a hemB mutant of S. epidermidis by gene replacement. (i) Cloning of the hemB gene.
The S. epidermidis hemB gene (975 bp, based on the sequence with accession no. SERP1232) (10) was amplified from chromosomal DNA of S. epidermidis O-47 by PCR carried out with Vent DNA polymerase (New England BioLabs, Beverly, MA) (Table 1). After an initial denaturation step (95°C for 4 min), 30 amplification cycles (95°C for 1 min, 60°C for 2 min, and 73°C for 2 min) were performed, with a final 10-min extension step at 73°C. The primers used were 5′-CTC CAC GGT ACC ATG AAA TTT GAT AGA CAT AGA AGA TTG C (forward primer, including nucleotides 13 to 41 of the hemB gene; the KpnI restriction site is underlined) and 5′-CTC CAC GTC GAC CTA TTT ATC TAA ATA ACG ACA GAT ATC (reverse primer, including nucleotides 948 to 975 of the hemB gene; the SalI restriction site is underlined). The fragment was ligated with pUC19, and E. coli DH5α was transformed with the ligation mixture. Ampicillin, isopropyl-β-d-thiogalactopyranoside (IPTG), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were used to select transformants. One representative plasmid was designated pNA1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, and/or property | Source or reference(s) |
|---|---|---|
| Strains | ||
| S. epidermidis O-47 | Biofilm positive | 15 |
| S. epidermidis hemB mutant | O-47 hemB::ermB SCV | This study |
| Complemented hemB mutant | O-47 hemB::ermB/pNA10 | This study |
| S. aureus SA113 | 8325, restriction deficient | ATCC 35556; 19 |
| S. carnosus TM 300 | Cloning host | |
| E. coli DH5α | supE44 ΔlacU169 (Φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 14 |
| E. coli GM161 | dam dcm | R. Brückner |
| E. coli TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | DSM 6056; 7 |
| Plasmids | ||
| pUC19 | Cloning vector | 48 |
| pEC4 | Composed of pUC19 and transposon Tn551 containing ermB erythromycin resistance cassette | 6 |
| pBT9 | E. coli-S. aureus shuttle vector | 49 |
| pCX19 | Derivate of pCX15 | 31, 47 |
| pNA1 | pUC19::hemB | This study |
| pNA2 | pUC19::hemB-ermB-hemB | This study |
| pNA3 | pBT9::hemB-ermB-hemB | This study |
| pNA10 | pCX19::hemB | This study |
(ii) Insertion of the ermB gene.
Plasmid pEC4 was isolated from E. coli GM161. The 1.4-kb fragment carrying the ermB gene was PCR amplified with primers with restriction sites (underlined) for StuI (5′-CAG CTC AGG CCT ATC GAT AAG AAA TAG ATT TAA AAA TTT CGC [forward primer] and 5′-ATA CTC AGG CCT ATC GAT ACA AAT TCC CCG TAG GCG CTA GGG [reverse primer]), digested with this enzyme, and ligated with StuI-linearized pNA1 propagated in E. coli DH5α. The StuI restriction site is located at the center (base 534) of the sequence of hemB. E. coli TG1 was transformed with the ligation mixture, and transformants were selected with erythromycin and ampicillin. One representative plasmid was designated pNA2. The hemB-ermB-hemB fragment was isolated from pNA2 as a 2.4-kb SalI-KpnI fragment, ligated with the shuttle vector pBT9, and cloned into E. coli TG1. Selection of the transformants was performed with ampicillin. One resulting plasmid which also conferred resistance to erythromycin and chloramphenicol was designated pNA3.
Insertional inactivation of hemB in S. epidermidis O-47.
Since the transformation efficiency of S. epidermidis O-47 with DNA from E. coli is very low because of its restriction-modification system, pNA3 was propagated in strain S. aureus SA113 and then introduced into S. epidermidis by electroporation. Transformants were selected with chloramphenicol (10 μg/ml); the transformation efficiency was 104 transformants/μg DNA.
For gene disruption experiments, one transformant was cultivated in TSB with chloramphenicol at 30°C overnight. The culture was diluted into 500 ml of fresh medium containing erythromycin (2.5 μg/ml) and grown at 42°C to an optical density at 578 nm (OD578)of about 3. Appropriate dilutions were plated onto TSA plates containing erythromycin (2.5 μg/ml) and grown at 43°C. Successful transfer of the ermB cassette into the S. epidermidis chromosome is indicated by the appearance of colonies that are chloramphenicol sensitive and erythromycin resistant. The successful recombination was verified by PCR; the PCR product of the mutant was 1.4 kb larger than the product of the parent strain, S. epidermidis O-47.
Complementation of the hemB mutant.
For complementation of the mutant, the hemB gene was PCR amplified from chromosomal DNA (see above) and cloned into S. carnosus with the vector pCX19 (with a copy number of 15 plasmids per cell) (31), yielding plasmid pNA10. Transformation of the S. epidermidis hemB mutant with pNA10 following selection on TSA supplemented with erythromycin and chloramphenicol restored the wild-type phenotype, while curing of the plasmid-complemented mutant strain of pNA10 reconstituted the small-colony phenotype.
Hemin supplementation.
In order to determine the growth rate and the amount of hemin appropriate for supplementing the hemB mutant, a range of concentrations of hemin (0.5, 1, and 2 μg/ml) was tested. For this, strains were inoculated into TSB to an OD578 of 0.05 and shaken at 160 rpm at 37°C for 24 h. Growth was determined by measuring the OD578 at 2-h intervals.
Catalase production.
Analysis of catalase production was performed by the standard procedure with colonies taken from (i) TSA and (ii) Columbia blood agar (4).
Biochemical characteristics and enzymatic activities.
In order to test the biochemical profile and enzymatic activities of the mutant in comparison to those of the wild-type strain and the plasmid-complemented mutant, strains were tested by using the ID 32 STAPH (BioMérieux, Marcy-l'Étoile, France) and API ZYM (BioMérieux) systems. The mutant was tested with and without addition of hemin (1 μg/ml). The tests were performed according to the manufacturer's recommendations, except that the ID 32 STAPH strips were incubated for 24 h, 48 h, and 72 h and the API ZYM strips were incubated for 4 h, 24 h, 48 h, and 72 h.
Susceptibility to aminoglycosides.
The MICs of gentamicin, kanamycin, amikacin, and tobramycin were determined with Etest strips, and those of gentamicin were also determined by the microdilution method. Results were read after 24 h, 48 h, and 72 h of incubation at 37°C.
Growth on Congo red agar.
A simple method for detecting biofilm formation described by Freeman et al. (9) and modified by Heilmann and Götz (16) was used first. With brain heart infusion agar containing Congo red stain at 0.8 g/liter (BDH Chemicals Ltd., Poole, United Kingdom), biofilm-positive strains yield a positive result (formation of black colonies with a dry crystalline surface appearance) while biofilm-negative strains mostly give a negative result. Test strains streaked onto Congo red agar plates were incubated at 37°C for 24 h, 48 h, and 72 h.
Tube adhesion assay.
Determination of biofilm formation on a glass surface was carried out according to Heilmann et al. (18) with incubation times of 24 h, 48 h, and 72 h. Biofilm formation was recorded as very strong adherence (+++), strong adherence (++), adherence (+), weak adherence [(+)], or equivocal or no adherence (−), according to the density of the adherent biofilm.
Semiquantitative assays of biofilm formation on polystyrene.
For quantification of biofilm-forming capacity, tests similar to those described previously were applied (15, 17, 18, 27) and, if necessary, adapted for SCV phenotype testing with different inocula, incubation times, and atmospheric conditions. S. epidermidis RP62A and S. carnosus TM300 were used as positive and negative controls, respectively. Non-SCV strains were cultivated overnight (or incubated for 24 h in the case of the hemB mutant) in TSB supplemented with 0.25% (wt/vol) glucose. Cultures were diluted in TSB supplemented with 0.25% (wt/vol) glucose to a final OD578 of 0.1 or 0.2, respectively, and 200 μl of the cell suspension was used to inoculate sterile, 96-well flat-bottom polystyrene microtiter plates (Greiner bio-one; Cellstar). After cultivation for 24 h, 48 h, or 72 h at 37°C, respectively, the wells were emptied and the contents were gently washed twice. The plates were air dried, and the remaining surface-adherent cells were stained with 0.1% safranin (Serva) for 30 s. Absorbance at 490 nm (A490) was measured with a Micro-ELISA Autoreader (SpectraMAX GeminiXS; Molecular Devices). Wells containing only sterile TSB served as a background control; their average A490 value was subtracted from all experimental readings. Each assay was performed in quadruplicate. In addition, the assay was repeated under different atmospheric conditions (aerobic versus microaerobic versus anaerobic atmosphere, OD578 of 0.1, 24 h of incubation).
Immunodot blot and immunofluorescence assays for detection of polysaccharide intercellular adhesin (PIA).
PIA was detected by using an immunodot blot assay as previously described (27). In brief, bacteria were grown in tissue culture plates (Nunc, Roskilde, Denmark) under static conditions at 37°C overnight. Cells were scraped off, collected by centrifugation, resuspended in phosphate-buffered saline, and subjected to sonication. After the final OD578 of the resulting suspension was adjusted to 1, the PIA extract was cleared by centrifugation and serial dilutions were spotted onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). Membranes were blocked by 0.5% (wt/vol) bovine serum albumin and subsequently incubated with a PIA-specific antiserum (29). After extensive washing, bound antibodies were detected with an anti-rabbit antiserum conjugated with peroxidase (Dianova, Hamburg, Germany). In addition, PIA was semiquantitatively detected by immunofluorescence assay as previously described (18, 27). S. epidermidis 1457 and 1457-M10 (29) served as positive and negative controls, respectively.
Real-time reverse transcription (RT)-PCR to study expression of icaA and asp23. (i) RNA isolation.
The levels of gene expression in the hemB mutant, the complemented mutant, and the wild-type strain were determined for the genes icaA and asp23 at comparable growth phases from early log phase to late stationary phase—comprising five time points—since expression of the genes tested is known to be growth phase dependent. Cells of the normal phenotype were harvested after 2.5 h (T1), 4.5 h (T2), 6.25 h (T3), 8 h (T4), and 10 h (T5), and cells of the corresponding hemB mutant were harvested after 4.5 h (T1), 6.25 h (T2), 10 h (T3), 20 h (T4), and 36 h (T5) as previously described for RNA isolation of the S. aureus hemB mutant (25, 35). Due to the slow growth of the S. epidermidis hemB mutant with a maximum OD578 of 0.4, two additional time points in the late stationary growth phase (20 h and 36 h) were additionally included.
A 10-ml bacterial cell suspension of the wild-type strain was immediately mixed with 10 ml of RNAprotect (QIAGEN, Hilden, Germany), vortexed for 5 s, incubated for 5 min, and pelleted by centrifugation for 10 min at 4,000 × g. Ten suspensions of the hemB mutant (each 10 ml) were pelleted by centrifugation for 10 min at 4,000 × g. Each pellet was immediately resuspended in 1 ml of RNAprotect (QIAGEN GmbH, Hilden, Germany). Afterwards, the suspensions were pooled, vortexed for 5 s, incubated for 5 min, and pelleted by centrifugation for 10 min at 4,000 × g. Each bacterial pellet was resuspended in 1 ml RNApro solution (Qbiogene, Heidelberg, Germany) and purified on a Matrix E column (Qbiogene). Cells were disrupted by mechanical lysis with a FastPrep Instrument (Qbiogene) for 30 s at a setting of 6.5 and for 30 s on ice, followed by 30 s at a setting of 6.5. Further RNA isolation was performed with an RNeasy Mini Kit (QIAGEN) according to the manufacturer's standard protocol. Contaminating DNA in the RNA preparations was eliminated by using the DNase step recommended in the manufacturer's (QIAGEN) protocol. The RNA quality and quantity were determined by A260 and A280 measurements (Eppendorf BioPhotometer) and agarose gel electrophoresis. Acquired A260/A280 ratios showed successful preparations. Purified RNA was stored at −70°C.
(ii) Multiplex real-time quantitative RT-PCR (qRT-PCR).
RNA was diluted to a concentration of 1 μg/ml with distilled, nuclease-free water and transcribed into cDNA with the QuantiTect RT kit (QIAGEN) by following the manufacturer's instructions. cDNA was synthesized from two or more independent RNA preparations extracted at corresponding time points. Subsequent real-time qRT-PCR was performed in duplicate by using 1 μl of cDNA as template DNA and applying LUX primers (Invitrogen, Karlsruhe, Germany).
Real-time qRT-PCR, in its multiplex adaptation for icaA and asp23, was carried out as relative quantification in comparison to the expression levels of a commonly used reference gene (guanylate kinase, gmk), which also served as an internal positive control (39). One primer of each LUX primer pair was labeled with the 6-carboxyfluorescein (FAM) and 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein (JOE) fluorescent dyes, respectively, as indicated. LUX primer sequences were as follows: for icaA, 5′-GTA CTT CTC AAG GCG GGC ATG AAG [FAM]AC and 5′-CGA TGC GAT TTG TTC AAA CAT T; for asp23, 5′-CGG TAA GCC TGT AGT TAC ATT GTT AC[FAM]G and 5′-GAC ATG AAA GGT GGC TTC ACA G; for gmk, 5′-CGC CGT CAT CTA AAC TTG GAG G[JOE]G and 5′-TTA GAA ATC GAA GTT GAA GGT GCT.
For PCR, a two-step protocol was applied with 45 cycles of 1 min of denaturation at 94°C and 1 min at 60°C for primer annealing and elongation. For increased performance, QuantiTect Multiplex PCR NoROX Master Mix (QIAGEN) was used. Amplifications were performed on an iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA); data were recorded with the iCycler iQ Real-Timer Detection Software, version 3.0a (Bio-Rad).
Data analysis was done with the Gene Expression Analysis Tool for the iCycler iQ real-time PCR detection system (Bio-Rad), the underlying mathematical basis of which is described elsewhere (40).
RESULTS
In order to study the SCV phenomenon in CoNS and to address questions concerning the impact of electron transport-defective S. epidermidis in the pathogenesis of staphylococcal infections, in particular those associated with medical devices, it was first necessary to generate a stable mutant. Therefore, we selected biofilm-positive S. epidermidis O-47 as the parent strain and constructed a genetically defined electron transport mutant with a stable SCV phenotype by interrupting one of the hemin biosynthetic genes, hemB, in S. epidermidis by inserting an ermB cassette into hemB. The SCV phenotype of the mutant was restored by complementation with intact hemB.
Characterization of the S. epidermidis hemB mutant. (i) Colony morphology and growth characteristics on solid media.
After 48 h of incubation at 37°C on TSA, colonies of the mutant displayed the features described for SCVs recovered from clinical specimens: pinpoint colonies that are approximately 10- to 15-fold smaller than those of the parent strain. Of particular importance, colonies showed strong adhesion to the agar surface and were difficult to detach with a loop, in contrast to the parent strain and the plasmid-complemented mutant. Interestingly, the SCV phenotype of the mutant was nearly restored to the wild-type phenotype by growth on Columbia blood agar, most probably due to uptake of hemin from red blood cells.
(ii) Growth in liquid culture and hemin supplementation.
A substantial reduction of the growth rate of the mutant in liquid medium (TSB) was also observed, with a maximal OD578 of 0.4. In order to determine the amount of hemin appropriate for supplementing the hemB mutant, several concentrations of hemin were added to a broth culture. The mutant supplemented with hemin at a concentration of 1 μg/ml showed a growth curve very similar to that of the parent strain.
(iii) Suspension of colonies in liquid medium.
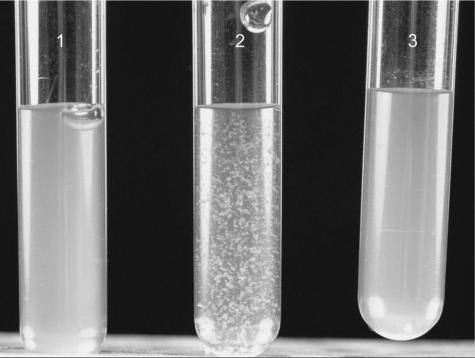
In contrast to colonies of the parent strain and the plasmid-complemented mutant, colonies of the mutant picked from TSA clumped and did not show a homogeneous suspension when inoculated into liquid medium (TSB or Mueller-Hinton bouillon), even after vigorous shaking for several minutes (Fig. 1). However, when mutant colonies were picked from Columbia blood agar (displaying the normal phenotype), they were evenly suspended in liquid medium, even with very brief, gentle shaking.
FIG. 1.
Suspension of the parent strain (S. epidermidis O-47), the hemB mutant, and the plasmid-complemented mutant (colonies were picked from TSA after growth for 48 h at 37°C) in normal saline (0.9%). Tube 2 shows the clumping of the mutant even after vigorous shaking for several minutes. Tubes 1 and 3 show the parent strain and the plasmid-complemented mutant, respectively.
(iv) Catalase production.
The hemB mutant was catalase positive when grown on Columbia blood agar, as were the parent strain and the plasmid-complemented mutant; however, the mutant was unable to produce catalase when grown on TSA. The prompt catalase reaction was not caused by the catalase activity of the red blood cells of the agar, which occurred with a clear delay.
(v) Basic physiological characteristics.
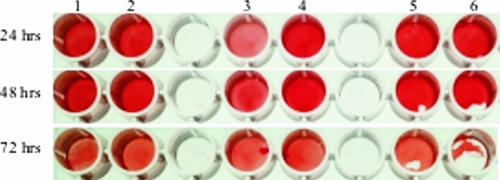
The biochemical profile and enzymatic activity of the strains were analyzed by the ID 32 STAPH and API ZYM systems. In contrast to the parent strain and the plasmid-complemented mutant, the hemB mutant revealed substantial changes in biochemical characteristics, such as negative glucose, maltose, lactose, turanose, and mannitol fermentation; reduced fructose and sucrose fermentation was also noted. Moreover, no nitrate reduction or N-acetylglucosamine utilization was observed, even after incubation for 48 h or 72 h. When tested for enzymatic activity by the API ZYM gallery, the mutant showed increased activity of several enzymes; e.g., esterase and esterase lipase activities were higher than those of the parent strain (Table 2). These differences were still detectable following incubation for 48 h or 72 h. Other enzymes, such as alkaline phosphatase and acid phosphatase, were produced in quantities similar to those of the parent strain and the plasmid-complemented mutant. Of interest, the activity of naphthol-AS-BI-phosphohydrolase was higher in the mutant and the plasmid-complemented mutant than in the parent strain after 4 h of incubation but not after 24 h. Addition of hemin and complementation by the hemB gene returned the biochemical and enzymatic profile of the mutant to that of the parent strain, except for some enzymatic reactions in which the complemented mutant revealed a profile similar to that of the mutant. Appropriate controls ruled out any nonspecific effects of the vector plasmid.
TABLE 2.
Enzymatic activities of parent strain S. epidermidis O-47, the hemB mutant, and the plasmid-complemented mutant tested by API ZYM strips
| Phenotypea |
S. epidermidis O-47
|
hemB mutant
|
Complemented mutant
|
|||
|---|---|---|---|---|---|---|
| 4 h | 24 h | 4 h | 24 h | 4 h | 24 h | |
| Alkaline phosphatase | + | +++ | + | +++ | + | +++ |
| Esterase (C4) | (+) | + | +++ | +++ | ++ | +++ |
| Esterase lipase (C8) | − | (+) | + | ++ | (+) | + |
| Acid phosphatase | +++ | +++ | +++ | +++ | +++ | +++ |
| Naphthol-AS-BI-phosphohydrolase | + | + | +++ | + | +++ | + |
Only positive enzymatic reactions (5/19) are shown. −, negative; (+), weakly positive; +, positive, ++, strongly positive; +++, very strongly positive (judged by eye).
(vi) In vitro activities of aminoglycosides.
With Etest strips, as well as the microdilution method, the MIC of gentamicin was >20-fold higher for the mutant (1.5 μg/ml) than for the wild-type strain (0.064 μg/ml). The MICs of kanamycin, amikacin, and tobramycin were up to 32-fold higher for the mutant (32 μg/ml) than for the wild-type strain (1 μg/ml). When hemin was added to the mutant, MICs identical to that for the parent strain were found. Also, the plasmid-complemented mutant showed MICs identical to that for the parent strain.
(vii) Determination of biofilm formation on Congo red agar, with the tube adherence assay, and on polystyrene microtiter plates.
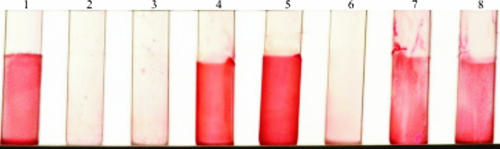
The biofilm-forming capacities of S. epidermidis O-47, its isogenic hemB mutant, and the plasmid-complemented mutant were first compared by using Congo red agar. The parent strain and the plasmid-complemented mutant formed black colonies with a metallic sheen after 24 h of incubation, indicating the ability to form a biofilm. In contrast, the hemB mutant showed this phenotype only if the incubation time was extended to 48 h or 72 h (data not shown). Furthermore, biofilm formation was determined on a glass surface with the tube adherence assay. The parent strain and the plasmid-complemented mutant formed a thick biofilm (documented as very strongly positive, +++), whereas the hemB mutant formed a weak biofilm (+). However, further extending the incubation time to 48 h or 72 h allowed the mutant to form a biofilm similar to that of the parent strain, indicative of primary binding to glass and intercellular adhesion (Fig. 2). Biofilm formation on polystyrene was determined in the (semi)quantitative biofilm assay. Taking into account the slow growth rate of the mutant, various assays were used to quantify biofilm-forming capacity. While the mutant produced much less biofilm than the parent strain and the complemented mutant under aerobic, anaerobic, or microaerobic conditions, the mutant formed significantly more biofilm (A490, 0.8 [OD578, 0.1] versus A490, 1.3 [OD578, 0.2])—similar to the amount produced by the parent strain or the complemented mutant—when a higher inoculum concentration (OD578, 0.2 [P = 0.036] or OD578, 0.4 [P = 0.007]) or a longer incubation time (48 h [P = 0.032]) was used (Fig. 3). Of interest, when the incubation time was extended to 72 h, (i) a decrease in biofilm formation or (ii) detachment was notable for all strains. When the mutant was supplemented with hemin (1 μg/ml), no differences in biofilm-forming capacity were detectable among the mutant, the complemented mutant, and the parent strain.
FIG. 2.
Qualitative determination of biofilm formation by the tube adherence assay. Tubes: 1, S. epidermidis RP62A (used as a positive control); 2, S. carnosus TM300 (used as a negative control); 3, background control containing only TSB; 4, parent strain S. epidermidis O-47; 5, plasmid-complemented mutant; 6 to 8, hemB mutant. Tubes 1 to 6 were incubated for 24 h, tube 7 was incubated for 48 h, and tube 8 was incubated for 72 h.
FIG. 3.
Semiquantitative determination of biofilm formation on polystyrene microtiter plates under different conditions. Biofilm formation was measured by determining OD490 with a Micro-ELISA Autoreader. The value for the background control was subtracted from the experimental readings. Values of ≥0.3 were considered biofilm positive. The effects of incubation time and inoculum size were tested in combination. Starting culture OD578 values of 0.1 (lanes 1, 3, and 5) and 0.2 (lanes 2, 4, and 6) were used, and the microtiter plates were incubated at 37°C for 24 h, 48 h, and 72 h. Lanes 1 and 2, S. epidermidis O-47; lanes 3 and 4, hemB mutant; lanes 5 and 6, complemented mutant.
(viii) Detection of PIA by immunofluorescence and immunodot blot assay.
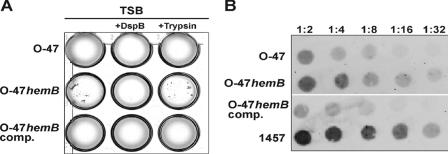
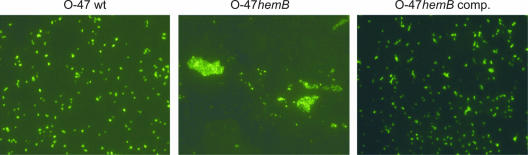
When grown under planktonic conditions, the hemB mutant formed markedly larger cell clusters than S. epidermidis O-47 and the complemented mutant (Fig. 4A). These clusters formed by the mutant were completely disintegrated by the specific β-1,6-hexosaminidase dispersin B (DspB) but were resistant to trypsin treatment. Indirect immunofluorescence assays with anti-PIA antiserum revealed that the large cell clusters formed by the hemB mutant contained amounts of cell-associated PIA similar to those contained by the substantially smaller aggregates formed by parent strain O-47 (Fig. 5).
FIG. 4.
(A) Planktonic growth of S. epidermidis O-47, the hemB mutant (O-47hemB), and the complemented mutant (O-47hemB comp.) in TSB containing 10 μg/ml hexosaminidase DspB and in TSB containing 100 μg/ml trypsin (21). Bacteria were grown overnight in 24-well tissue culture plates in a shaker (120 rpm) at 37°C. In an independent experiment, the enzymes were added to the medium after overnight growth. This measure also led to the disintegration of cell clusters by DspB but not by trypsin (data not shown), giving further evidence that expression of PIA is essential for cluster formation by O-47hemB. (B) Semiquantitative detection of cell wall-associated PIA in S. epidermidis O-47, the hemB mutant, and the complemented mutant. Whereas PIA is detected in S. epidermidis O-47 and the complemented mutant at dilutions of up to 1:8 and 1:4, respectively, a much stronger signal was found in the hemB mutant at dilutions of up to 1:16. PIA-positive reference strain S. epidermidis 1457 served as a control.
FIG. 5.
Detection of PIA production by S. epidermidis O-47 (wild type [wt]), the hemB mutant (O-47hemB), and the complemented hemB mutant (O-47hemB comp.) by immunofluorescence assay. Most strikingly, in contrast to the wild type, the hemB mutant forms large cell clusters that are not seen with the complemented mutant.
The results of PIA expression by S. epidermidis O-47 and the small-colony-forming mutant in the immunofluorescence assay suggested that larger amounts of PIA might be produced by the mutant because of the formation of larger cell clusters. To this end, PIA expression by S. epidermidis O-47, the hemB mutant, and the complemented mutant was semiquantitatively assessed by using a dot blot assay with PIA-specific antiserum. Indeed, the mutant expressed larger amounts of PIA than did the corresponding wild-type strain and the complemented mutant (Fig. 4B).
(ix) Real-time qRT-PCR assay for expression of icaA and asp23.
Since the alternative sigma factor σB is a positive regulator of icaADBC transcription, we analyzed the expression of icaA, as well as the expression of the σB reporter gene asp23. At all time points covering comparable growth phases of the mutant and the parent strain, icaA was up-regulated in the hemB mutant (Table 3). At T1, an up-regulation of 10.9-fold was observed, increasing to 22.2-fold at T2. Surprisingly, at time points T3 and T5, no transcript of icaA was found in the wild-type strain, while the reference gene gmk was expressed. In contrast, the icaA transcript was detectable in the corresponding hemB mutant during all growth phases.
TABLE 3.
Expression factors of the icaA gene of the hemB mutant compared to the wild-type strain at time points T1 to T5
| Time point | Expression factor (mean ± SD)a
|
|
|---|---|---|
| icaA | asp23 | |
| T1 | 10.9 ± 3.0 | 20.8 ± 2.6 |
| T2 | 22.2 ± 6.7 | 20.8 ± 6.1 |
| T3 | NDb | 0.8c ± 0.6 |
| T4 | 4.4 ± 1.3 | 2.6 ± 0.4 |
| T5 | NDb | 0.26c ± 0.04 |
Calculated factors are means from at least two independent RNA preparations from the hemB mutant and the wild type.
ND; no transcript of icaA detected in the wild-type strain.
An expression factor of <1 indicates down-regulation.
Furthermore, asp23 was up-regulated as much as 20.8-fold at T1 and T2 in the hemB mutant, as shown by RT-PCR. At T3 and T5, a slight down-regulation was detectable in the mutant, while at T4 asp23 was also found to be up-regulated.
The transcripts of the investigated genes icaA and asp23 in the complemented mutant returned to wild-type levels, with icaA and asp23 expression factors of 1.03- and 0.83-fold at T1 (2.5 h) and 1.13- and 0.70-fold at T4 (8 h), respectively.
DISCUSSION
In the past decade, S. epidermidis emerged as one of the most important pathogens in nosocomial infections, particularly related to infections of foreign bodies (41, 44). Of special interest, device-related infections due to staphylococcal SCVs were also described (1, 36, 37, 45). Most of the infections caused by these hemin-auxotrophic SCVs of S. aureus, S. epidermidis, or other CoNS occurred several months after the placement of a pacemaker (36, 37, 45).
While the ability of S. epidermidis to form a biofilm is known to be the major pathogenic feature of this species and no corresponding information was available for clinical SCVs, which often reveal an unstable phenotype, a defined and stable S. epidermidis mutant displaying the SCV phenotype was constructed by inactivating the hemB gene. This gene reveals 89.2% DNA sequence similarity to the corresponding hemB gene in S. aureus and 96% identity on the protein level (10). Heme is the prosthetic group of cytochromes, which play an essential role in electron transport (13). In this study, the S. epidermidis hemB mutant showed properties very similar to those of the SCVs recovered from clinical specimens and the S. aureus hemB mutant constructed previously (2, 5, 32, 42, 45), with tiny colonies growing at a slow rate. In comparison to the parental strain and the complemented mutant, many biochemical and physiological characteristics were also changed. Of interest, the mutant was catalase negative when grown on TSA due to its inability to synthesize the prosthetic heme group. While the parent strain and the complemented mutant were able to use both full anaerobic fermentation and citric acid cycle-linked cellular respiration, the mutant was not able to ferment glucose and other sugars, which does not exclude their utilization through alternative biochemical pathways (25, 42). Changes in the ability to use different carbon sources might be due to the interruption of the electron transport chain, which makes the hemB mutant unable to use oxygen or nitrate as a terminal electron acceptor in the terminal step of oxidative phosphorylation (25). Furthermore, the hemB mutant showed increased lipase activity compared to the wild type and the complemented mutant. It was previously shown that the activity of lipases is directly regulated through the alternative sigma factor σB (20, 22). The fact that we found the transcription of the σB reporter asp23 up-regulated in the hemB mutant may explain the increased lipase activity due to activation of these enzymes through σB (22, 38). The decreased susceptibility of the hemB mutant to aminoglycosides might be caused by the reduced electrochemical gradient as a result of the interrupted electron transport chain, as previously observed for S. aureus (2).
Both biofilm formation and the SCV phenotype may contribute to the recurrence and persistence of staphylococcal infections; bacteria are either embedded in large, adherent biofilms on the surfaces of implanted foreign bodies or may persist intracellularly as SCVs in nonprofessional phagocytes, such as epithelial or endothelial cells, and thus evade the host immune response and the action of antimicrobial agents. As both phenomena might explain the poor clinical and bacteriologic response to standard antimicrobial regimens and the difficulty in clearing the infection despite the use of antibiotics with proven in vitro activity, we were particularly interested in the ability of SCVs to produce a biofilm.
In this study, we observed the formation of large cell clusters by the hemB mutant when it was grown under planktonic conditions whereas the parent strain formed a homogeneous suspension, indicating enhanced expression of intercellular adhesive properties in the mutant strain. These large clusters of the hemB mutant, containing amounts of PIA similar to those of the parent strain, are most likely stabilized by PIA, as they were completely disintegrated by the specific β-1,6-hexosaminidase dispersin B (DspB) but were resistant to trypsin treatment (21). Involvement of proteinaceous intercellular adhesins in the formation of these cell clusters is therefore quite unlikely, especially as exclusive action of PIA in intercellular adhesion of S. epidermidis strains forming PIA-dependent biofilms, which were completely independent of any synergistically involved proteins, has been shown previously (33). By assessing PIA expression by S. epidermidis O-47, the hemB mutant, and the complemented mutant with a dot blot assay with PIA-specific antiserum, we found that the mutant expressed larger amounts of PIA than the corresponding wild-type strain and the complemented mutant, which reasonably explains the phenotype observed.
PIA synthesis and biofilm accumulation are integrated in a regulatory network that, in response to changing environmental conditions, modulates biofilm formation via control of the expression of icaADBC and by icaADBC-independent factors (28, 34). Compared to the wild-type strain, a marked increase in icaA expression was found of the mutant in all growth phases. Most stunningly, icaA was also expressed in later growth phases when expression of the operon in the parent strain was obviously shut down. This observation may support the notion of impaired glucose utilization in the hemB mutant as transcription of icaADBC was similarly up-regulated in the stationary growth phase in S. epidermidis 1457 grown in TSB lacking glucose (8).
Several explanations may account for the increased PIA expression in the mutant. As described previously, the hemB mutant is not able to use the respiratory chain and therefore cannot use oxygen or nitrate as a terminal electron acceptor. This leads to decreasing activity of the Krebs cycle and an increased acetate level, which results in activation of the ica operon (46). Furthermore, we found evidence that the alternative sigma factor σB is upregulated in the hemB mutant, which plays a central role in the control of the expression of the icaADBC operon in response to various stimuli (23, 24).
In conclusion, interrupting a hemin biosynthetic gene of S. epidermidis resulted in an SCV phenotype comparable to that of the hemin-auxotrophic SCVs recovered from patients with persistent infections. Markedly larger cell clusters and the ability of the hemB mutant to form a biofilm are related to the augmented expression of PIA. Thus, both the SCV phenotype and biofilm formation might be regarded as different strategies by which bacteria survive in their host.
Acknowledgments
We sincerely thank Daniela Kuhn and Angela Eggemann for excellent technical assistance.
This study was supported in part by research grants from the Deutsche Forschungsgemeinschaft (EI 247/7-1 and Collaborative Research Center 492, project B9) and from the German Federal Ministry for Education and Science (BMBF) in the context of the Pathogenomic Network Plus (PTJ-BIO/0313801B).
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Baddour, L. M., and G. D. Christensen. 1987. Prosthetic valve endocarditis due to small-colony staphylococcal variants. Rev. Infect. Dis. 9:1168-1174. [DOI] [PubMed] [Google Scholar]
- 2.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 3.Becker, K., D. Harmsen, A. Mellmann, C. Meier, P. Schumann, G. Peters, and C. von Eiff. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S rDNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, K., P. Schumann, J. Wüllenweber, M. Schulte, H. P. Weil, E. Stackebrandt, G. Peters, and C. von Eiff. 2002. Kytococcus schroeteri sp. nov., a novel gram-positive actinobacterium isolated from a human clinical source. Int. J. Syst. Evol. Microbiol. 52:1609-1614. [DOI] [PubMed] [Google Scholar]
- 5.Brouillette, E., A. Martinez, B. J. Boyll, N. E. Allen, and F. Malouin. 2004. Persistence of a Staphylococcus aureus small-colony variant under antibiotic pressure in vivo. FEMS Immunol. Med. Microbiol. 41:35-41. [DOI] [PubMed] [Google Scholar]
- 6.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 7.Carter, P., H. Bedouelle, and G. Winter. 1985. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 13:4431-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman, D. J., F. R. Falkiner, and C. T. Keane. 1989. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Götz, F., B. Kreutz, and K. H. Schleifer. 1983. Protoplast transformation of Staphylococcus carnosus by plasmid DNA. Mol. Gen. Genet. 189:340-342. [Google Scholar]
- 12.Götz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 13.Granick, S., and S. I. Beale. 1978. Hemes, chlorophylls, and related compounds: biosynthesis and metabolic regulation. Adv. Enzymol. Relat. Areas Mol. Biol. 46:33-203. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Götz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heilmann, C., and F. Götz. 1998. Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentbl. Bakteriol. 287:69-83. [DOI] [PubMed] [Google Scholar]
- 17.Heilmann, C., M. Hussain, G. Peters, and F. Götz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 18.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 19.Iordanescu, S., and M. Surdeanu. 1976. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J. Gen. Microbiol. 96:277-281. [DOI] [PubMed] [Google Scholar]
- 20.Jäger, S., D. Mack, H. Rohde, M. A. Horstkotte, and J. K. Knobloch. 2005. Disintegration of Staphylococcus epidermidis biofilms under glucose-limiting conditions depends on the activity of the alternative sigma factor σB. Appl. Environ. Microbiol. 71:5577-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kies, S., M. Otto, C. Vuong, and F. Götz. 2001. Identification of the sigB operon in Staphylococcus epidermidis: construction and characterization of a sigB deletion mutant. Infect. Immun. 69:7933-7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knobloch, J. K., S. Jäger, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 72:3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 185:6928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. C. 1995. Electrotransformation of staphylococci. Methods Mol. Biol. 47:209-216. [DOI] [PubMed] [Google Scholar]
- 27.Mack, D., K. Bartscht, C. Fischer, H. Rohde, C. de Grahl, S. Dobinsky, M. A. Horstkotte, K. Kiel, and J. K. Knobloch. 2001. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzymol. 336:215-239. [DOI] [PubMed] [Google Scholar]
- 28.Mack, D., A. P. Davies, L. G. Harris, H. Rohde, M. A. Horstkotte, and J. K. Knobloch. 2007. Microbial interactions in Staphylococcus epidermidis biofilms. Anal. Bioanal. Chem. 387:399-408. [DOI] [PubMed] [Google Scholar]
- 29.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 31.Novick, R. P. 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43:537-565. [DOI] [PubMed] [Google Scholar]
- 32.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 33.Rohde, H., E. C. Burandt, N. Siemssen, L. Frommelt, C. Burdelski, S. Wurster, S. Scherpe, A. P. Davies, L. G. Harris, M. A. Horstkotte, J. K. Knobloch, C. Ragunath, J. B. Kaplan, and D. Mack. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711-1720. [DOI] [PubMed] [Google Scholar]
- 34.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883-1895. [DOI] [PubMed] [Google Scholar]
- 35.Seggewiss, J., K. Becker, O. Kotte, M. Eisenacher, M. R. Khoschkhoi Yazdi, A. Fischer, P. McNamara, N. Al Laham, R. Proctor, G. Peters, M. Heinemann, and C. von Eiff. 2006. Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 188:7765-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert, H., D. Oltmanns, K. Becker, H. Wisplinghoff, and C. von Eiff. 2005. Staphylococcus lugdunensis pacemaker-related infection. Emerg. Infect. Dis. 11:1283-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert, H., H. Wisplinghoff, P. Schnabel, and C. von Eiff. 2003. Small colony variants of Staphylococcus aureus and pacemaker-related infection. Emerg. Infect. Dis. 9:1316-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senn, M. M., M. Bischoff, C. von Eiff, and B. Berger-Bächi. 2005. σB activity in a Staphylococcus aureus hemB mutant. J. Bacteriol. 187:7397-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. Van Eldere. 2003. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J. Infect. Dis. 188:730-737. [DOI] [PubMed] [Google Scholar]
- 40.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1-research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Eiff, C., C. R. Arciola, L. Montanaro, K. Becker, and D. Campoccia. 2006. Emerging Staphylococcus species as new pathogens in implant infections. Int. J. Artif. Organs 29:360-367. [DOI] [PubMed] [Google Scholar]
- 42.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Götz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Eiff, C., P. McNamara, K. Becker, D. Bates, X. H. Lei, M. Ziman, B. R. Bochner, G. Peters, and R. A. Proctor. 2006. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 188:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 45.von Eiff, C., P. Vaudaux, B. C. Kahl, D. Lew, S. Emler, A. Schmidt, G. Peters, and R. A. Proctor. 1999. Bloodstream infections caused by small-colony variants of coagulase-negative staphylococci following pacemaker implantation. Clin. Infect. Dis. 29:932-934. [DOI] [PubMed] [Google Scholar]
- 46.Vuong, C., J. B. Kidder, E. R. Jacobson, M. Otto, R. A. Proctor, and G. A. Somerville. 2005. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187:2967-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wieland, K. P., B. Wieland, and F. Götz. 1995. A promoter-screening plasmid and xylose-inducible, glucose-repressible expression vectors for Staphylococcus carnosus. Gene 158:91-96. [DOI] [PubMed] [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 49.Youngman, P., H. Poth, B. Green, K. York, G. Olmedo, and K. Smith. 1989. Methods for genetic manipulation, cloning, and functional analysis of sporulation genes in Bacillus subtilis, p. 65-87. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of prokaryotic development. American Society for Microbiology, Washington, DC.