Abstract
Salmonella enterica strains survive and propagate in macrophages by both circumventing and resisting the antibacterial effectors normally delivered to the phagosome. An important aspect of Salmonella resistance is the production of periplasmic superoxide dismutase to combat phagocytic superoxide. S. enterica serovar Typhimurium strain 14028 produces two periplasmic superoxide dismutases: SodCI and SodCII. Both enzymes are produced during infection, but only SodCI contributes to virulence in the animal. Although 60% identical to SodCII at the amino acid level with very similar enzymatic properties, SodCI is dimeric, protease resistant, and tethered within the periplasm via a noncovalent interaction. In contrast, SodCII is monomeric and protease sensitive and is released from the periplasm normally by osmotic shock. We have constructed an enzymatically active monomeric SodCI enzyme by site-directed mutagenesis. The resulting protein was released by osmotic shock and sensitive to protease and could not complement the loss of wild-type dimeric SodCI during infection. To distinguish which property is most critical during infection, we cloned and characterized related SodC proteins from a variety of bacteria. Brucella abortus SodC was monomeric and released by osmotic shock but was protease resistant and could complement SodCI in the animal. These data suggest that protease resistance is a critical property that allows SodCI to function in the harsh environment of the phagosome to combat phagocytic superoxide. We propose a model to account for the various properties of SodCI and how they contribute to bacterial survival in the phagosome.
Cu/Zn superoxide dismutases (SODs) are metalloproteins that dismute toxic superoxide radicals to H2O2 and O2 through the alternate oxidation and reduction of the copper(II) ion in the active site. Among bacteria, Cu/Zn SODs (referred to as SodCs) are located in the periplasms of certain gram-negative bacteria (2, 25) and anchored to the surfaces of some gram-positive bacteria (13). Salmonella enterica serovar Typhimurium strain 14028 produces two Cu/Zn SODs-SodCI and SodCII. SodCII is the ortholog of Escherichia coli SodC, while SodCI is encoded by the Gifsy-2 prophage. The ability of these two enzymes to contribute to virulence has been studied extensively (12, 14, 15, 24, 34, 36). Uzzau et al. (36) showed that only SodCI is required for the full virulence of serovar Typhimurium and that SodCII does not contribute to virulence even in the absence of SodCI. Our previous work has confirmed this result, and we have shown that this disparity in virulence phenotypes is due mainly to some differences at the protein level rather than in the regulation of the genes (24). Indeed, SodCI under the control of the more weakly in vivo-induced sodCII promoter (20) was fully capable of complementing wild-type SodCI in an animal infection. These and other data suggested that any differences in enzymatic activity or stability of the active site were insufficient to explain the differential roles of SodCI and SodCII in virulence.
We did, however, note two significant differences between SodCI and SodCII: dimerization and tethering. First, SodCI, like most Cu/Zn SODs, is a dimer composed of two identical monomers, with a total molecular mass of 32 kDa (31). In contrast, SodCII, like its ortholog in E. coli, is a 16-kDa monomer. Second, we have previously shown that SodCI is not released from the periplasm by osmotic shock (24). Thus, SodCI is “tethered” within the periplasm by some interaction. SodCII is quantitatively released by osmotic shock. We hypothesized that the tethering of SodCI to the periplasm may be crucial for the virulence contribution of the enzyme.
SodCI is known to protect the bacterium from phagocytic superoxide during infection, and this is evident from the fact that the attenuation of a sodCI null mutant is not observed in phox−/− mice that are devoid of the respiratory burst (12). However, it has yet to be shown that the dismutase activity of SodCI is essential for this effect. To test this directly, we constructed active-site mutations of SodCI and tested the effects of these mutations on virulence. We then attempted to determine the mechanism and the significance of SodCI tethering within the periplasm. We show that the association of SodCI with the periplasm occurs through some noncovalent interaction that requires its dimeric conformation. Further, the dimeric conformation of SodCI is not required for SOD activity but is required for resistance to proteinase K. Finally, in mouse virulence assays, the monomeric SodCI is unable to replace wild-type SodCI function, potentially on account of either the loss of tethering or the loss of resistance to proteases. To further understand the structure-function relationship of SodCI, we analyzed homologs of SodCI from other pathogens and the ability of these proteins to replace SodCI function during infection. These data suggest that protease resistance is a critical factor in the ability to combat superoxide in the harsh environment of the phagosome. We propose a model to account for the synergistic effects of phagocyte antimicrobial factors and how the regulation and structural properties of SodCI contribute to resistance.
MATERIALS AND METHODS
Strains and plasmid construction.
Bacterial strains and plasmids are described in Table 1. All serovar Typhimurium strains used in this study are isogenic derivatives of strain 14028 (American Type Culture Collection). Insertion-deletion mutants of sod genes were obtained by λ Red-mediated recombination (11). The deletion and the presence of the appropriate antibiotic resistance marker were confirmed by PCR analysis. The constructs obtained by this method were then transduced into a clean wild-type (strain 14028) background using phage P22 HT105/1 int-201-mediated transduction (28). The sodCII gene was cloned into the vector pWSK29 (38) using the engineered sites BamHI and XbaI, giving pRK101. The sodCI plasmid pMC101 has been described previously (24). Site-directed mutagenesis of sodCI was performed on pMC101 using the Stratagene QuikChange site-directed mutagenesis kit. The amino acids are numbered with respect to the mature protein obtained after processing by signal peptidase I.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant genotype or characteristicd | Deletion or cloned endpointsb | Source or referencec |
|---|---|---|---|
| Strains | |||
| 14028 | Wild type | ATCC | |
| JS227 | ΔailT::Km | 1129388-1129938 | 22 |
| JS457 | ΔsodCI-104::Cm | 1130586-1129969 | 24 |
| JS459 | ΔsodA101 ΔsodB102 ΔsodCI-104::Cm | 24 | |
| JS460 | ΔsodA101 ΔsodB102 ΔsodCII-105::Cm | 24 | |
| JS672 | ΔsodCI::Tet | 1129358-1130612 | |
| JS673 | sodCI(H49C) ΔailT::Km | ||
| JS674 | sodCI(H49R) ΔailT::Km | ||
| JS675 | (sodCII1-25sodCI26-157)hyb ΔailT::Km | ||
| JS676 | sodCI Y87E Y107E ΔailT::Km | ||
| JS677 | ΔsodCI::sodCBa ΔailT::Km | ||
| JS678 | ΔsodA101 ΔsodB102 ΔsodCI::Tet ΔsodCII-105::Cm | ||
| JS679 | ΔsodA101 ΔsodB102 (sodCII1-25sodCI26-157)hyb ΔailT::Km ΔsodCII-105::Cm | ||
| JS680 | ΔsodA101 ΔsodB102 sodCI Y87E Y109E ΔailT::Km ΔsodCII-105::Cm | ||
| JS681 | ΔsodA101 ΔsodB102 ΔsodCI::sodCBa ΔailT::Km ΔsodCII-105::Cm | ||
| JS682 | ΔsodA101 ΔsodB102 ΔsodCI::sodCK-12 ΔailT::Km ΔsodCII-105::Cm | ||
| JS683 | ΔsodA101 ΔsodB102 ΔsodCI::sodCIII ΔailT::Km ΔsodCII-105::Cm | ||
| JS684 | ΔsodA101 ΔsodB102 ΔsodCI::sodC;Ps ΔailT::Km ΔsodCII-105::Cm | ||
| AS391 | E. coli AB1157 (F−thr-1 leuB6 proA2 his-4 thi-1 argE2 lacY1 galK2 rspL supE44 ara-14 xyl-15 mtl-1 tsx-33) (sodA::MudPR13)25 (sodB-Km)1-Δ2 sodC::Spec | 21 | |
| Plasmids | |||
| pMC101 | pWKS30 sodCI | 1129594-1130988 | |
| pRK101 | pWSK29 sodCII | 1516038-1516589 | |
| pRK104 | pWKS30 sodCI(C−5T) | ||
| pRK105 | pWKS30 sodCI(Y109E) | ||
| pRK106 | pWKS30 sodCI(Y87E Y109E) | ||
| pBK101 | pWSK29 (CI1-26CII27-153)hyb | ||
| pBK102 | pWSK29 (CII1-25CI26-157)hyb | ||
| pEM101 | pWKS30 sodCK-12 | ||
| pEM102 | pWKS30 sodCPs | ||
| pEM103 | pWKS30 sodCIII | ||
| pMEK15 | pBBR 1MCS4 sodCBa | 19 |
All Salmonella strains are isogenic derivatives of S. enterica serovar Typhimurium strain 14028.
Numbers indicate the base pairs that are deleted or cloned (inclusive) as defined for the S. enterica serovar Typhimurium LT2 genome sequence (National Center for Biotechnology Information).
This study, unless otherwise specified.
sodCK-12, sodC in E. coli K-12; sodCPs, sodC in P. syringae; sodCBa, sodC in B. abortus.
Media and growth of strains.
Bacterial cultures were grown in Luria-Bertani (LB) medium (10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter) with 15 g of agar per liter for solid medium. The following antibiotics were used in the given concentrations: ampicillin and kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; and tetracycline, 25 μg/ml. IPTG (isopropyl-β-d-thiogalactopyranoside) was used to induce the overexpression of SOD proteins from plasmids when used in E. coli. To ensure the activity of Cu/Zn SOD overexpressed from plasmids, the growth medium was supplemented with 0.25 mM CuSO4.
Cloning of sodCI homologs.
The sodC genes of Escherichia coli MG1655 (K-12 derivative), Pseudomonas syringae pv. syringae B728a, and serovar Typhimurium strain LT2 (sodCIII) were cloned into pWKS30 (38), giving plasmids pEM101, pEM102, and pEM103, respectively. The plasmid pMEK15 expressing sodC of Brucella abortus was a generous gift from R. Martin Roop II and is a derivative of the plasmid pBBR1MCS-4 (19). For enzymatic studies, all the plasmids were moved into an E. coli K-12 strain devoid of cytoplasmic and periplasmic SODs (AS391) (Table 1).
Construction of hybrid SodCI-SodCII proteins.
For the construction of (SodCII1-25 SodCI26-157)hyb (SodCI with the first 25 amino acids of SodCII), the sodCII open reading frame (ORF) was PCR amplified with a forward sodCII cloning primer and a hybrid reverse primer (R sodCII Hy). This reverse primer at the 3′ end has the sodCII sequence corresponding to amino acids 21 to 25 and at the 5′ end has the sodCI sequence corresponding to amino acids 27 to 32. The sodCI ORF was PCR amplified with a forward hybrid primer (F sodCI Hy), which has the sodCI sequence corresponding to amino acids 27 to 32 at the 3′ end and the sodCII sequence corresponding to amino acids 21 to 25 at the 5′ end. The reverse primer for this reaction was a standard sodCI cloning primer. The 5′ end of the F sodCI Hy primer and the 3′ end of the R sodCII Hy primer have 16 complementary bases. Equal amounts of the above-described sodCI and sodCII PCR products were used as templates in a standard PCR using Pfx polymerase with forward primer F sodCII RK and reverse primer R sodCI to give the hybrid product (SodCII1-25 SodCI26-157)hyb. The hybrid primers allow annealing of the products, resulting in the formation of the hybrid gene. An analogous hybrid, (SodCI1-26 SodCII27-153)hyb, was constructed using primers F sodCI RK and R sodCI Hy to PCR amplify the first 26 amino acids of SodCI, and F sodCII Hy and R sodCII RK were used to PCR amplify SodCII starting from the 26th amino acid. The standard primer pairs FsodCI RK and RsodCI RK and FsodCII RK and RsodCII RK have sites for restriction enzymes XbaI and BamHI for cloning into the vector pWSK29.
Recombining sodC ORFs into the sodCI locus of Salmonella.
The sodC genes were recombined back into the sodCI locus using extension/annealing PCR combined with λ Red-mediated recombination (11). The sodC gene of interest was PCR amplified using the forward primer a and the reverse primer b. The 5′ end of the a primer has 22 bases of the sodCI gene ending at the ATG codon, and the b primer has 22 bases of identity starting precisely after the last amino acid of the sodCI ORF. The remainder of the sequences of the a and b primers are specific to the sodC gene to be recombined into the sodCI locus. These primers PCR amplify the ORF of the sodC gene starting at the ATG codon and ending precisely at the last amino acid. A kanamycin resistance cassette inserted 114 bp downstream of the termination codon of sodCI was PCR amplified using the primer c-sodCI, which shares 25 bases of identical sequence with the reverse primer b and the reverse primer d-Ail, which directs amplification starting 683 bases downstream of the sodCI ORF. The sodC PCR product and the kanamycin insertion cassette, which have 25 bases of overlapping sequence, were annealed and stitched together via PCR. The annealed product was used as a template in a PCR with the universal sodCI forward primer e and the reverse primer f. The forward primer e has 5′ extensions of homology to the sodCI locus and shares with the forward primer a 15 bases ending 6 bases before the ATG. The reverse primer f shares 15 bases of homology with the kanamycin reverse primer d-Ail. The final product allows precise replacement of the chromosomal sodCI gene with the new sodC gene starting at the ATG codon and associated with a downstream kanamycin cassette that we have previously shown has no effect on virulence (24). The recipient strain has a tetracycline insertion associated with a deletion of the sodCI locus through the position of the kanamycin cassette created with the λ Red-mediated recombinase method and harbors the pKD46 plasmid. Therefore, selection for kanamycin resistance demands inheritance of the sodC gene of interest.
Preparation of cellular fractions.
Whole-cell extracts were prepared in 50 mM potassium phosphate, pH 7.8, using a French pressure cell. The whole-cell lysates were clarified by centrifugation at 13,000 × g for 10 min at 4°C. Periplasmic extracts were prepared by osmotic shock (23). Briefly, 25 ml of overnight cultures was centrifuged at 4°C and washed in 50 mM potassium phosphate buffer, pH 7.4. The cell pellet was resuspended and incubated in 5 ml plasmolysis buffer (50 mM Tris, 2.5 mM EDTA, 20% [wt/vol] sucrose [pH 7.4]) at room temperature for 10 min. After centrifugation at 4°C, the cell pellet was resuspended in ice-cold doubly deionized water that was 1/10 the volume of the original culture, and the cells were allowed to sit on ice for 15 min. The supernatant obtained after subsequent centrifugation was considered to be the periplasmic fraction. In some experiments, the pellet remaining after osmotic shock was resuspended in 0.6 M KCl solution at room temperature, the cells were centrifuged, and the supernatant was assayed for SodCI activity. Periplasmic extracts were also obtained by the lysozyme-EDTA method (3). Cultures grown overnight were centrifuged, and cell pellets were resuspended in a 1/10 volume of ice-cold solution containing 30 mM Tris-HCl (pH 8.0), 1 mM EDTA, 20% (wt/vol) sucrose, and 1 mg/ml lysozyme. After incubation on ice for 10 min, the cells were centrifuged, and the supernatant was considered the periplasmic fraction.
Enzyme assays.
SOD activity was measured by the xanthine oxidase-cytochrome c method (29). The protein content of the cell extracts was determined using the Coomassie blue dye-based assay from Pierce (Rockford, IL). SodCI was assayed in a Salmonella sodA sodB sodCII background, and SodCII was assayed in a Salmonella sodA sodB sodCI background. Enzymes overexpressed from plasmids were assayed in a Salmonella (JS678) or E. coli (AS391) strain with all SODs deleted (SOD−), thus allowing the measurement of a single Cu/Zn SOD from the plasmid.
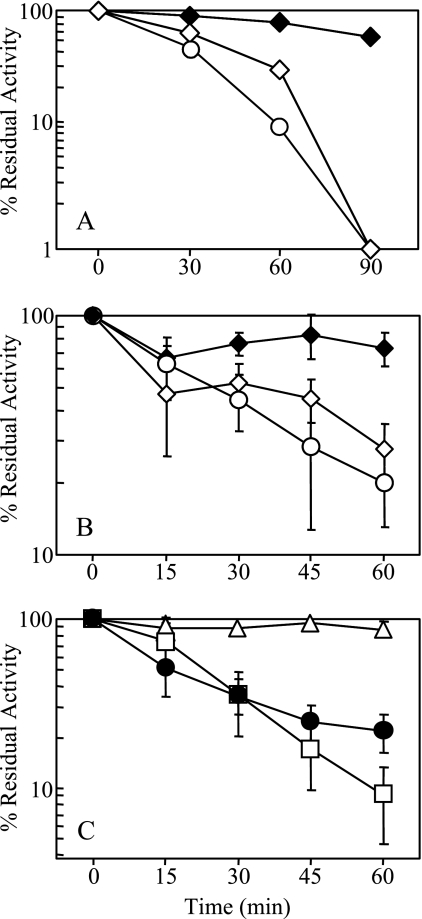
Proteinase K treatment of the purified proteins SodCI, SodCII, and SodCI Y87E Y109E was carried out using the method described by Gabbianelli et al. (18). The enzymes (at a concentration of 0.01 mg/ml) were incubated in the presence of 0.2 mg/ml proteinase K (Sigma) in 20 mM Tris-HCl, pH 6.8, at 37°C. At the time points indicated in Fig. 2, samples were removed and tested for residual SOD activity by the xanthine oxidase-cytochrome c method.
FIG. 2.
Proteinase K treatment of Cu/Zn SODs. Samples were removed at the indicated time points and assayed for residual activity. The activity obtained in the untreated sample was considered 100% (0 min). Data are from representative but repeatable experiments. (A) ♦, SodCI; ⋄, SodCI monomer; ○, SodCII. Whole-cell extracts from strains producing a single SOD (JS460, JS680, or JS459) were diluted 10-fold in 20 mM Tris-HCl, pH 6.8, and incubated at 37°C with 0.25 mg/ml proteinase K. (B) ♦, SodCI; ⋄, SodCI monomer; ○, SodCII. Purified proteins at 0.01 mg/ml were treated with 0.2 mg/ml proteinase K. (C) •, SodCIII; □, E. coli SodC; ▵, B. abortus SodC. Whole-cell extracts from strains producing a single SOD (JS683, JS682, or JS681) were diluted 10-fold and treated with 0.25 mg/ml proteinase K.
Proteinase K treatment of the B. abortus, P. syringae pv. syringae, E. coli K-12, and Salmonella serovar Typhimurium LT2 Cu/Zn SODs was performed with whole-cell extracts prepared from an E. coli SOD− strain (AS391) containing the plasmid pEM101, pEM102, pEM104, or pMEK15. Cells were suspended in 50 mM Tris-HCl, pH 7.0, and lysed using the French pressure cell. The whole-cell extracts, unless otherwise indicated, were then diluted 10-fold in 20 mM Tris-HCl, pH 6.8. Proteinase K was added to these diluted extracts to a final concentration of 0.25 mg/ml. At the indicated time points, samples were removed and tested for residual SOD activity.
Size exclusion chromatography to determine the sizes of Cu/Zn SODs.
Size exclusion fast protein liquid chromatography (FPLC) was performed to determine whether the SodC enzymes were dimers or monomers. All the enzymes were overexpressed in the E. coli SOD− background. Whole-cell extracts were prepared as described above, and the membrane fraction was removed by centrifugation at 141,370 × g in a Beckman ultracentrifuge for 1 hour at 4°C. The soluble fraction was then dialyzed against 20 mM Tris-HCl, 0.15 M NaCl, pH 7.0. The protein samples were filtered through a 0.25-μm syringe filter, loaded onto a Superdex 75 10/300 GL gel filtration column, and eluted with the same buffer, with a flow rate of 0.4 ml/minute. To determine the elution pattern of the SodC enzymes, the column was first calibrated with a low-molecular-weight gel filtration calibration kit (Amersham Biosciences). Fractions (0.2 ml) were collected and assayed for SOD activity. Wild-type SodCI eluted with an apparent molecular mass of 32 kDa, and SodCII eluted with a molecular mass of about 16 kDa.
Protein purification.
SodCI was purified based on the method described by Pesce et al. (31). Briefly, E. coli SOD− cells expressing pMC101 were grown for 16 h in LB supplemented with 0.25 mM CuSO4 and 0.05 mM ZnSO4. Periplasmic fractions were prepared using the lysozyme-EDTA method. The proteins in the periplasmic fractions were concentrated twofold using Amicon YM10 membrane filters, dialyzed against 10 mM potassium phosphate buffer (pH 7.0), and loaded onto a DE52 Sepharose column. Fractions were eluted with the same buffer and collected at 4-minute intervals at a flow rate of 1 ml/minute. Flowthrough fractions with SOD activity were concentrated, dialyzed against 20 mM Tris-HCl-0.15 M NaCl (pH 7.0), loaded onto a HiLoad 16/60 Superdex 75 gel filtration column (Pharmacia), and eluted with the same buffer at a flow rate of 0.25 ml/minute. SodCI eluted as a single peak with a molecular mass of about 32 kDa, and the SodCI Y87E Y109E mutant eluted as a single peak with an apparent molecular mass of about 16 kDa.
SodCII purification was carried out based on the methods of Battistoni and Rotilio (5) and Benov et al. (7) from E. coli SOD− cells containing the plasmid (pRK101). Periplasmic extracts were prepared using the lysozyme-EDTA method. The purification procedure was identical to that used for SodCI, except for the elution buffers. After being concentrated through Amicon YM10 filters, the extracts were dialyzed against 15 mM phosphate buffer (pH 8.4) and loaded onto a DE52 Sepharose column that had been equilibrated with the same buffer. Fractions were eluted in the same buffer, and flowthrough fractions containing SOD activity were concentrated, dialyzed against 20 mM Tris-HCl-0.15 M NaCl (pH 7.5), loaded onto the gel filtration column, and eluted with the same buffer. SodCII eluted as a single peak with an apparent molecular mass of 16 kDa.
Mouse virulence assays.
Mutant and wild-type strains were grown overnight in LB for 16 h and diluted in 0.15 M sterile NaCl. For competition assays, female BALB/c mice (Harlan Sprague-Dawley, Inc.) in groups of 4 to 10 were injected intraperitoneally with a 1:1 ratio of the mutant and wild-type bacteria (approximately 500 total bacteria). Inocula were plated on LB and then replica plated onto the appropriate selective media to determine the actual number and percentage of mutant and wild-type bacteria used for the infection. The infection was allowed to progress for 4 to 5 days, after which the animals were sacrificed. The spleens were harvested and homogenized, and dilutions were plated on LB agar. Colonies were replica plated onto selective medium to determine the percentage of mutant bacteria recovered. A competitive index was calculated as follows: (percentage of strain A recovered/percentage of strain B recovered)/(percentage of strain A inoculated/percentage of strain B inoculated). Student's t test was used for statistical analysis. In each case, the strains were rebuilt by phage P22-mediated transduction, and the mouse virulence assay was repeated to confirm that the phenotype was the result of the designated mutation.
RESULTS
The dismutase activity of SodCI is required for the virulence phenotype.
The loss of SodCI, but not of SodCII, confers a significant virulence defect in serovar Typhimurium strain 14028 (24, 36). We have previously demonstrated that the discrepancy in the virulence contributions of SodCI and SodCII is due primarily to a difference at the protein level rather than at the level of regulation. The two enzymes have similar specific activities, and it seems unlikely that subtle differences in enzyme kinetics are sufficient to explain the striking difference in virulence contributions (24).
To understand the unique property of SodCI, we first wanted to test the assumption that dismutase activity is required for its contribution to virulence. It was possible that SodCI might contribute some function to the pathogen aside from SOD activity per se. For example, the Cu/Zn SOD (SOD1) of Saccharomyces cerevisiae is required for zinc and copper homeostasis (33), and this property appears to be distinct from its antioxidant activity. The Cu/Zn SOD of Haemophilus parainfluenzae has a His-rich N terminus that can sequester copper and transfer the metal to the active site of another Cu/Zn SOD dimer, thus behaving as a copper chaperone (4). SodC of Haemophilus ducreyi, aside from its role as an antioxidant, binds heme via distinct residues in the dimeric interface, facilitating the acquisition of this essential nutrient from the host (30).
We tested the above-described hypothesis by generating metal-binding mutants of SodCI and testing their effects on virulence. Based on the fact that the active sites of all Cu/Zn SODs are highly conserved (9) and using yeast SOD1 and human SOD active-site mutants as models, we used site-directed mutagenesis to generate two mutants. Earlier studies have shown that an H49C mutation eliminates SOD activity but allows copper to bind in the active site (27), while an H49R mutation eliminates both copper-binding and SOD activity (26). The mutations were introduced into cloned sodCI, and the resulting proteins were assayed for SOD activity. As expected, the specific activities of H49C and H49R were below the detection limit of the SOD assay, even when the proteins were overexpressed from plasmids. In both cases, the mutant proteins were produced and were stable at wild-type levels, as evidenced by our ability to partially purify the proteins and to visualize them on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Table 3). The mutations were recombined into the chromosomal sodCI locus, and the resulting strains were assayed in mouse competition assays. Both active-site mutants were attenuated in virulence compared to the wild-type strain (Table 2). In laboratory cultures there was no difference in the growth rates of the wild-type strain and the strains expressing the mutant proteins. Thus, the SOD activity of SodCI is required for its virulence contribution. These results are consistent with the data obtained from the infection of phox−/− mice, showing that the sodCI phenotype is seen only in the presence of superoxide production from the respiratory burst (12).
TABLE 3.
Specific activities and release of SodC enzymes by osmotic shock
| Enzymea | Strainb | Sp act (U/mg)c | SOD activity (U/ml/OD600) (%)c
|
|
|---|---|---|---|---|
| French press | Osmotic shock (%) | |||
| Multicopy enzymes in E. coli | ||||
| SodCI | E. coli pMC101 | 53.6 | 23.3 | 0.21 (0.9) |
| SodCII | E. coli pRK101 | ND | 11.0 | 10.3 (94) |
| SodCI C−5T | E. coli pRK104 | ND | 13.9 | 0.08 (0.6) |
| (SodCII1-25 SodCI26-157)hyb | E. coli pBK101 | 68.1 ± 9.7 | 6.4 ± 0.4 | 0.004 ± 0.0 (0.06) |
| (SodCI1-26 SodCII27-153)hyb | E. coli pBK102 | 24.4 ± 0.3 | 2.5 ± 0.3 | 2.7 ± 0.1 (108) |
| SodCI Y87E Y109E | E. coli pRK106 | 48.7 ± 10.4 | 22.1 ± 7.8 | 15.3 ± 0.0 (69) |
| Single-copy enzymes in Salmonella | ||||
| SodCI | Serovar Typhimurium | 1.0 ± 0.3 | 0.09 ± 0.02 | 0.005 ± 0.00 (5.6) |
| SodCII | Serovar Typhimurium | 3.6 ± 0.6 | 0.36 ± 0.01 | 0.30 ± 0.08 (83) |
| (SodCII1-25 SodCI26-157)hyb | Serovar Typhimurium | 0.5 ± 0.3 | ND | ND |
| SodCI Y87E Y109E | Serovar Typhimurium | 0.9 ± 0.1 | 0.11 ± 0.00 | 0.07 ± 0.02 (64) |
All strains produce only the indicated SOD.
Strains used are either E. coli AS391 containing the indicated plasmid or serovar Typhimurium strains JS460, JS459, JS679, and JS680.
Data are from single but repeatable experiments (n = 3) where means of the results ± standard deviations are given. Numbers in parentheses indicate the percentage of enzyme released by osmotic shock, and the amount of enzyme released by a French press is considered 100%. ND, not determined; OD600, optical density at 600 nm.
TABLE 2.
Competition assays of strains expressing various SodC proteins
| Strain A genotypea | Strain B genotypeb | Median CIc | No. of miced | Pe |
|---|---|---|---|---|
| ΔsodCIf | wt | 0.13 | 16 | <0.0005 |
| sodCI(H49C) | wt | 0.059 | 6 | <0.0005 |
| sodCI(H49R) | wt | 0.21 | 6 | 0.001 |
| (sodCII1-25sodCI26-157)hyb | wt | 0.481 | 9 | NS |
| sodCI(Y87E Y109E) | wt | 0.26 | 7 | 0.008 |
| sodCI(Y87E Y109E) | ΔsodCI | 0.84 | 6 | NS |
| PsodCI::sodCBa | ΔsodCI | 3.3 | 6 | 0.03 |
| PsodCI::sodCBa | wt | 1.80 | 6 | NS |
Strains used were 14028, JS457, JS673, JS674, JS675, JS676, and JS677.
wt, wild type.
Competitive index (CI) = [output (strain A/strain B)/inoculum (strain A/strain B)].
Competition assays were performed with intraperitoneal injection of BALB/c mice.
Student's t test was used to compare the output and the inoculum. NS, not significant.
Data for the ΔsodCI mutant are from Krishnakumar et al. (24).
SodCI is noncovalently tethered within the periplasm.
We have previously shown that, unlike other known periplasmic proteins, SodCI is not released by osmotic shock. We describe this phenomenon as the protein being “tethered” within the periplasm. Even a 10-fold overproduction of SodCI in E. coli or Salmonella does not allow the release of this enzyme by osmotic shock (24). When we consider the amount of enzyme present in whole-cell extracts prepared by a French press as 100%, only a very small amount (<1%) of SodCI was released by osmotic shock when overproduced in E. coli (Table 3). In comparison, >90% of SodCII activity was released by osmotic shock in this experiment. Thus, SodCI is not tethered to some Salmonella-specific factor, nor is any supposed tethering partner of SodCI saturable at this level.
Like SodC of Mycobacterium tuberculosis, the signal sequence of SodCI has a potential signal peptidase II motif, Leu-X-X-Cys (13), suggesting that SodCI could undergo lipid modification. There is no indication of such modification from the crystal structure of SodCI (31), which suggests that the mature protein is released by cleaving at the Ala-X-Ala motif by signal peptidase I. We have also shown previously that SodCI, released by a French press, is not bound to membranes (24). However, to explicitly test whether SodCI is processed by signal peptidase II, we mutated the putative signal peptidase II site at the −5 position with respect to the mature protein, altering the cysteine residue in the potential lipoprotein motif LISC to threonine. We found no increase in the amount of mutant SodCI released by osmotic shock compared to that of the wild-type protein (Table 3). The mutant remained fully active and tethered within the periplasm, showing that the tethering of SodCI does not involve lipid modification.
We have noted that a significant amount of SodCI activity can be obtained by preparing periplasmic extracts using the lysozyme-EDTA method (24). To characterize this property further, we tested whether SodCI could be released by other treatments. Table 4 shows that, in addition to that released after treatment with lysozyme-EDTA, a significant fraction of SodCI was released when osmotically shocked cells were washed with 0.6 M KCl. Together, these results are consistent with SodCI being tethered within the periplasm by some noncovalent ionic interaction.
TABLE 4.
Release of SodCI by various methodsa
| Method | % of SodCI released | % of SodCI remaining cell associated |
|---|---|---|
| French press | 100 | NA |
| Osmotic shock | 0.15 | NA |
| Osmotic shock + 0.6 M KCl wash | 51 | 56 |
| Lysozyme-EDTA | 62 | 37 |
The strain used was AS391 pMC101. The amount of enzyme released by a French press is considered 100%. NA, not applicable.
The N terminus of SodCI is not related to tethering or virulence.
SodCI and SodCII are 60% identical at the protein level. The two proteins differ most in the first 26 amino acids, where they share only 28% identity. To see whether this region of SodCI was related to tethering or to virulence, we constructed hybrid SodCI-SodCII proteins in which the first 25 (or 26) amino acids of the proteins were exchanged. The hybrids (SodCII1-25 SodCI26-157)hyb and (SodCI1-26 SodCII27-153)hyb were assayed for SOD activity and the ability to be released by osmotic shock in both E. coli and Salmonella. Both proteins had significant SOD activity when overproduced from plasmids (Table 3). (SodCII1-25 SodCI26-157)hyb appeared to have about two- to threefold more activity than (SodCI1-26 SodCII27-153)hyb. The hybrid (SodCII1-25 SodCI26-157)hyb remained tethered within the periplasm like SodCI. Thus, the first 26 amino acids of SodCI are not necessary for tethering. (SodCI1-26 SodCII27-153)hyb, like SodCII, was released by osmotic shock, indicating that the first 26 amino acids of SodCI were not sufficient to attach the protein within the periplasmic compartment.
The (SodCII1-25 SodCI26-157)hyb gene was recombined back into the sodCI locus for virulence studies. The hybrid protein (SodCII1-25 SodCI26-157)hyb had specific activity similar to that of wild-type SodCI when expressed in a single copy (Table 3). Virulence studies were conducted by competing the strain expressing the hybrid protein against wild-type 14028. The strain expressing the (SodCII1-25 SodCI26-157)hyb protein was not significantly different in virulence from the wild-type strain (Table 2). Thus, we find that the first 26 amino acids of SodCI, although significantly different from those in SodCII, are not important for tethering or virulence.
The dimeric conformation of SodCI is required for tethering.
One potentially important structural difference between SodCI and SodCII is that, like most Cu/Zn SODs, SodCI is a dimer with a molecular mass of 32 kDa, whereas SodCII is a monomer of 16 kDa. This is confirmed by our gel filtration analyses of these proteins and comparison with molecular weight standards (Fig. 1A). It seemed possible that the dimeric conformation of SodCI was important for tethering and/or virulence. To address this idea directly, we constructed a monomeric SodCI enzyme. Examination of the three-dimensional crystal structure of SodCI (9, 31) suggested that two Tyr residues, Y87 and Y109, might be critical for dimerization. The side chains of these residues appear to extend into the dimeric interface. We mutated these residues to Glu, which is the corresponding amino acid in the monomeric SodCII enzyme. The Y109E mutant was constructed first, and the resulting plasmid was used as a template for the Y87E mutant.
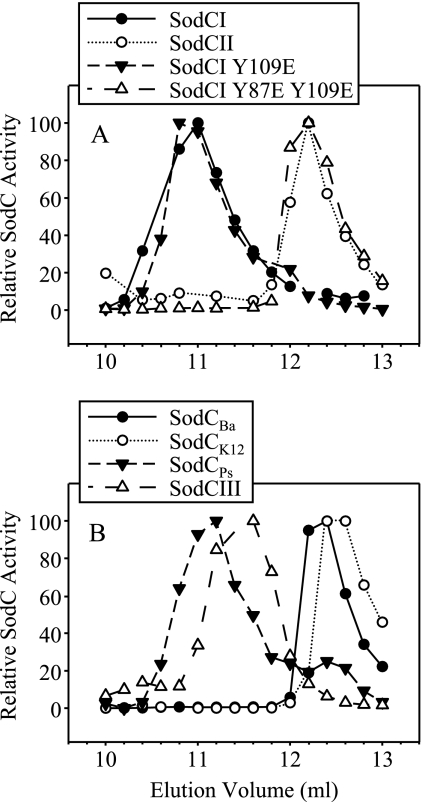
FIG. 1.
Size exclusion chromatography of Cu/Zn SODs. Whole-cell extracts from strains producing a single SOD were separated on a Superdex 75 gel filtration column, and fractions (0.2 ml) were assayed for SOD activity. For each sample, the activities were plotted relative to the fraction with the highest activity. The activities produced in these strains are given in Tables 3 and 5. (A) The strain used was AS391 with pMC101, pRK101, pRK105, or pRK106. (B) The strain used was AS391 with pMEK15, pEM101, pEM102, or pEM103. Ba, B. abortus; Ps, P. syringae.
The mutations had no effect on the SOD activity of the enzyme when it was overproduced in E. coli (Table 3). To determine whether either of these mutations affected the dimeric conformation of SodCI, size exclusion FPLC was performed. The Y109E single mutant retained the dimeric structure, as its elution pattern was identical with that of wild-type SodCI (Fig. 1A). This mutant was not released by osmotic shock and remained tethered within the periplasm (data not shown). The Y109E Y87E double mutant eluted as a monomer with a molecular mass of about 16 kDa (Fig. 1A). The double mutant (SodCI monomer) was also significantly released by osmotic shock when overproduced from a plasmid in E. coli (Table 3) and Salmonella. Thus, the dimeric configuration of SodCI is apparently required for tethering the protein within the periplasm.
Proteinase K sensitivity of SodCI, SodCII, and the SodCI monomer.
Our earlier work showed that SodCII is produced during infection but is inadequate to confer virulence. Uzzau et al. (36) have also shown that the SodCI protein accumulates in more abundance than SodCII in intracellular bacteria during infection. The apparent absence of SodCII protein during infection could reflect its degradation by either bacterial or phagocytic proteases. Gabbianelli et al. (18) have shown that purified SodCI is resistant to proteinase K. SodC from E. coli was inactivated in the same experiment to 25% of its original activity. We wanted to see whether this phenomenon occurred with SodCII, the ortholog of E. coli SodC. First we tested the effect of proteinase K on SodCI and SodCII in whole-cell extracts. We found that while SodCII was inactivated to less than 10% of its original activity upon treatment with 0.25 mg/ml proteinase K, SodCI remained mostly unaffected (Fig. 2A). Next, we used purified proteins to confirm this phenomenon. Figure 2B shows the effect of 0.2 mg/ml proteinase K on purified Cu/Zn SODs (0.01 mg/ml). After 60 min of incubation with proteinase K, SodCI retained about 73% of its original activity, while only about 27% of the original activity of SodCII remained. Our results are similar to those seen by Gabbianelli et al. with regard to the inactivation of SodCI with proteinase K. We show here that SodCII, like its ortholog in E. coli, is sensitive to degradation by proteinase K. Importantly, the SodCI monomer, unlike its parent wild-type protein, was inactivated by proteinase K, both when purified and in crude extracts, behaving like SodCII (Fig. 2). The simplest explanation for these results is that protease-sensitive sites that are buried in the dimer became exposed in monomeric SodCI.
The SodCI monomer does not confer virulence.
We tested the ability of monomeric SodCI, which is released by osmotic shock and is protease sensitive, to complement wild-type SodCI in animal infections. First we wanted to compare the specific activity of this protein when expressed from the sodCI locus with that of wild-type SodCI. For this purpose, the double mutant SodCI Y109E Y87E was recombined back into the sodCI locus in an otherwise SOD− background, and the enzyme activity was measured. The specific activity of the SodCI monomer was comparable to that of the wild-type enzyme (Table 3). We also confirmed that the mutant protein was released by osmotic shock when expressed as a single copy from the sodCI locus (Table 3).
To test the effect of this mutant on virulence, a strain carrying the SodCI monomeric allele in an otherwise wild-type background was competed against the wild-type Salmonella 14028 strain. In mouse virulence assays, the mutant was fourfold attenuated compared to the wild-type strain (Table 2). To more explicitly test the effect of SodCI Y87E Y109E on virulence, we competed the strain expressing the SodCI monomer against the sodCI deletion mutant. We found that the two strains competed evenly, indicating that the sodCI monomer strain was as attenuated as a SodCI null strain. In laboratory cultures there was no difference in the growth rates of the wild-type strain and the strain expressing the mutant proteins. Taken together, these data show that the SodCI Y87E Y109E protein is enzymatically active, monomeric, released by osmotic shock, protease sensitive, and incapable of conferring virulence in the animal model of infection.
Comparison of SodCI to its closely related homologs.
A search of the protein database using SodCI as the query sequence reveals closely related homologs in a small group of pathogens. This group of about 12 sequences, approximately 60% identical to each other, also includes SodCII. Thus, this small list of proteins includes SodCI, capable of protecting an organism from phagocytic superoxide, and SodCII, which does not contribute to virulence. Therefore, we wanted to examine the properties of these closely related homologs of SodCI and determine their ability to replace SodCI function during pathogenesis.
We analyzed four of these Cu/Zn SODs in more detail: (i) E. coli K-12 SodC, which is the ortholog of SodCII; (ii) SodCIII, encoded by the Fels-2 phage found in serovar Typhimurium LT2 but not in strain 14028 (16); (iii) SodC of Brucella abortus, which is known to contribute to Brucella virulence (19, 35); and (iv) SodC of Pseudomonas syringae, which has not been studied in detail. The above-described four Cu/Zn SODs were compared to SodCI with regard to (i) SOD activity, (ii) the ability to be released by osmotic shock, (iii) quaternary structure, and (iv) sensitivity to proteinase K. We wanted to determine whether there were any correlations among these traits and virulence.
In each case, the sodC gene was PCR amplified and cloned into a low-copy-number expression vector. The Cu/Zn SOD from serovar Typhimurium LT2 is referred to as SodCIII (16). The clones were verified by DNA sequencing. The specific activities of the enzymes were measured by overexpressing the proteins from plasmids in an E. coli SOD− background. The activity of most enzymes was as good as or greater than that of SodCI. The exception was SodC of P. syringae, which had 10-fold-less specific activity (Table 5). We then determined whether these enzymes were released by osmotic shock. It is well established that SodC of E. coli is released by osmotic shock (6, 8), and our results confirmed this. Approximately 60% of the activity of SodC of B. abortus was also consistently recovered in the osmotic shock fraction (Table 5). The SOD activity of SodC of P. syringae was low compared to those of other Cu/Zn SODs, but only 2 to 3% of this activity was consistently released by osmotic shock. The amount of SodCIII released by osmotic shock was intermediate between the amounts of SodCII and SodCI released, at approximately 25%.
TABLE 5.
Specific activities of SodC enzymes and release by osmotic shock
| Enzymee | Straina | Mean sp act ± SD (U/mg)b | % of SodC released by osmotic shockc |
|---|---|---|---|
| Multicopy enzymes in E. coli | |||
| SodCI | E. coli pMC101 | 69.4 ± 2.0 | 0.4 |
| SodCBa | E. coli pMEK15 | 404.6 ± 61.9 | 60 |
| SodCK-12 | E. coli pEM101 | 51.6 ± 25.9 | 71 |
| SodCIII | E. coli pEM103 | 241.9 ± 16.0 | 26 |
| SodCPs | E. coli pEM102 | 5.6 ± 0.2 | 2 |
| Single-copy enzymes in Salmonella | |||
| SodCI | Serovar Typhimurium | 5.7 ± 1.9 | 6.5 |
| SodCBa | Serovar Typhimurium | 2.1 ± 0.4 | 109 |
| SodCK-12 | Serovar Typhimurium | 0.1 ± 0.0 | 100 |
| SodCIII | Serovar Typhimurium | 0.2 ± 0.1 | 41 |
| SodCPs | Serovar Typhimurium | BDd | BD |
All strains produce the indicated SOD only. Strains used were either E. coli AS391 containing the indicated plasmid or serovar Typhimurium strains JS460, JS681, JS682, JS683, and JS684.
Specific activity after cell lysis by a French press (n = 2). Data are from single but repeatable experiments.
As in Table 3, the amount of enzyme released by French press is considered 100%.
BD, below detectable level.
SodCBa, SodC in B. abortus; SodCK-12, SodC in E. coli K-12; SodCPs, SodC in P. syringae.
By comparing the protein sequences of known Cu/Zn SODs, we can predict whether they are dimers or monomers based on the presence of Y (or W) or E at the appropriate positions (see above) (17). The Cu/Zn SOD from E. coli is known to exist as a stable monomer (32), and the Brucella enzyme is also predicted to be monomeric based on sequence analysis (10, 17). The sequence analysis of SodCIII and the Cu/Zn SOD from P. syringae suggests that they exist as dimers. The results of size exclusion FPLC confirmed these predictions (Fig. 1B), although the SodCIII protein consistently eluted at a slightly higher volume than other dimeric SodCs.
We next compared these proteins for sensitivity to proteinase K. E. coli SodC, a monomeric protein, was inactivated to less than 10% of its initial activity, as previously shown by Gabbianelli et al. (18). SodCIII, a dimer, was also sensitive to proteinase K inactivation and retained only 20% of its original activity. The most resistant Cu/Zn SOD in our analysis was the monomeric Cu/Zn SOD of B. abortus, which retained 87% of its activity after 60 min of protease treatment (Fig. 2C). SodC of P. syringae did not give consistent results, possibly due to its low activity even when overexpressed from plasmids. Thus, while most SodC proteins are apparently sensitive to protease treatment, SodCI and SodC of B. abortus are significantly resistant.
Brucella Cu/Zn SOD can replace SodCI function during infection.
We correlated the above properties of these Cu/Zn SODs with their ability to replace SodCI function during pathogenesis. In order to perform mouse virulence studies, the sodC ORFs were recombined into the sodCI locus on the Salmonella chromosome. First, we measured the SOD activities of these enzymes when they were expressed in single copies from the chromosome under the regulatory control of the sodCI promoter. These SOD measurements were carried out in an otherwise SOD− background.
In contrast to the SOD activity measurements of the enzymes from plasmids, the specific activities of SodCIII and E. coli K-12 SodC were <10% of that of wild-type SodCI when expressed from PsodCI in Salmonella, reaching the lower limit of accuracy of our SOD assay (Table 5). The specific activity of SodC of P. syringae was below the detection limit of the SOD assay. The only SodCI homolog that had specific activity close to that of SodCI was SodC of B. abortus. SodC of B. abortus, expressed in a single copy, was quantitatively released by osmotic shock.
In mouse virulence assays, the strains expressing SodCIII, P. syringae SodC, or E. coli SodC were 5- to 10-fold attenuated when competed against the wild-type strain (not shown). However, due to the low activity in the strains expressing SodCIII, P. syringae SodC, or E. coli SodC, we could not draw any meaningful conclusions from these virulence assays. To determine whether B. abortus SodC could replace SodCI function during pathogenesis, we competed a sodCI null mutant against a strain expressing B. abortus SodC from the sodCI locus. The strain expressing the Cu/Zn SOD of B. abortus outcompeted the sodCI-null mutant by threefold (Table 2). To further confirm this result, the strain expressing B. abortus SodC was competed against the wild-type strain. The B. abortus SodC-producing strain was fully virulent. This shows that SodC of the gram-negative pathogen B. abortus can replace SodCI function during pathogenesis.
DISCUSSION
Evidence suggests that SodCI protects Salmonella from phagocytic superoxide during growth in macrophages. Null mutations in sodCI attenuate all tested serovars of Salmonella enterica, and this virulence defect is seen only in animal models that are capable of initiating a respiratory burst (12, 14, 15, 24, 33, 34, 36). SodCII, on the other hand, is not required for the virulence of S. enterica serovar Typhimurium strain 14028, even in the absence of SodCI (24, 36). We have previously shown that differential regulation of these enzymes does not explain the discrepancy in virulence phenotypes but rather that some physical difference between the two proteins allows SodCI, but not SodCII, to contribute to virulence. By analyzing the structure-function relationship of SodCI and by comparing it to those in Cu/Zn SOD homologs from other organisms, we sought to determine the features of SodCI that allow it to confer virulence. First, we confirmed that the SOD activity of SodCI was essential for pathogenesis. We then attempted to identify any unique property of SodCI that might be required to confer virulence. During the course of this study, we noted three important biochemical differences between SodCI and SodCII: (i) SodCI is tethered within the periplasm through some noncovalent association, while SodCII is not; (ii) SodCI is a homodimer with a molecular mass of 32 kDa, while SodCII is a monomer of 16 kDa; and (iii) SodCI is resistant to proteinase K, while SodCII is sensitive. We attempted to correlate the above-named properties with pathogenesis.
Tethering.
Our previous (24) and current data show that SodCI is held within the periplasm by noncovalent interactions. To our knowledge, SodCI (and perhaps some homologs) is the only periplasmic protein that is tethered in this fashion. There are two models to explain this tethering of SodCI within the periplasm: (i) SodCI interacts with a specific partner and (ii) SodCI undergoes polymerization that prevents its release from the periplasm. If SodCI does interact with a specific partner, then this partner is not saturated by a 10-fold overproduction of SodCI. The other possibility is that SodCI associates with nonspecific components of the cell envelope through the interaction of charged surfaces; we have shown that a significant amount of SodCI is released by washing cells that have been subjected to osmotic shock with a high-salt solution. However, an electrostatic patch on the surface of SodCI that could account for this tethering is not obvious. The second model that we propose is the polymerization of SodCI, allowing it to form a large complex. If it does, this oligomer must be greater than 100 kDa, since E. coli PhoA, for example, a 100-kDa dimer, is freely released by osmotic shock (data not shown). This second model is consistent with the fact that formation of the dimer is necessary for tethering.
We do not understand the mechanism by which dimeric formation contributes to tethering. In order to determine whether dimerization was sufficient to cause tethering of a Cu/Zn SOD, we attempted to construct a dimeric SodCII. By comparing the dimeric interface of SodCI to the analogous regions of SodCII and other dimeric enzymes, we identified the amino acids of SodCII that when mutated could potentially allow intersubunit interaction. These residues were mutated to the corresponding amino acids in SodCI (E86Y, E108Y, E28P-G29Y, G88D, Q89K, and [an insertion] 87N). The enzyme with seven mutations in the potential dimeric interface remained a monomer, as observed by size exclusion FPLC (data not shown). Thus, we were unable to construct a dimeric SodCII protein. It is possible that there are several other residues (probably around the dimeric interface) that are crucial to the formation of a stable dimer. By analyzing the properties of several closely related SodCs, we have shown that SodC of Brucella abortus is capable of replacing SodCI's function during infection. This enzyme is released by osmotic shock, thereby showing that tethering is not an essential attribute for a periplasmic SOD to confer virulence in serovar Typhimurium.
Protease sensitivity.
In addition to having differences in tethering, SodCI and SodCII differ in protease sensitivity. Monomeric SodCI is both released by osmotic shock and protease sensitive. We presume that the latter trait results from the exposure of protease sites normally buried in the dimer interface, but we have not explicitly tested this hypothesis. However, this mutant SodCI can no longer contribute to virulence. Like wild-type SodCI, the Cu/Zn SOD of B. abortus, which can complement SodCI in a virulence assay, is resistant to inactivation by proteinase K. Thus, there appears to be a correlation between protease resistance and the ability to contribute to virulence.
Salmonella survives in a modified vacuole in macrophages. Although the various killing effectors normally delivered to the phagolysosome are lessened by an alteration in vesicular trafficking mediated by the SPI2 type 3 secretion system (37), resistance to various lethal substances is important for bacterial survival. Our current model for SodCI function takes into account three important factors in the Salmonella-containing vacuole: antimicrobial peptides, proteases, and phagocytic superoxide. Antimicrobial peptides could partially disrupt the outer membrane of Salmonella. If the outer membrane is disrupted, periplasmic proteins, including SodCII, could be released into the phagosome, where they could be degraded by proteases. It is also possible that proteases gain access to the periplasm after disruption of the outer membrane. Superoxide production leads to oxidative damage, although the targets are not known.
Salmonella has complex systems to combat these antimicrobial substances. The PhoPQ regulon, critical for macrophage survival, includes genes whose products modify the outer membrane, conferring resistance to antimicrobial peptides. Indeed, it has been suggested that the inner membrane sensor, PhoQ, directly senses antimicrobial peptides to induce the system (1). This implies that the peptides gain access across the outer membrane. We have recently shown that sodCI is a member of the PhoPQ regulon and is induced in the macrophage phagosome (20). SodCI is both tethered within the periplasm and protease resistant, thereby remaining intact and present to combat phagocytic superoxide. Our work suggests that resistance to proteases is more critical than tethering. However, we lack a SodC protein that is tethered but protease sensitive. Having such a mutant would allow us to test whether tethering alone can protect against proteolytic degradation. Further work is required to both test and refine this model.
Acknowledgments
This work was supported by Public Health Service grant AI063230 from the National Institute of Allergy and Infectious Diseases.
We thank Marty Roop for plasmid pMEK15.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, M. H. Le, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 2.Battistoni, A. 2003. Role of prokaryotic Cu,Zn superoxide dismutase in pathogenesis. Biochem. Soc. Trans. 31:1326-1329. [DOI] [PubMed] [Google Scholar]
- 3.Battistoni, A., A. P. Mazzetti, R. Petruzzelli, M. Muramatsu, G. Federici, G. Ricci, and B. M. Lo. 1995. Cytoplasmic and periplasmic production of human placental glutathione transferase in Escherichia coli. Protein Expr. Purif. 6:579-587. [DOI] [PubMed] [Google Scholar]
- 4.Battistoni, A., F. Pacello, A. P. Mazzetti, C. Capo, J. S. Kroll, P. R. Langford, A. Sansone, G. Donnarumma, P. Valenti, and G. Rotilio. 2001. A histidine-rich metal binding domain at the N terminus of Cu,Zn-superoxide dismutases from pathogenic bacteria: a novel strategy for metal chaperoning. J. Biol. Chem. 276:30315-30325. [DOI] [PubMed] [Google Scholar]
- 5.Battistoni, A., and G. Rotilio. 1995. Isolation of an active and heat-stable monomeric form of Cu,Zn superoxide dismutase from the periplasmic space of Escherichia coli. FEBS Lett. 374:199-202. [DOI] [PubMed] [Google Scholar]
- 6.Benov, L., L. Y. Chang, B. Day, and I. Fridovich. 1995. Copper, zinc superoxide dismutase in Escherichia coli: periplasmic localization. Arch. Biochem. Biophys. 319:508-511. [DOI] [PubMed] [Google Scholar]
- 7.Benov, L. T., W. F. Beyer, Jr., R. D. Stevens, and I. Fridovich. 1996. Purification and characterization of the Cu,Zn SOD from Escherichia coli. Free Radic. Biol. Med. 21:117-121. [DOI] [PubMed] [Google Scholar]
- 8.Benov, L. T., and I. Fridovich. 1994. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 269:25310-25314. [PubMed] [Google Scholar]
- 9.Bordo, D., D. Matak, K. Djinovic-Carugo, C. Rosano, A. Pesce, M. Bolognesi, M. E. Stroppolo, M. Falconi, A. Battistoni, and A. Desideri. 1999. Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase. J. Mol. Biol. 285:283-296. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. L., S. Park, R. W. Thornburg, L. B. Tabatabai, and A. Kintanar. 1995. Structural characterization of the active site of Brucella abortus Cu-Zn superoxide dismutase: a 15N and 1H NMR investigation. Biochemistry 34:12265-12275. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'orazio, M., S. Folcarelli, F. Mariani, V. Colizzi, G. Rotilio, and A. Battistoni. 2001. Lipid modification of the Cu,Zn superoxide dismutase from Mycobacterium tuberculosis. Biochem. J. 359:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Bäumler, U. Ochsner, T. Testerman, S. Bearson, J.-C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrant, J. L., A. Sansone, J. R. Canvin, M. J. Pallen, P. R. Langford, T. S. Wallis, G. Dougan, and J. S. Kroll. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol. Microbiol. 25:785-796. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 17.Forest, K. T., P. R. Langford, J. S. Kroll, and E. D. Getzoff. 2000. Cu, Zn superoxide dismutase structure from a microbial pathogen establishes a class with a conserved dimer interface. J. Mol. Biol. 296:145-153. [DOI] [PubMed] [Google Scholar]
- 18.Gabbianelli, R., M. D'Orazio, F. Pacello, P. O'Neill, L. Nicolini, G. Rotilio, and A. Battistoni. 2004. Distinctive functional features in prokaryotic and eukaryotic Cu, Zn superoxide dismutases. Biol. Chem. 385:749-754. [DOI] [PubMed] [Google Scholar]
- 19.Gee, J. M., M. W. Valderas, M. E. Kovach, V. K. Grippe, G. T. Robertson, W.-L. Ng, J. M. Richardson, M. E. Winkler, and R. M. Roop II. 2005. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect. Immun. 73:2873-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golubeva, Y. A., and J. M. Slauch. 2006. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutase SodCI is a member of the PhoPQ regulon and is induced in macrophages. J. Bacteriol. 188:7853-7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gort, A. S., D. M. Ferber, and J. A. Imlay. 1999. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 32:179-191. [DOI] [PubMed] [Google Scholar]
- 22.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imlay, K. R. C., and J. A. Imlay. 1996. Cloning and analysis of sodC, encoding the copper-zinc superoxide dismutase of Escherichia coli. J. Bacteriol. 178:2564-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnakumar, R., M. Craig, J. A. Imlay, and J. M. Slauch. 2004. Differences in enzymatic properties allow SodCI but not SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium strain 14028. J. Bacteriol. 186:5230-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroll, J. S., P. R. Langford, K. E. Wilks, and A. D. Keil. 1995. Bacterial [Cu,Zn]-superoxide dismutase: phylogenetically distinct from the eukaryotic enzyme, and not so rare after all! Microbiology 141:2271-2279. [DOI] [PubMed] [Google Scholar]
- 26.Liu, H., H. Zhu, D. K. Eggers, A. M. Nersissian, K. F. Faull, J. J. Goto, J. Ai, J. Sanders-Loehr, E. B. Gralla, and J. S. Valentine. 2000. Copper(2+) binding to the surface residue cysteine 111 of His46Arg human copper-zinc superoxide dismutase, a familial amyotrophic lateral sclerosis mutant. Biochemistry 39:8125-8132. [DOI] [PubMed] [Google Scholar]
- 27.Lu, Y., J. A. Roe, C. J. Bender, J. Peisach, L. Banci, I. Bertini, E. B. Gralla, and J. S. Valentine. 1996. New type 2 copper-cysteinate proteins. Copper site histidine-to-cysteine mutants of yeast copper-zinc superoxide dismutase. Inorg. Chem. 35:1692-1700. [DOI] [PubMed] [Google Scholar]
- 28.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 29.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 30.Pacello, F., P. R. Langford, J. S. Kroll, C. Indiani, G. Smulevich, A. Desideri, G. Rotilio, and A. Battistoni. 2001. A novel heme protein, the Cu,Zn-superoxide dismutase from Haemophilus ducreyi. J. Biol. Chem. 276:30326-30334. [DOI] [PubMed] [Google Scholar]
- 31.Pesce, A., A. Battistoni, M. E. Stroppolo, F. Polizio, M. Nardini, J. S. Kroll, P. R. Langford, P. O'Neill, M. Sette, A. Desideri, and M. Bolognesi. 2000. Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene. J. Mol. Biol. 302:465-478. [DOI] [PubMed] [Google Scholar]
- 32.Pesce, A., C. Capasso, A. Battistoni, S. Folcarelli, G. Rotilio, A. Desideri, and M. Bolognesi. 1997. Unique structural features of the monomeric Cu,Zn superoxide dismutase from Escherichia coli, revealed by X-ray crystallography. J. Mol. Biol. 274:408-420. [DOI] [PubMed] [Google Scholar]
- 33.Sansone, A., P. R. Watson, T. S. Wallis, P. R. Langford, and J. S. Kroll. 2002. The role of two periplasmic copper- and zinc-cofactored superoxide dismutases in the virulence of Salmonella choleraesuis. Microbiology 148:719-726. [DOI] [PubMed] [Google Scholar]
- 34.Sly, L. M., D. G. Guiney, and N. E. Reiner. 2002. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutases SodCI and SodCII are required for protection against the phagocyte oxidative burst. Infect. Immun. 70:5312-5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatum, F. M., P. G. Detilleux, J. M. Sacks, and S. M. Halling. 1992. Construction of Cu-Zn superoxide dismutase deletion mutants of Brucella abortus: analysis of survival in vitro in epithelial and phagocytic cells and in vivo in mice. Infect. Immun. 60:2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uzzau, S., L. Bossi, and N. Figueroa-Bossi. 2002. Differential accumulation of Salmonella [Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol. Microbiol. 46:147-156. [DOI] [PubMed] [Google Scholar]
- 37.Vazquez-Torres, A., and F. C. Fang. 2001. Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect. 3:1313-1320. [DOI] [PubMed] [Google Scholar]
- 38.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]