Abstract
Successful pathogens must be able to protect themselves against reactive nitrogen species generated either as part of host defense mechanisms or as products of their own metabolism. The regulatory protein NsrR (a member of the Rrf2 family of transcription factors) plays key roles in this stress response. Microarray analysis revealed that NsrR represses nine operons encoding 20 genes in Escherichia coli MG1655, including the hmpA, ytfE, and ygbA genes that were previously shown to be regulated by NsrR. Novel NsrR targets revealed by this study include hcp-hcr (which were predicted in a recent bioinformatic study to be NsrR regulated) and the well-studied nrfA promoter that directs the expression of the periplasmic respiratory nitrite reductase. Conversely, transcription from the ydbC promoter is strongly activated by NsrR. Regulation of the nrf operon by NsrR is consistent with the ability of the periplasmic nitrite reductase to reduce nitric oxide and hence protect against reactive nitrogen species. Gel retardation assays were used to show that both FNR and NarL bind to the hcp promoter. The expression of hcp and the contiguous gene hcr is not induced by hydroxylamine. As hmpA and ytfE encode a nitric oxide reductase and a mechanism to repair iron-sulfur centers damaged by nitric oxide, the demonstration that hcp-hcr, hmpA, and ytfE are the three transcripts most tightly regulated by NsrR highlights the possibility that the hybrid cluster protein, HCP, might also be part of a defense mechanism against reactive nitrogen stress.
The ability of both pathogenic and free-living bacteria to protect themselves against reactive nitrogen species (RNS) generated either as products of their own metabolism or as part of the innate immune response is critical to their survival. A primary source of RNS is the highly reactive nitric oxide (NO), which is generated by phagocytes in response to infection, by the chemical transformation of nitrite formed either in the host or as the product of bacterial nitrate reduction, and by bacteria, either as an intermediate in denitrification or as a by-product of nitrite reduction to ammonia by fermentative bacteria (30).
In Nitrosomonas europaea, transcription of the gene encoding a copper nitrite reductase is regulated by a “nitrite-sensitive repressor,” NsrR, that is a member of the Rrf2 family of transcription factors (3). The experimental data are consistent with the possibility that the N. europaea NsrR senses NO made as a product of nitrite reduction, which may also be the case for an NsrR homologue in Rhodobacter capsulatus (9). A recent bioinformatic analysis of Rrf2 family regulators and their predicted binding sites led to the proposal that NsrR homologues in many other bacteria regulate their defense against RNS (40). This predicted role for NsrR has recently been confirmed for Escherichia coli, Bacillus subtilis, and the obligate human pathogen Neisseria gonorrhoeae (4, 33, 34). Four E. coli promoters were predicted to be regulated by the NsrR homologue (encoded by the gene formerly designated yjeB) in response to RNS (40). These predictions were confirmed experimentally for the hmpA, ytfE, and ygbA promoters (4). The fourth promoter predicted to be regulated by NsrR was Phcp, the promoter of a two-gene operon encoding the hybrid cluster protein, HCP, and its reductase, HCR. In our recent microarray study of the FNR, NarL, and NarP regulons of E. coli, members of the NsrR regulon were up-regulated in cultures growing anaerobically in the presence of nitrate and nitrite, presumably as a consequence of NO formation. Genes showing this pattern of expression included hmpA, ytfE, ygbA, and hcp (the known and predicted members of the NsrR regulon) and a number of others, raising the possibility that the NsrR regulon might be more extensive than that proposed on the basis of bioinformatic analysis alone (11). The first aim of the current study was therefore to exploit our genome-wide microarrays and a repressor titration approach to obtain a first evaluation of the possible extent of the NsrR regulon.
HCPs are iron-sulfur proteins that contain two iron-sulfur clusters: one is either a conventional [2Fe-2S] or a cubane [4Fe-4S] cluster; the other is a hybrid [4Fe-4S-2O] cluster (2). HCPs are related to the carbon monoxide dehydrogenase protein family (16, 31) and are widely distributed among obligate anaerobes and facultatively anaerobic bacteria. High-resolution crystal structures reveal a potential redox center that can accommodate a small or linear substrate, but their physiological roles remain controversial (31). Two reports implicated HCP in hydroxylamine reduction as part of the nitrate assimilation pathway in Rhodobacter capsulatus (9) or as a defense against hydroxylamine toxicity in E. coli (46). However, the latter authors were careful not to discount an alternative role for HCP, noting that, although effective detoxification enzymes typically have very high affinities for their substrates, the Km for hydroxylamine of E. coli HCP is high. The catalytic efficiency of E. coli HCP for hydroxylamine reduction increases with increasing pH, with an optimum at the nonphysiological pH of 9 (46). Synthesis of E. coli HCP is induced during anaerobic growth, especially in the presence of nitrite (17, 45). Furthermore, HCP from Salmonella enterica serovar Typhimurium has been implicated in defense against RNS generated from acidified nitrite (27). This raises the possibility that HCP synthesis might be regulated as part of the response to RNS and, therefore, that it might protect E. coli against a reactive species other than hydroxylamine. A second aim of this work was therefore to establish how transcription from Phcp is regulated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotide primers.
Escherichia coli strains and plasmids used during this study are listed in Tables 1 and 2. The sequences of the oligonucleotide primers are available on request.
TABLE 1.
Bacterial strains used in this work
| Bacterial strain | Relevant genotype | Reference or source |
|---|---|---|
| MG1655 | Parent strain for microarray experiments, F−Lac− prototroph | 11 |
| JCB1001 | Δfnr, derivative of MG1655 | 11 |
| JCB1002 | ΔnarXL, derivative of MG1655 | 11 |
| JCB1003 | ΔnarXL ΔnarP, derivative of MG1655 | 11 |
| RK4353 | ΔlacU169 araD139 rpsL gyrA | 41 |
| JCB5000 | RK4353 Δhcp | This work |
| JCB5010 | RK4353 ΔnsrR | This work |
| JOEY19 | MC1000 ytfE-lacZ | 4 |
| JOEY61 | JOEY19 ΔnsrR | 4 |
| JOEY103 | MC1000 hcp-lacZ | This work |
| JOEY104 | JOEY103 ΔnsrR | This work |
| JOEY105 | JOEY19 fnr::cat | This work |
| JOEY106 | JOEY61 fnr::cat | This work |
| JOEY126 | JOEY103 fnr::cat | This work |
| JOEY127 | JOEY104 fnr::cat | This work |
| RV | Δlac | 24 |
TABLE 2.
Plasmids used in this work
| Plasmid | Description | Reference or source |
|---|---|---|
| pAA182 | 11.2-kb Ampr promoter-probe vector with ColE1 origin of replication, carrying the lac operon without the lac promoter (see Fig. 2) | 24 |
| pCP20 | 14 | |
| pGIT1 | 205-bp ytfE promoter fragment in pSTBlue-1 | 4 |
| pGIT8 | 205-bp ytfE promoter fragment with ΔA deletion in pSTBlue | 4 |
| pNF383 | hcp regulatory region (383 bp) cloned into pAA182 to create the hcp-lacZ fusion | This work |
| pnrf53 | nrfA promoter cloned into the promoter-probe vector pRW50 | 5 |
| pKD3 | Ampr and Cmr marker used as a PCR template to replace a gene of interest with a cat cassette | 14 |
| pKD46 | Ampr marker with temperature-sensitive origin of replication (active at 30°C), expressing λ redβ, γ, and exo genes (the products of which enable homologous recombination) under control of the parCB promoter | 14 |
| pRW50 | Broad-host-range lacZ fusion vector for cloning promoters on EcoRI-HindIII fragments containing the RK2 origin of replication and encoding tetracycline resistance | 29 |
Growth conditions.
Unless otherwise specified, the strains were grown at 37°C either aerobically in Lennox broth (LB; 10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) or anaerobically in minimal salts (MS) medium [4.5 g/liter KH2PO4, 10.5 g/liter K2HPO4, 1 g/liter (NH4)2SO4, 0.05 g/liter MgCl2, 10 μM ammonium-molybdate, 1 μM sodium-selenate, and 1 ml/liter of E. coli sulfur-free salts] (37), supplemented with 5% (vol/vol) LB and, as appropriate, 20 mM NaNO3 or 2.5 mM NaNO2. The carbon and energy source for anaerobic cultures was either 0.4% (wt/vol) glucose, or 0.4% (wt/vol) glycerol and 20 mM sodium fumarate. For the β-galactosidase assays whose results are reported in Table 4, cultures were grown in a minimal medium previously described (4). The source of NO was 0.05 mM spermine NONOate in aerobic cultures or 5 mM nitrite in anaerobic cultures (4).
TABLE 4.
Regulation of hcp transcription by FNR, NsrR, and sources of nitric oxidea
| Promoter | Background | β-Galactosidase activity under the indicated growth conditions
|
|||
|---|---|---|---|---|---|
| Aerobic | Aerobic with NONO | Anaerobic | Anaerobic with nitrite | ||
| ytfE | Wild type | 27 ± 1.2 | 943 ± 53 | 34 ± 3 | 1,200 ± 195 |
| nsrR | 4,450 ± 317 | 5,155 ± 188 | 9,580 ± 553 | 8,615 ± 477 | |
| fnr | 37 ± 5 | 936 ± 30 | 239 ± 26 | 1,740 ± 115 | |
| nsrR fnr | 6,140 ± 150 | 4,460 ± 200 | 17,600 ± 1,050 | 19,900 ± 650 | |
| hcp | Wild type | 8 ± 1 | 20 ± 2 | 49 ± 6 | 1,064 ± 34 |
| nsrR | 147 ± 5 | 152 ± 4 | 4,880 ± 364 | 6,180 ± 786 | |
| fnr | 8 ± 2 | 13 ± 2 | 9 ± 2 | 34 ± 2 | |
| nsrR fnr | 114 ± 3 | 109 ± 5 | 165 ± 5 | 161 ± 3 | |
The bacteria for the microarray experiments were grown anaerobically in the MS medium described above, supplemented with 20 mM trimethylamine-N-oxide (11), in the presence or absence of 2.5 mM sodium nitrite. RNA was extracted from four biological replicates, as described previously, from samples of bacteria harvested at the early exponential phase of growth. The pool of RNA used as a control in every experiment was isolated from eight independent cultures of E. coli strain MG1655 transformed with pGIT8 (a plasmid with a 1-bp deletion in the NsrR binding site of the ytfE promoter; see Table 2) and grown anaerobically in the absence of nitrite.
Whole-genome array analysis.
Preparation of mRNA, labeling, hybridization, and analysis of array data were performed as described previously (11), with the following amendments. RNA was extracted from 15-ml samples from four independent cultures of each transformant growing in the presence or absence of nitrite. Each 15-ml sample was mixed with 30 ml of RNAprotect bacterial reagent (QIAGEN Ltd.), and an RNeasy Midikit was used to prepare total RNA according to the manufacturer's instructions (QIAGEN Ltd.). Any contaminating DNA was removed by using a DNase column kit (QIAGEN Ltd.). Total RNA was transcribed to Cy3- and Cy5-labeled cDNA, hybridized onto Corning Ultra GAPI glass slides with the 6,112 70-mer oligonucleotides of the Operon Array-Ready E. coli set 1.0 (Operon) as described previously (11). The slides were washed in Advalytix hybridization and wash stations, according to the manufacturer's instructions, and scanned, and the data were analyzed using Genepix and Genespring software as previously described (11).
Identification of NsrR binding sites.
The NsrR binding sites at the hcp, hmpA, ygbA, and ytfE promoters were used to construct a position weight matrix using Consensus (RSAT tools, http://rsat.scmbb.ulb.ac.be/rsat/). These sites were first identified by Rodionov et al. (40). Bodenmiller and Spiro (4) later defined the NsrR binding site as being 23 bp in length, and these 23-bp binding sites were used to construct the matrix. The E. coli genome was searched with this matrix using genomic-scale PATSER (RSAT tools). Only potential NsrR binding sites with a score of >3 within 200 bp of transcription start sites were considered to be significant.
Construction of reporter fusions.
The 400-nucleotide hcp promoter region between nucleotides 912971 and 913339 in E. coli genomic DNA was amplified by PCR from genomic DNA using primers hcp-prom1 and hcp-prom2, which create unique EcoRI and HindIII restriction sites at the 5′ and 3′ ends, respectively, of the amplified fragment. Plasmid pNF383 was constructed by ligating the EcoRI/HindIII-digested PCR fragment into pAA182 that had been digested with the same enzymes (Table 2). The hcp-lacZ fusion was crossed onto λRS45 by homologous recombination, and the resulting phage was used to isolate monolysogens (confirmed by PCR). The construction of the ytfE-lacZ fusion has been described previously (4).
Construction of hcp mutants.
The hcp gene in strain RK4353 was first replaced by a chloramphenicol resistance cassette (cat) to give the strain JCB4999 (Table 1) (14). The hcp::cat mutation was transferred to other strains by bacteriophage P1 transduction. Purified transductants were checked by PCR, transformed with pCP20, and grown at 30°C to isolate derivatives, such as JCB5000, that had lost the cat cassette.
Gel retardation assays.
The hcp promoter fragment from pNF383 was gel purified, end labeled with [γ-32P]ATP, and incubated at 37°C for 30 min with different concentrations of purified FNR and NarL proteins, and protein-DNA complexes were separated by electrophoresis on a 6% nondenaturing polyacrylamide gel containing 2% glycerol for 3 to 4 h, following the protocol described previously (6). Gels were analyzed using a Bio-Rad molecular imager FX and Quantity One software (Bio-Rad).
Chemical and biochemical assays.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (43). For β-galactosidase assays, strains were grown and assayed as described by Jayaraman et al. (24) or according to Miller (32). Protein concentrations were determined by the Lowry method.
Effects of hydroxylamine and other RNS on growth.
Test tube cultures of each test strain were grown without aeration in 10 ml of MS medium supplemented with 0.8% (vol/vol) glycerol, 20 mM sodium fumarate, 2.5% (vol/vol) LB, and various concentrations of hydroxylamine in the range of 0.1 to 1.0 mM. The inoculum was 60 μl of an exponential-phase aerobic culture in LB. The tubes were incubated at 37°C, and the density of each culture was determined at intervals. When relevant, the β-galactosidase activity of each culture was assayed.
RESULTS
Microarray analysis of the E. coli NsrR regulon.
The nsrR gene of E. coli MG1655 is located immediately upstream of, and is cotranscribed with, the rnr gene encoding RNase R (10). The expression of this transcription unit is cold-shock inducible (10), which may well involve secondary structures in the mRNA in the vicinity of the nsrR coding region. Deletion of the nsrR gene might therefore perturb rnr expression by a simple consequence of polarity or by disruption of mRNA structural elements involved in posttranscriptional regulation. The consequence would likely be secondary effects on global RNA stability; hence, a comparison of the transcriptomes of an nsrR mutant and its parent would be difficult to interpret. To avoid such potential artifacts, we exploited the observation that regulation by NsrR is extremely sensitive to repressor titration by NsrR binding sites provided on a multicopy plasmid (4). Transformation of an nsrR+ strain with plasmid pGIT1, which carries the ytfE promoter in multicopy, titrates out NsrR to phenocopy an NsrR− mutation. As a control, pGIT8 carries the same promoter fragment, but a 1-bp deletion mutation in the NsrR binding site eliminates repressor titration in vivo (4). Strain MG1655 transformed with either pGIT1 or pGIT8 was grown anaerobically under the carefully controlled conditions described previously, so that growth rates were almost identical (11), and RNA was isolated from early-exponential-phase cultures grown in the presence or absence of nitrite. For each growth condition, RNA was isolated from four independent cultures, and to validate comparisons between different plasmids and growth conditions, a pool of RNA from the control strain (MG1655 transformed with pGIT8 and grown in the absence of nitrite) was included in every microarray to provide a common reference. Nine transcripts encoding 20 genes were either more abundant in cultures in which NsrR was titrated out by the ytfE promoter than in the control cultures or more abundant in cultures grown in the presence than in the absence of nitrite (Table 3). Operons observed to be differentially expressed include not only all four of the transcripts, hmpA, ytfE, ygbA, and hcp-hcr, known or predicted to be NsrR-regulated (4, 40), but also other transcripts predicted to be regulated by nitrite or RNS generated from nitrite, for example, nitric oxide (11). Particularly interesting is the demonstration that genes encoding the periplasmic nitrate and nitrite reductases, Nap and Nrf, but not the cytoplasmic nitrate and nitrite reductases, NarGHI and NirBD, are also part of the NsrR regulon. At least in the case of Nrf, there is a documented role in the defense against RNS, since Nrf is both a nitrite and an NO reductase (36). Also repressed by NsrR was the promoter of the yeaR-yoaG genes of unknown function, previously implicated in the metabolism of RNS (Table 3) (see Table 5 of reference 11).
TABLE 3.
Genes differentially regulated in strains carrying pGIT1 and pGIT8a
| Gene | Product | Transcript expression ratio inc:
|
Operon structure | Coregulation byd:
|
NsrR sitee | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| pGIT8 Ø2 vs pGIT1 Ø2 | pGIT8 Ø2 vs pGIT8 with NO2− | pGIT8 Ø2 vs pGIT1 with NO2− | FNR | NarL | NarP | |||||
| NsrR repressed | ||||||||||
| hcr | NADH oxidoreductase | 12.20 | 4.47 | 13.09 | hcp-hcr | Af,g | Ag | +6 wrt mapped TSS | 17 | |
| hcp | Hybrid cluster protein | 15.65 | 5.28 | 9.93 | ||||||
| yccM | Predicted 4Fe-4S membrane protein | 4.89 | 3.19 | 4.18 | yccM | Af | +69 wrt predicted TSS | |||
| uspF | Nucleotide binding protein | 2.09 | 1.63 | 1.78 | uspF | |||||
| yeaR | Hypothetical protein | 2.57 | 3.57 | 3.12 | yeaR-yoaG | R | A | A | −20 wrt predicted TSS | |
| ccmG | Cytochrome c biogenesis system | 2.75 | 2.17 | 2.64 | napFDAGHBC-ccmABCDEFGH | Af,g | Rg | Ag | −30 wrt mapped TSS | 13 |
| ccmF | Cytochrome c biogenesis system | 2.08 | 1.73 | 2.34 | ||||||
| napB | Periplasmic nitrate reductase | 2.07 | 3.49 | 3.75 | ||||||
| napH | Periplasmic nitrate reductase | 2.13 | 3.39 | 3.86 | ||||||
| napD | Periplasmic nitrate reductase | 2.02 | 3.49 | 2.80 | ||||||
| napF | Periplasmic nitrate reductase | 2.41 | 3.99 | 3.00 | ||||||
| hmpAb | Flavohemoglobin | 13.07 | 8.50 | 14.44 | hmpA | Rf,g | A | +1 wrt mapped TSS | 12 | |
| ygbAb | Hypothetical protein | 4.58 | 3.20 | 3.86 | ygbA | Rf | −7 wrt mapped TSS | |||
| nrfA | Periplasmic nitrite reductase | 5.28 | 7.37 | 6.66 | nrfABCDEFG | Af,g | Rg | Ag | −63 wrt mapped TSS | 6 |
| nrfB | Periplasmic nitrite reductase | 3.54 | 4.31 | 4.67 | ||||||
| nrfC | Periplasmic nitrite reductase | 4.51 | 5.30 | 6.19 | ||||||
| nrfD | Periplasmic nitrite reductase | 3.68 | 3.98 | 3.99 | ||||||
| nrfE | Periplasmic nitrite reductase | 4.72 | 5.15 | 6.44 | ||||||
| nrfF | Periplasmic nitrite reductase | 2.65 | 3.29 | 2.47 | ||||||
| ytfEb | RNS-induced conserved protein | 31.91 | 7.84 | 33.18 | ytfE | Rg | A | −12 wrt mapped TSS | 25, 26 | |
| NsrR activated | ||||||||||
| insB_1 | IS1 protein InsB | 0.40 | 1.29 | 0.60 | ||||||
| yafE | Predicted S-adenosylmethionine-dependent methyltransferase | 0.36 | 0.36 | 0.35 | yafDE | |||||
| yafU | Predicted inner membrane protein | 0.23 | 0.44 | 0.16 | yafU | |||||
| ykfJ | Hypothetical protein | 0.43 | 0.52 | 0.68 | ykfJ[prfH] | |||||
| mmuP | Predicted ABC transporter | 0.37 | 0.57 | 0.27 | mmuPM | A | ||||
| insB_2 | IS1 protein InsB | 0.41 | 1.46 | 0.63 | ||||||
| yagA | Hypothetical protein | 0.23 | 0.56 | 0.18 | yagA | |||||
| insB_3 | IS1 protein InsB | 0.40 | 1.07 | 0.59 | ||||||
| ykgF | Predicted amino acid dehydrogenase | 0.40 | 0.42 | 0.49 | ykgEFG | R | A | |||
| betB | Betaine aldehyde dehydrogenase | 0.47 | 0.70 | 0.54 | betIBA | R | A | |||
| cyoE | Cytochrome bo terminal oxidase | 0.44 | 0.94 | 0.47 | cyoABCDE | Rf | A | |||
| cyoC | Cytochrome bo terminal oxidase | 0.36 | 0.96 | 0.51 | ||||||
| cyoB | Cytochrome bo terminal oxidase | 0.29 | 0.67 | 0.38 | ||||||
| cyoA | Cytochrome bo terminal oxidase | 0.26 | 0.79 | 0.35 | ||||||
| ompF | Outer membrane porin | 0.43 | 0.63 | 0.40 | ompF | |||||
| insB_4 | IS1 protein InsB | 0.35 | 1.18 | 0.54 | ||||||
| dadA | d-Amino acid dehydrogenase | 0.49 | 0.76 | 0.49 | dadAX | |||||
| ydbC | Putative oxidoreductase | 0.03 | 0.03 | 0.01 | ydbC | |||||
| insB_5 | IS1 protein InsB | 0.40 | 1.25 | 0.61 | ||||||
| ygeF | Hypothetical protein | 0.43 | 0.94 | 0.49 | ygeF | |||||
| insB_6 | IS1 protein InsB | 0.38 | 1.30 | 0.61 | ||||||
| yiiL | l-Rhamnose mutarotase | 0.17 | 0.23 | 0.16 | rhaBAD-yiiL | A | ||||
| yjbB | Putative α-helix protein | 0.35 | 0.42 | 0.23 | yjbB | A | ||||
| yjiV | Hypothetical protein | 0.30 | 0.26 | 0.45 | yjiU | |||||
This table lists genes that display at least a twofold difference in transcript abundance between anaerobically grown cultures of E. coli strain MG1655 harboring pGIT1, a multicopy plasmid with a copy of the ytfE promoter to which NsrR binds, and pGIT8, the same plasmid with a mutation which abolishes NsrR binding. Also shown are the ratios of transcript abundance in cultures carrying either pGIT8 or pGIT1 grown in the presence of 5 mM nitrite compared to those in cultures carrying pGIT8 in the absence of nitrite.
Gene previously shown to be regulated by NsrR (4).
Ø2, anaerobic growth conditions.
Observed differential regulation in strains carrying fnr, narXL, and narXLP in Constantinidou et al. (11). A, activation; R, repression.
NsrR sites located by position weight matrix search of the E. coli MG1655 genome using a matrix generated from the NsrR binding sites at the hcp, hmpA, ygbA, and ytfE promoters. Predicted transcription start sites (TSS) are taken from RegulonDB (http://regulondb.ccg.unam.mx/index.html). wrt, with respect to.
An FNR binding site is located in the promoter region of this gene.
Regulation has been independently documented.
A further 22 transcripts fulfilled the statistical criteria to be considered to be activated by NsrR (Table 3). In the case of the ydbC promoter, the differences in transcript levels between the two transformants were at least as great as those for the NsrR-repressed genes, making PydbE a prime candidate for direct activation by NsrR. At least one other Rrf2 family transcription factor, IscR, can also function both as a repressor and as a transcription activator (19, 47). The microarray data for the other promoters that are apparently activated by NsrR must be evaluated cautiously, however, because few of these transcripts encode proteins known or suspected to be involved in the response to RNS, and most of the differences were much smaller (typically two- to threefold apparent activation) than the 30-fold repression of ytfE, the 14-fold repression of hmpA, and the up-to-15-fold repression of hcp. It is likely that most of these other apparent transcription activation effects of NsrR are indirect consequences of the NsrR titration. Nevertheless, whether the effect is direct or secondary, it is interesting that the genes encoding the cytochrome bo oxidase, but not the cytochrome bd oxidase, appear to be modestly activated by NsrR. Recent microarray analysis and real-time quantitative PCR data revealed that the cydAB mRNA was the transcript most strongly induced as part of the nitrosative stress response when Staphylococcus aureus was exposed to S-nitroso N-acetyl dl-penicillamine (39).
In summary, the microarray data established that NsrR plays a central role in E. coli defense against RNS and implicated additional genes of unknown function in this response. The prediction that Phcp is part of the NsrR regulon (40) was confirmed. Subsequent experiments were designed to confirm these and other key results from the microarray analysis.
Binding of FNR and NarL at the hcp promoter.
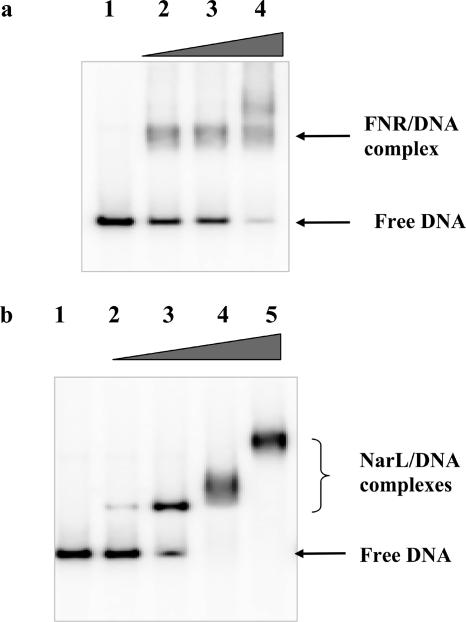
We recently reported microarray data for E. coli strain MG1655 that showed that transcription from Phcp is induced during anaerobic growth (11). Anaerobic induction was totally dependent upon a functional FNR protein, partially dependent on the nitrate-sensing two-component regulatory system NarXL, and independent of the alternative nitrate response protein NarP. To determine whether the effects of FNR and NarL on hcp transcription are direct or indirect (for example, due to inactivation of NsrR by nitrite or nitric oxide generated during anaerobic growth), the hcp promoter fragment from pNF383 was incubated with different concentrations of purified FNR and NarL proteins. Gel retardation assays showed that both proteins formed multiple complexes; therefore, the effects of FNR and NarL on transcription are likely to be direct (Fig. 1). This correlates with the previously noted presence of FNR and NarL binding sites in the promoter located so that FNR would function, unusually, as a class 1 activator and NarL as a class 2 activator (8, 17).
FIG. 1.
Binding of FNR and NarL to the hcp promoter. The hcp regulatory region was prepared by digesting plasmid pNF383 with EcoRI and HindIII. The hcp regulatory region was labeled with radioactive [γ-32P]ATP and mixed with FNR or NarL at a range of concentrations, indicated by the gray triangles. The reaction mixtures were incubated at 37°C for 30 min and run on polyacrylamide electrophoresis image gels for 3 to 4 h. The gels were fixed and dried, and the phosphorscreen image was developed. Free DNA and complexes of DNA with FNR and NarL are denoted. (a) Gel retardation assay with FNR. Lane 1, no FNR; lane 2, 0.25 μM FNR; lane 3, 0.5 μM FNR; lane 4, 1.0 μM FNR. (b) Gel retardation assays with NarL. Lane 1, no NarL; lane 2, 0.4 μM NarL; lane 3, 0.8 μM NarL; lane 4, 1.6 μM NarL; lane 5, 3.2 μM NarL.
Regulation of hcp transcription in response to nitric oxide and NsrR but not by hydroxylamine.
There was a striking discrepancy between the apparent NarP dependence of hcp transcription revealed in preliminary experiments with a multicopy reporter plasmid, pNF353 (17), and the microarray data, in which RNA expressed from a single chromosomal copy of the hcp-hcr operon was analyzed (11). As multicopy plasmids titrate out the effects of NsrR (4) (Table 3), a single-copy chromosomal hcp::lacZ fusion was constructed to investigate the role of NsrR at this promoter. The previously characterized ytfE::lacZ fusion was used as a control (Table 4). During aerobic growth, the very low level of transcription from the hcp promoter was induced 2.5-fold by spermine NONOate in the parental strain but only 1.6-fold in an fnr mutant. The hcp gene was constitutively expressed at a higher level in the nsrR mutant, and this constitutive level was lower in the fnr nsrR double mutant.
During anaerobic growth, the background level of hcp transcription was sixfold higher than during aerobic growth and was induced a further 21-fold by nitrite. Increased transcription during anaerobic growth was strongly dependent upon FNR (Table 4). The highest expression level, 6,180 units, was detected during anaerobic growth of the nsrR mutant in the presence of nitrite, but the activity was slightly lower, 4,880 units, in the absence of nitrite, presumably reflecting the smaller effect of NarL and NarP under these conditions. These data clearly indicate that the hcp promoter is activated by FNR and repressed by NsrR. In the case of the ytfE promoter, repression by NsrR is the dominant regulatory mechanism, though there is also some evidence for anaerobic repression by FNR, as reported previously (25). The effect of the fnr mutation may well be indirect in this case, since FNR probably controls the expression of genes that are involved in the reduction of nitrite to NO. In the absence of both FNR and NsrR, ytfE promoter activity was significantly higher in anaerobic cultures, but the underlying mechanism is not known.
No evidence was obtained for the induction of hcp transcription by hydroxylamine when the promoter activity was assayed using the multicopy plasmid pNF383 in strain RK4353 or in a pcnB derivative in which the plasmid copy number is decreased to about one (data not shown). Furthermore, growth of both the parental strain and the hcp mutant, JCB5000, was totally inhibited by 1 mM hydroxylamine, unaffected by 0.2 mM hydroxylamine, and partially inhibited by 0.3 or 0.5 mM hydroxylamine; there were no significant differences in hydroxylamine toxicity between the two strains.
NsrR: sixth protein to regulate transcription factor at PnrfA.
Transcription initiation at the nrfA promoter is activated by FNR in the absence of oxygen and induced further by NarL and NarP in response to low concentrations of nitrate or to nitrite (35, 44). FNR-dependent transcription is also repressed by two nucleoid-associated factors, Fis and IHF, which bind to multiple sites within the promoter (6). Thus, PnrfA is a complex promoter with at least five proteins directly controlling transcription initiation. However, there is a residual response to nitrite even in a narL narP double mutant (38), and the possibility that a sixth factor might regulate transcription initiation at PnrfA was apparent from detailed experimental analysis of this promoter (5-7, 11). Two approaches were used to confirm that NsrR is this sixth factor that regulates PnrfA.
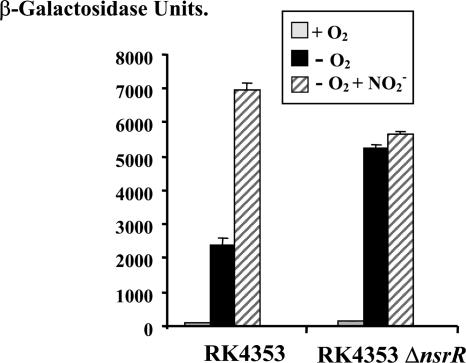
Plasmid pnrf53, which includes DNA from 209 bases upstream of the transcription start and a further 131 bases downstream cloned into the low-copy lacZ expression vector pRW50 to generate a PnrfA::lacZ transcriptional fusion (44), was transformed into strain RK4353 and its nsrR null mutant, strain JCB5010. Purified transformants were grown either aerobically or anaerobically in the presence or absence of nitrite, and the β-galactosidase activities were determined. Anaerobic expression from PnrfA was increased 2.2-fold by the disruption of nsrR (Fig. 2), consistent with the observation from microarray data that NsrR is a weak repressor of PnrfA. Nitrite induction was not observed in the nsrR null strain, indicating that NsrR might also play a role in the response of PnrfA to nitrite, possibly indirectly due to the formation of NO from nitrite.
FIG. 2.
NsrR represses anaerobic expression from the nrf promoter. The figure shows the β-galactosidase activities of RK4353 and its ΔnsrR derivative strain JCB5010, carrying pRW50 containing the pnrf53 promoter fragment. Bacteria were grown aerobically and anaerobically in MS medium and nitrite was added to a final concentration of 2.5 mM where indicated. The β-galactosidase activities are expressed as nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed min−1 (mg dry cell mass)−1. Each level of activity shown is the average of three independent determinations ± standard deviation.
In view of the ability of multicopy plasmids to titrate out the repressor activity of NsrR and the possibility that a mutation in nsrR might have secondary polar effects on the downstream rnr gene encoding RNase R (10), the repressor titration experiments were repeated with the Lac− strain RV, doubly transformed with pnrf53 and either pGIT1 or pGIT8. Bacteria were grown anaerobically with fumarate, trimethylamine-N-oxide, and either glucose or glycerol as the main carbon source, and the β-galactosidase activities were determined. The ability of the NsrR binding site on the multicopy plasmid pGIT1, but not on pGIT8, to titrate out the NsrR repressor and hence derepress PnrfA transcription was measured during anaerobic growth in the absence of nitrate and nitrite. The data from independent duplicate cultures were completely consistent with NsrR repression of PnrfA [1,200 and 1,270 nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed·min−1·(mg bacterial dry mass)−1 for the pGIT1 transformant compared with 180 and 230 units for the pGIT8 transformant]. However, the double transformants were unable to grow anaerobically with glycerol rather than glucose as the primary carbon source. Even with glucose, the growth of the double transformants was extremely slow compared with the growth of bacteria transformed with one of the pGIT plasmids alone and was variable between experiments.
DISCUSSION
The exploitation of the ability of a multicopy plasmid to titrate out NsrR and hence relieve NsrR repression enabled us to demonstrate that NsrR is a global regulator of the response to RNS in E. coli (4). This approach is similar to the ferric uptake regulator titration assay (42), with the inclusion of a microarray analysis to identify the genes derepressed by NsrR titration. Some of the small increases in transcript abundance detected when NsrR is active were almost certainly (but it was not independently confirmed) due to secondary effects, but the primary effects of NsrR as a repressor of operons involved in the relief of reactive nitrogen stress were largely confirmed by independent evidence. Previous studies have reported that hmpA transcription is induced during NO- or S-nitrosoglutathione-induced stress (18, 26), and NO also induces synthesis of the di-iron protein YtfE (4, 25, 26). We recently predicted that there is a previously undocumented nitrite-responsive transcription factor that regulates the expression of the nrf operon encoding the periplasmic nitrate reductase, as well as genes of unknown function that include ytfE and yeaR-yoaG (11). All of these transcripts were more abundant in the transformant in which the pool of NsrR was depleted than in the control transformant, validating our repressor titration approach and indicating that NsrR is the additional transcription factor.
One interesting result from the microarray analysis was the observation that the complete periplasmic pathway for nitrate reduction to ammonia is regulated by NsrR. Pathogenic bacteria must be able to defend themselves from RNS originating from four sources: products of their own metabolism; products of other bacteria that share their ecological niche (for example, NO generated by lactic acid bacteria in the gastrointestinal tract); NO generated as part of host defense mechanisms; and products of nonspecific chemical reactions. Most of the above threats originate outside enteric bacteria, so it is appropriate that they can be neutralized by the NO reductase activity previously documented for the periplasmic nitrite reductase Nrf (36). If so, how does E. coli protect itself against RNS that enter or are made in the cytoplasm?
A major protection mechanism against NO in the cytoplasm is provided by flavorubredoxin and its reductase, NorVW, which are synthesized in response to NO activation of the transcription activator NorR (15, 20, 22). The three operons most strongly up-regulated in response to NsrR titration are hmpA, ytfE, and hcp-hcr (Table 3). Both Hmp and YtfE are clearly established as cytoplasmic components of the RNS response (25, 26, 28), suggesting that the same might be true for HCP. Microarray analysis of the E. coli FNR, NarXL, and NarQP regulons and supporting transcription fusion data revealed that hcp expression is regulated in parallel with the cytoplasmic NADH-dependent nitrite reductase Nir (11). We inferred that this implied a function for HCP in detoxifying a product generated when the NarXL two-component system is activated, possibly an RNS generated as a side product of nitrite reduction to ammonia by Nir. The hcr product has been shown to be an NADH-dependent HCP reductase that presumably functions to provide electrons for the reduction of the HCP substrate (45), which was tentatively identified to be hydroxylamine (46). However, there is a hydroxylamine reductase activity associated with the cytoplasmic NADH-dependent nitrite reductase NirBD that is at least as effective as that of HCP (23). While it is possible that HCP is a back-up mechanism to provide protection against hydroxylamine toxicity, conditions under which its role is significant remain to be revealed, so other physiological roles for HCP must be considered. Even when it is expressed from a single chromosomal copy of the hcp gene, HCP accumulates as an abundant protein (45). Possibly HCP simply binds hydroxylamine stoichiometrically to prevent it from inhibiting bacterial metabolism until it can be reduced by Nir to ammonia (23). A precedent for such a detoxification mechanism is cytochrome c′, which protects pathogenic neisseria by binding nitric oxide (21, 43). A further possibility meriting consideration is that HCP repairs NO- or hydroxylamine-induced damage to iron-sulfur centers, for example, the Fe-S centers of NirB.
Recently an entirely different role was proposed for HCP, namely, that it functions as a peroxidase (1). This suggestion was based upon observations that HCP is oxidized by hydrogen peroxide, that an hcp mutant is more sensitive to hydrogen peroxide than its parent, and that hcp transcription is regulated by OxyR. However, other links between RNS and the OxyR regulon have been reported (18), and the phenotype of the hcp mutant was rather weak. We therefore suggest that the physiological substrate reduced by HCP remains to be determined but is more likely a reactive nitrogen compound (other than hydroxylamine) than a reactive oxygen species.
The microarray analysis confirmed our previous suggestion that transcription of the two-gene operon yeaR-yoaG is subject to NsrR repression. This operon is therefore another candidate for encoding proteins that protect E. coli against RNS, especially in anaerobic environments where FNR might be inactivated by severe NO damage (11, 12, 18). Future studies must focus on the biochemical functions of YeaR, YoaG, YtfE, and yet again, HCP.
Acknowledgments
We thank Lesley Griffiths for excellent technical support, Martha Justino for strain JCB4401, and W. M. van Dongen for helpful discussions and access to unpublished data.
We thank the Darwin Trust of Edinburgh and the United Kingdom Medical Research Council for research studentships for N.F. and D.S., respectively; the Biotechnology and Biological Sciences Research Council for funding via grants EGA16107 and P20180 to J.C.; and the National Science Foundation for funding via grant MCB0702858 to S.S.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Almeida, C. C., C. V. Romao, P. F. Lindley, M. Teixeira, and L. M. Saraiva. 2006. The role of hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 281:32445-32450. [DOI] [PubMed] [Google Scholar]
- 2.Arendsen, A. F., J. Hadden, G. Card, A. S. McAlpine, S. Bailey, V. Zaitsev, P. F. Lindley, M. Kröckel, A. X. Trautwein, M. C. Feiters, J. M. Charnock, C. D. Garner, S. J. Marritt, A. J. Thomson, I. M. Kooter, M. K. Johnson, W. A. M. van den Berg, W. M. A. M. van Dongen, and W. R. Hagen. 1998. The prismane protein resolved: X-ray structure at 1.7 Å and multiple spectroscopy of two novel 4Fe clusters. J. Biol. Inorg. Chem. 3:81-95. [Google Scholar]
- 3.Beaumont, H. J. E., S. I. Lens, W. N. M. Reijnders, H. V. Westerhoff, and R. J. M. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54:148-158. [DOI] [PubMed] [Google Scholar]
- 4.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning, D. F., C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2002. Independent regulation of the divergent Escherichia coli nrfA and acsP1 promoters by a nucleo-protein assembly at a shared regulatory region. Mol. Microbiol. 43:687-701. [DOI] [PubMed] [Google Scholar]
- 6.Browning, D. F., D. C. Grainger, C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2005. Integration of three signals at the Escherichia coli nrf promoter: a role for Fis protein in catabolite repression. Mol. Microbiol. 57:496-510. [DOI] [PubMed] [Google Scholar]
- 7.Browning, D. F., D. C. Grainger, C. M. Beatty, A. J. Wolfe, J. A. Cole, and S. J. W. Busby. 2006. The Escherichia coli K-12 NarL and NarP proteins insulate the nrf promoter from the effects of integration host factor. J. Bacteriol. 188:7449-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busby, S. J. W., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 9.Cabello, P., C. Pino, M. F. Olmo-Mira, F. Castillo, M. D. Roldan, and C. Moreno-Vivian. 2004. Hydroxylamine assimilation by Rhodobacter capsulatus E1F1: requirement of the hcp gene (hybrid cluster protein) located in the nitrate assimilation nas gene region for hydroxylamine reduction. J. Biol. Chem. 279:45485-45494. [DOI] [PubMed] [Google Scholar]
- 10.Cairrao, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 50:1349-1360. [DOI] [PubMed] [Google Scholar]
- 11.Constantinidou, C. C., J. L. Hobman, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the fumarate and nitrate reduction regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL and NarQP as Escherichia coli adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4808. [DOI] [PubMed] [Google Scholar]
- 12.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thompson, J. Green, and R. K. Poole. 2002. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin, A., E. C. Ziegelhoffer, P. J. Kiley, and V. Stewart. 1998. Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Autréaux, B., N. P. Tucker, R. Dixon, and S. Spiro. 2005. A non-heme iron centre in the transcription factor NorR senses nitric oxide. Nature 437:769-772. [DOI] [PubMed] [Google Scholar]
- 16.Dobbek, H., V. Svetlitchnyi, L. Gremer, R. Huber, and O. Meyer. 2001. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster. Science 293:1281-1285. [DOI] [PubMed] [Google Scholar]
- 17.Filenko, N. A., D. F. Browning, and J. A. Cole. 2005. Transcriptional regulation of a hybrid cluster (Prismane) protein. Biochem. Soc. Trans. 33:195-197. [DOI] [PubMed] [Google Scholar]
- 18.Flatley, J., J. Barrett, S. T. Pullan, M. N. Hughes, J. Green, and R. K. Poole. 2005. Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J. Biol. Chem. 280:10065-10072. [DOI] [PubMed] [Google Scholar]
- 19.Giel, J. L., D. Rodionov, M. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60:1058-1076. [DOI] [PubMed] [Google Scholar]
- 20.Gomes, C. M., J. B. Vicente, A. Wasserfallen, and M. Teixeira. 2000. Spectroscopic studies and characterization of a novel electron-transfer chain from Escherichia coli involving a flavorubredoxin and its flavoprotein reductase partner. Biochemistry 39:16230-16237. [DOI] [PubMed] [Google Scholar]
- 21.Huston, W. M., E. C. Lowe, C. S. Butler, and J. W. Moir. 2005. Purification and characterization of cytochrome c′ from Neisseria meningitidis. Biochem. Soc. Trans. 33:187-189. [DOI] [PubMed] [Google Scholar]
- 22.Hutchins, M. I., N. Mandhana, and S. Spiro. 2002. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 184:4640-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, R. H., J. A. Cole, and A. Cornish-Bowden. 1981. The steady-state kinetics of the NADH-dependent nitrite reductase from Escherichia coli K12: nitrite and hydroxylamine reduction. Biochem. J. 199:171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayaraman, P. S., T. C. Peakman, S. J. Busby, R. V. Quincey, and J. A. Cole. 1987. Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the Fnr protein and nitrite. J. Mol. Biol. 196:781-788. [DOI] [PubMed] [Google Scholar]
- 25.Justino, M. C., C. C. Almeida, V. L. Goncalves, M. Teixeira, and L. M. Saraiva. 2006. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 257:278-284. [DOI] [PubMed] [Google Scholar]
- 26.Justino, M. C., J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2006. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280:2636-2643. [DOI] [PubMed] [Google Scholar]
- 27.Kim, C. C., D. Monack, and S. Falkow. 2003. Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect. Immun. 71:3196-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. O., Y. Orii, D. Lloyd, M. N. Hughes, and R. K. Poole. 1999. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445:389-394. [DOI] [PubMed] [Google Scholar]
- 29.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N. R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol. Lett. 95:271-276. [DOI] [PubMed] [Google Scholar]
- 30.Lundberg, J. O., E. Weitzberg, J. A. Cole, and N. Benjamin. 2004. Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2:593-602. [DOI] [PubMed] [Google Scholar]
- 31.Macedo, S., D. Aragao, E. P. Mitchell, and P. Lindley. 2003. Structure of the hybrid cluster protein (HCP) from Desulfovibrio desulfuricans ATCC 27774 containing molecules in the oxidized and reduced states. Acta Crystallogr. Sect. D 59:2065-2071. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Nakano, M. M., H. Geng, S. Nakano, and K. Kobayashi. 2006. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 188:5878-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overton, T. W., R. Whitehead, Y. Li, L. A. S. Snyder, N. J. Saunders, H. Smith, and J. A. Cole. 2006. Coordinated regulation of the Neisseria gonorrhoeae truncated denitrification pathway by the oxygen-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J. Biol. Chem. 281:33115-33126. [DOI] [PubMed] [Google Scholar]
- 35.Page, L., L. Griffiths, and J. A. Cole. 1990. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch. Microbiol. 154:349-354. [DOI] [PubMed] [Google Scholar]
- 36.Poock, S. R., E. R. Leach, J. W. B. Moir, J. A. Cole, and D. J. Richardson. 2002. Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 277:23664-23669. [DOI] [PubMed] [Google Scholar]
- 37.Pope, N. R., and J. A. Cole. 1984. Pyruvate and ethanol as electron donors for nitrite reduction by Escherichia coli K12. J. Gen. Microbiol. 130:1279-1284. [DOI] [PubMed] [Google Scholar]
- 38.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61:927-939. [DOI] [PubMed] [Google Scholar]
- 40.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, V., and C. H. McGregor. 1982. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J. Bacteriol. 151:788-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 43.Turner, S. M., J. W. B. Moir, L. Griffiths, T. W. Overton, H. Smith, and J. A. Cole. 2005. Mutational and biochemical analysis of cytochrome c′, a nitric oxide-binding lipoprotein important for adaptation of Neisseria gonorrhoeae to oxygen-limited growth. Biochem. J. 388:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyson, K. L., J. A. Cole, and S. J. W. Busby. 1994. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for NarP- and NarL-dependent regulation. Mol. Microbiol. 13:1045-1055. [DOI] [PubMed] [Google Scholar]
- 45.van den Berg, W. A., W. R. Hagen, and W. M. van Dongen. 2000. The hybrid-cluster protein (“prismane protein”) from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe-2S] and [4Fe-2S-2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe-2S]. Eur. J. Biochem. 267:666-676. [DOI] [PubMed] [Google Scholar]
- 46.Wolfe, M. T., J. Heo, J. S. Garavelli, and P. W. Ludden. 2002. Hydroxylamine reductase activity of the hybrid cluster protein from Escherichia coli. J. Bacteriol. 184:5898-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeo, W.-S., J.-H. Lee, K.-C. Lee, and J.-H. Roe. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61:206-218. [DOI] [PubMed] [Google Scholar]