Abstract
Spores of Bacillus megaterium QM B1551 germinate in response to a number of trigger compounds, including glucose, proline, leucine, and inorganic salts. An approximate 6-kb region of the 165-kb plasmid was found to harbor a tricistronic receptor operon, gerU, and a monocistronic receptor component, gerVB. The gerU operon was observed to complement the germination response in plasmidless strain PV361 to glucose and leucine, with KBr acting as a cogerminant. Proline recognition is conferred by the monocistronic gerVB gene, the presence of which also improves the germination response to other single-trigger compounds. A chimeric receptor, GerU*, demonstrates interchangeability between receptor components and provides evidence that it is the B protein of the receptor that determines germinant specificity. Introduction of the gerU/gerVB gene cluster to B. megaterium KM extends the range of germinants recognized by this strain to include glucose, proline, and KBr in addition to alanine and leucine. A chromosomally encoded receptor, GerA, the B component of which is predicted to be truncated, was found to be functionally redundant. Similarly, the plasmid-borne antiporter gene, grmA, identified previously as being essential for germination in QM B1551, did not complement the germination defect in the plasmidless variant PV361. Wild-type spores carrying an insertion-deletion mutation in this cistron germinated normally; thus, the role of GrmA in spore germination needs to be reevaluated in this species.
Bacterial spores rely upon a number of small molecules, including amino acids, purine ribosides, sugars, and ions, to indicate that environmental conditions are favorable for the resumption of growth. The rapid transition from extreme dormancy to loss of resistance properties and resumption of metabolism displayed by spores upon exposure to germinant molecules is regulated by a process known as spore germination (22, 23, 32). Germinant interaction with cognate receptors located within the inner spore membrane (11, 26) stimulates changes in the permeability of this membrane to various metal ions and Ca-dipicolinic acid (DPA), which are effluxed from the spore core, while permitting partial hydration of the spore core. Enzymatic hydrolysis of the spore's thick layer of cortical peptidoglycan and protective outer coat layers is then initiated, permitting complete hydration and the resumption of vegetative metabolism. Since strict germination events precede macromolecular synthesis, the germination apparatus must be present in the dormant spore (22, 32).
A substantial amount of genetic evidence has been presented that strongly suggests that orthologous proteins belonging to the GerA family form the receptors through which the spore senses its environment (21, 23, 25). GerA structural genes are arranged in tricistronic operons, which are expressed during sporulation in the developing forespore (7). Multiple copies of these receptor operons reside within the genomes of all Bacillus species examined to date. Hydropathy profiling indicates that products of the A and B genes are integral membrane proteins—consistent with their being receptors for environmental stimuli—while the C component encodes a relatively hydrophilic product that is probably anchored to the membrane via the addition of a diacylglycerol moiety. While the genetic organization of the A, B, and C cistrons can vary between species, the conserved nature of the operon structure suggests that each receptor is a complex of all three proteins, and experiments have shown that mutation of any cistron within the operon results in inactivation of the respective receptor (22, 25). Genetic evidence based on studies using mutant Bacillus subtilis constructs has been presented suggesting that different receptors can physically interact (1, 3) while genetic evidence for the physical interaction of the respective A, B, and C proteins that form the receptor complex has also been presented (13, 24).
While the GerA receptor in B. subtilis can function independently to trigger the spore germination response to l-alanine, most germinant receptors appear to work in concert to induce the germination cascade. The best understood example of this is probably the B. subtilis germination response to a mixture of asparagine, glucose, fructose, and potassium ions, which requires both the GerB and GerK receptors (1, 4, 15). The response to single germinants can also require interaction between two or more receptors, such as the inosine germination response in Bacillus cereus 569, which has a strict requirement for the products of the gerI and gerQ operons (2). Other examples of this phenomenon have been elucidated in Bacillus anthracis, where each known germination pathway requires at least two distinct receptors (8). Whether receptors that function cooperatively physically interact, perhaps as localized receptor clusters within the inner membrane, has yet to be established. Similarly, whether receptors that function cooperatively to trigger germination in response to single germinants recognize different parts of the molecule also remains to be determined. Other unknowns include the identification of proteins that interact with the receptors, which may include a response regulator protein that processes receptor stimuli as part of the signal transduction process. Since the redistribution of ions constitutes the earliest detectable biochemical event associated with germination (35), ion transporter or DPA channels appear likely candidates to fulfill this function.
The current study employs molecular techniques to investigate a number of genes implicated in the germination response of Bacillus megaterium QM B1551 (ATCC 12872), a strain that has been used extensively in the past to study biochemical and physiological aspects of spore germination (12, 19, 27, 29). Spores of B. megaterium QM B1551 germinate rapidly in response to a number of trigger compounds, including glucose, proline, leucine, and, unusually, a variety of inorganic salts. Approximately 11% of the B. megaterium QM B1551 genome is carried on seven discrete plasmids, which range in size from 5.4 kb to 165 kb (18). A previous study determined that B. megaterium QM B1551 strains cured of the 165-kb plasmid fail to germinate in response to single germinants, suggesting that a gene or genes encoding essential components of the germination apparatus must reside on this plasmid (33). A candidate gene, grmA, encoding a putative sodium-proton antiporter, was subsequently identified via transposon mutagenesis experiments as being essential for the QM B1551 germination response to single-trigger germinants (37). Sequence analysis of purified plasmid DNA has revealed that the grmA gene resides on either the 71-kb or 165-kb plasmids (P. S. Vary, personal communication). A homologue of this protein, GerN, was also identified as being important in the inosine, but not alanine, germination response of B. cereus 569, suggesting that transporters may be associated with specific receptor complexes (38). Genes predicted to encode distant GrmA-type protein homologues, yhaU and yjbQ, do not appear to be involved in spore germination in B. subtilis.
In this communication we provide evidence that the B. megaterium QM B1551 spore germination response to single-trigger compounds is mediated by a plasmid-borne GerA type receptor operon. Uniquely in Bacillus species studied to date, a third component—a plasmid-borne monocistronic gene predicted to encode a GerAB type protein—is shown to contribute to germinant specificity while also improving the rate of germination in response to other trigger compounds. Surprisingly, the GrmA antiporter was found not to be required for germination, either in wild-type spores carrying a defective copy of the structural gene or in strain PV361 complemented with the appropriate receptor genes.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains employed in this study are listed in Table 1. B. megaterium strains were routinely cultured on LB agar or broth at 30°C, containing antibiotics where appropriate (tetracycline, 1.25 μg/ml; kanamycin, 5 μg/ml). Bacillus protoplasts were transformed as described previously (20), except selection for transformants was accomplished by direct selection at 30°C on antibiotic-containing RHAF agar plates (12.5 μg/ml tetracycline or 5 μg/ml kanamycin). Escherichia coli NovaBlue (Novagen), which was used to prepare all plasmid constructs, was cultured in LB medium at 37°C supplemented with 75 μg/ml carbenicillin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| B. megaterium strains | ||
| QM B1551 | Wild-type strain | P. S. Varya |
| PV361 | Plasmidless variant of QM B1551 | 40 |
| PV203 | 9-, 71-, and 165-kb plasmids | P. S. Vary |
| KM | Wild-type strain | K. Johnstone |
| QM B1551 transformants | ||
| GC413 | gerAA::pUCTV2 Kanr Tetr | This study |
| GC414 | ΔgrmA::Kanr | This study |
| PV361 transformants | ||
| GC417 | gerAA::pUCTV2 Kanr Tetr | This study |
| GC420 | pUCTV-grmA Tetr | This study |
| GC421 | pUCTV-gerU Tetr | This study |
| GC422 | pUCTV-gerVB Tetr | This study |
| GC423 | pUCTV-(gerU-gerVB) Tetr | This study |
| GC424 | pUCTV-(gerUA-gerUC) Tetr | This study |
| GC428 | pUCTV-gerU* Tetr | This study |
| KM transformants | ||
| GC425 | ΔgerAA::Kanr | This study |
| GC426 | pUCTV-(gerU-gerVB) Tetr | This study |
| Plasmids | ||
| pUCTV2 | Shuttle plasmid with TS Bacillus ori, Tetr | 42 |
| pGEM3Z | E. coli cloning vector Ampr | Promega |
| pDG792 | Kanr cassette | BGSC |
Original source J. C. Vary.
Spore preparation.
B. megaterium spores were prepared in 200-ml aliquots of supplemented nutrient broth (SNB) (6) in 2-liter flasks (30°C at 200 rpm). Strains carrying freely replicating plasmid constructs were supplemented with 1.25 μg/ml tetracycline to provide selective pressure for maintenance of the plasmid. Spores were harvested after 48 h and then cleaned by repeated rounds of centrifugation and sterile cold-water washing before being stored on ice. All spore suspensions used in this work were observed to be free of vegetative cells and debris. Germination experiments were typically conducted within 48 h of harvesting to counter any changes in germination properties that might develop upon longer-term storage.
Construction of Bacillus megaterium mutant strains. (i) Disruption of gerA.
A plasmid developed with a view to obtaining an insertion-deletion in the gerAA cistron was constructed. Essentially, a 1322-bp region of gerAA (positions 301 to 1623 in the gerAA coding sequence; GenBank accession number U61380) was amplified by PCR using primers that contained EcoRI and BamHI restriction sites in the forward and reverse primers, respectively (all primer sequences are available upon request). This product was digested and ligated with pGEM3Z restricted with the same enzymes, and the ligation mixture was used to transform E. coli, from which a recombinant plasmid, pGEM-gerAA, was isolated. An inverse PCR was performed using primers incorporating XhoI and SacI restriction sites creating a deletion in gerAA between positions 1040 and 1211 of the original coding sequence. The digested and purified PCR product was ligated with a kanamycin resistance cassette, excised from pDG792 with the same enzymes, and the ligation mixture used to transform E. coli, from which plasmid pGEM-ΔgerAA::Kan was isolated. The entire ΔgerAA::Kan construct was PCR amplified with EcoRI-tagged primers and cloned into the EcoRI site of pUCTV2 after restriction digestion and ligation with the appropriate enzymes. Plasmid pUCTV-ΔgerAA::Kan was isolated from a transformant of E. coli. This plasmid was used to transform B. megaterium strains QM B1551, PV361, and KM to kanamycin resistance via polyethylene glycol-mediated protoplast transformation. Transformants were recovered on RHAF agar plates containing 5 μg/ml kanamycin and incubated at 30°C. Purified transformant colonies were then incubated overnight at 42°C (the nonpermissive temperature for plasmid replication) on LB agar plates containing 5 μg/ml kanamycin to permit selection of colonies that had integrated the plasmid into the chromosome at the cloned locus via homologous recombination. The correct integration and subsequent disruption of gerAA in single crossover candidate colonies were confirmed by PCR, employing primer pairs specific to the 5′ end of gerAA, which is outside the cloned locus on the plasmid, and to a region of gerAA downstream from the inserted kanamycin cassette. Despite repeated attempts, double-crossover colonies, which had lost the tetracycline resistance phenotype and wild-type gene at 42°C, could be isolated only from strain KM (GC425), as confirmed by PCR. Single-crossover mutants, however, which do not have an intact copy of gerAA, were demonstrated to be genetically stable when cultured at 30°C in the absence of antibiotic. This was established by conducting a series of PCRs, employing primers to specifically denote the absence of intact gerAA and replicating plasmid (to detect excision events), using DNA extracted from spore samples germinated in complex medium (SNB) for 1 h as a template.
(ii) grmA constructs.
In order to construct a grmA insertion-deletion mutant strain, a 1,754-bp region (from positions 33 to 1787 of the known sequence; GenBank accession number U17283), which encompassed the entire gene and included flanking sequences, was amplified by PCR using primers that incorporated SacI and HindIII sites at the 5′ and 3′ ends, respectively. This product was digested and ligated between the SacI and HindIII sites of pGEM3Z and used to transform E. coli, from which the plasmid pGEM-grmA was isolated. An inverse PCR was then performed, using NcoI- and XhoI-tagged primers, to introduce a deletion between positions 800 and 967 of the known sequence. Digested and purified PCR product was ligated with a kanamycin resistance cassette excised from pDG792 with the appropriate enzymes. The ligation mixture was used to transform E. coli, from which the plasmid pGEM-ΔgrmA::Kan was isolated. The ΔgrmA::Kan cassette was then amplified by PCR using primers to incorporate EcoRI sites and then digested and ligated with EcoRI-cut pUCTV2. Plasmid pUCTV-ΔgrmA::Kan was isolated from transformant E. coli. This plasmid was used to transform B. megaterium QM B1551 to tetracycline and kanamycin resistance via polyethylene glycol-mediated protoplast transformation. Selection of colonies that had integrated the plasmid at the cloned locus was achieved by culturing transformants overnight at the nonpermissive temperature for plasmid replication on LB agar plates containing kanamycin (5 μg/ml). A transformant that had undergone double homologous recombination (Tets Kanr), GC414, carrying insertion-deletion mutations in the grmA gene, was isolated after further incubation at 42°C on LB agar plates without antibiotic. The correct construction of this strain was confirmed by PCR and sequencing. To test whether the grmA gene could complement the germination defect in strain PV361, a 1,754-bp region that encompassed the entire open reading frame including putative regulatory sequences was PCR amplified with primers incorporating EcoRI sites. This product was ligated between the EcoRI sites of pUCTV2, and the recombinant plasmid pUCTV-grmA was isolated from a transformant E. coli. This plasmid was used to transform PV361 to tetracycline resistance, creating strain GC420.
(iii) gerU and gerVB complementation.
A number of plasmids incorporating different fragments of the 6-kb region that encompasses the gerU operon and gerVB gene were constructed for complementation analyses in strain PV361. A 5,922-bp fragment (from positions 78 to 6000 of the known sequence) encompassing the gerU operon and gerVB was amplified by PCR with primers incorporating MfeI restriction sites. This product was cloned into the EcoRI site of pUCTV2 using appropriate enzymes, and the ligation mixture was used to transform E. coli, from which plasmid pUCTV-(gerU-gerVB) was isolated. This plasmid was used to transform strains PV361 and KM to tetracycline resistance, giving strains GC423 and GC426, respectively.
The gerU operon was cloned into pUCTV2 by PCR amplification of the known sequence between positions 78 and 4561 using primers that incorporate MfeI sites. This product was digested and ligated with EcoRI-cut pUCTV2 and used to transform E. coli to give plasmid pUCTV-gerU. This plasmid was used to transform PV361 to tetracycline resistance, giving strain GC421.
The gerVB gene was cloned into pUCTV2 by PCR amplification of the known sequence between positions 4511 to 6000 using primers incorporating SacI sites. This product, which included putative up- and downstream regulatory sequences, was ligated with SacI-digested pUCTV2 and used to transform E. coli. Plasmid pUCTV-gerVB, isolated from a transformant E. coli, was used to transform PV361 to tetracycline resistance, giving strain GC422.
An overlap PCR technique was used to create the gerUA-UC-VB chimeric fusion operon. PCR was used to prepare two fragments of DNA, the first encompassing the coding and putative upstream regulatory sequences for gerUA and gerUC (but not gerUB) and the second encompassing the gerVB gene and potential rho-independent terminator sequence. These products, which included 20 bp of overlapping sequence at the 3′ and 5′ ends, respectively, were purified and mixed to provide template for a subsequent round of PCR using primers with MfeI sites, which resulted in the creation of a fragment of DNA encompassing the gerUA, gerUC, and gerVB genes, arranged as a GerA-type receptor operon with appropriate regulatory sequences. This product, subsequently referred to as gerU*, was digested with MfeI, ligated with EcoRI-digested pUCTV2, and used to transform E. coli. Recombinant plasmid pUCTV-gerU* was used to transform PV361 to tetracycline resistance, giving strain GC428. The gerUA-gerUC fragment was also ligated with pUCTV2 to create plasmid pUCTV-(gerUA-gerUC), which was used to transform PV361 to tetracycline resistance, giving strain GC424.
Germination assays.
Spores at 5 to 10 mg/ml (dry weight) in water were heat shocked at 60°C for 10 min and then cooled on ice. Spores were routinely germinated at 30°C at an optical density at 600 nm (OD600) of between 0.9 to 1.0 in 5 mM Tris-HCl buffer (pH 7.8) with a 10 mM concentration of germinant (50 mM for KBr). Control experiments with heat-shocked spores in buffer alone were included for each experiment. Spore germination was monitored by measurement of the OD600 of the suspension over a 40-min period using a 1-ml cuvette in a Hewlett Packard 8452A diode array spectrophotometer. All values reported are the averages of at least duplicate experiments utilizing independent batches of spores. Where presented, maximum rates of spore germination are given relative to the OD600 loss observed for QM B1551 spores incubated with 10 mM glucose, where a 65% loss of OD600 correlates to approximately 100% spore germination, as determined by loss of heat resistance. In some experiments we also measured spore germination by determining the amount of DPA released. Essentially, 1-ml aliquots of germinating culture were passed through a 0.22-μm-pore-size syringe filter, and the DPA released into the supernatant was determined by absorption at 270 nm (3). Germination was also routinely followed by phase-contrast microscopy.
Molecular biology methods and bioinformatic analyses.
All PCR procedures were performed using standard methodologies using KOD Hot Start polymerase (Novagen). Vectorette PCR was performed using the Vectorette II Starter Pack (Sigma Genosys) according to the manufacturer's instructions. Vectorette libraries prepared from ClaI- and MfeI-digested genomic DNA were employed to sequence the 5′ end of the gerU operon. DNA sequencing was performed by the Department of Genetics sequencing facility (University of Cambridge). DNA assembly and analysis were performed using GCG version 11 (Accelrys) and the ExPASy proteomics server (Swiss Institute of Bioinformatics). The Institute for Genomic Research Comprehensive Microbial Resource web tool (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi) was used to search the genomes of sequenced Bacillus species for potential monocistronic receptor genes.
Nucleotide sequence accession number.
A 6,000-bp region of the 165-kb plasmid, including the gerU operon and gerVB, has been submitted to the GenBank (accession number EF362724).
RESULTS
The putative sodium-proton antiporter GrmA is not required for spore germination.
Using a transposon mutagenesis approach, a previous study identified a homologue of the Enterococcus hirae sodium-proton antiporter gene as being required for germination in QM B1551 spores in response to all single-trigger compounds tested (37). A PCR approach employing primers specific to grmA and template DNA purified from QM B1551 and PV361 suggested that the grmA gene is carried on a plasmid. This finding was verified by previously unpublished sequence information made available to us, which confirmed that grmA resides on either the 71-kb or 165-kb plasmid. We cloned the entire grmA open reading frame and putative regulatory sequences into pUCTV2 and transformed plasmidless PV361 to tetracycline resistance (strain GC420) in order to determine whether grmA could complement this strain's defective germination phenotype. We also constructed a QM B1551 derivative with an insertion-deletion in grmA (strain GC414; ΔgrmA::Kanr), using a temperature-dependent allelic-exchange strategy, in order to reassess the role of the GrmA protein in B. megaterium spore germination. Both the QM B1551 grmA knockout strain and PV361 carrying a copy of grmA on a multicopy plasmid sporulated normally when cultured in SNB.
When spores from these respective strains were assayed for their germination response to each of the single-trigger compounds, the wild-type strain carrying the grmA mutation (GC414) germinated normally, where as the PV361 derivative carrying intact grmA (GC420) failed to germinate (Fig. 1). The germination response of the grmA mutant strain contradicts previous observations (37) and is difficult to explain. Spores from our study were prepared using the same medium as reported in the previous study; therefore, differences in microbial culture conditions and spore preparation procedures, which can exert significant effects on the expression level of germination associated genes (10), appear unlikely. Perhaps significant variation exists between strains deposited with different culture collections, and this may contribute to differences in phenotype. Clearly, it would be of interest to transfer our grmA disruption vector to the strain employed in the original study, while characterization of the role of GerN in other strains of B. cereus would also be of interest. Regardless, the role of GrmA-type proteins in spore germination requires further consideration and investigation.
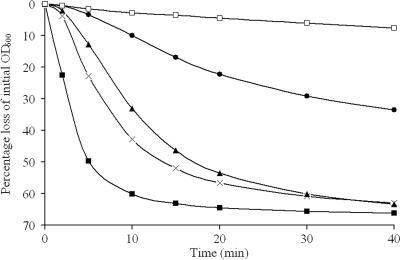
FIG. 1.
Germination of B. megaterium QM B1551 grmA (GC414) and PV361 complemented with grmA (GC420) in 5 mM Tris-HCl (pH 7.8), plus a 10 mM concentration of the respective germinant (KBr, 50 mM). GC414 responses to glucose (▪), proline (×), leucine (▴), and KBr (•) are shown. The GC420 (□) response to only a mixture of germinants (GPLK) is shown for clarity. Results are averages of two independent spore preparations (standard deviation is <5% of the mean).
Plasmid-borne receptor homologues are required for germination in response to single germinants.
In the course of this study we were generously provided with unpublished sequence information derived from a mixture of the 71-kb and 165-kb plasmids (P.S. Vary, personal communication). One 10.6-kb contig of sequence was of particular interest as it contained an incomplete GerA-type receptor operon, which was truncated at the 5′ end of the contig. We subsequently sequenced the missing region of the A component of this tricistronic operon using a Vectorette PCR approach. The entire operon, which we have named gerU, comprises three open reading frames, which by homology with known receptor genes, are organized as gerUA, gerUC, and gerUB (Fig. 2). The gerUA gene (positions 525 to 2147 of the deposited sequence) is predicted to encode a 59.9-kDa protein containing 540 amino acid residues, with a central region containing four to six transmembrane alpha-helical segments (TMS). A possible alternative ATG start codon is positioned upstream (position 483) but is preceded by a poorly positioned and relatively weak potential ribosome binding site. The gerUC gene (positions 2137 to 3366), the initiation codon of which overlaps with the 3′ end of gerUA, is predicted to be a 45.1-kDa, 409-residue prelipoprotein, containing an N-terminal signal protein followed by a consensus sequence for diacylglycerol addition to a cysteine residue. The third open reading frame, gerUB (positions 3388 to 4488), is predicted to encode a 41-kDa, 366-residue protein with 10 TMS. The gerUB gene is followed by a region of dyad symmetry typical of a rho-independent terminator of transcription (positions 4580 to 4597). The entire gerU operon is preceded by −35 and −10 promoter sequences (positions 409 to 436) that are in reasonable agreement with the consensus sequence for transcription by σG-associated RNA polymerase, and each gene is preceded by an appropriately spaced ribosome binding site. All three gerU genes show a high degree of similarity to the corresponding chromosomal gerA cistrons (77, 74, and 81% amino acid sequence identity, respectively), suggesting a common evolutionary origin.
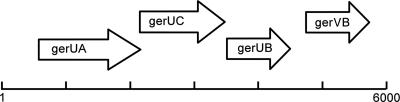
FIG. 2.
Genetic organization of the B. megaterium QM B1551 gerU operon and gerVB loci. The gerU operon is unusual in that the typical spore-receptor tricistronic operon structure is followed by a monocistronic transcription unit, gerVB, which possibly evolved via duplication and divergence from the gerUB gene. All four genes show close homology to the chromosomal gerA receptor operon.
The gerU operon is followed by an open reading frame (positions 4701 to 5801) that shares the same DNA strand and orientation as gerU and that is predicted to encode a 366-residue, 40.9-kDa protein with 10 TMS. This gene, which we have designated gerVB in deference to its location and similarity to gerUB (80% amino acid sequence identity), is flanked by a putative σG promoter (positions 4641 to 4671) and strong ribosome binding site and a potential rho-independent terminator of transcription (positions 5836 to 5857), suggesting that gerVB is transcribed as a monocistronic cistron. The proximity and similarity of gerVB to gerUB probably suggests that gerVB arose from duplication and subsequent divergence from gerUB, as opposed to horizontal gene transfer.
PCR analyses conducted on gel-extracted plasmids purified from strain PV203, which carries the 165-kb, 71-kb, and 9-kb plasmids, suggest that the gerU and gerVB genes reside on the 165-kb plasmid (data not shown). These data are consistent with PCR analyses conducted on a number of colonies that were inadvertently cured of the 165-kb plasmid during temperature-dependent mutagenesis procedures and which were also observed to have lost germinability to single-trigger compounds.
We decided to pursue an allelic-exchange mutagenesis strategy in order to attempt to determine whether these plasmid-encoded loci contribute to the germination response of the spore. This experimental approach proved to be extremely problematic since, for a number of pUCTV2 derivatives carrying cloned gerU inserts, transformants could not be recovered. The physiological reason for this effect was not fully established, although we suspect that transcriptional run-on from the antibiotic cassette embedded within the cloned target loci was causing problems. Modified vectors that were successfully transformed repeatedly integrated at the incorrect locus, while curing of the 165-kb plasmid occurred in a number of cases, leading to loss of the germination phenotype (data not shown). In view of these difficulties, we then decided to employ a genetic complementation approach, transforming the plasmidless host strain PV361 with vectors carrying copies of the gerU operon, gerVB, and gerU-gerVB before assessing the germination phenotype of transformant spores. Tetracycline-resistant transformants carrying these plasmid constructs sporulated normally when cultured in SNB.
A number of conclusions can be drawn from germination assay data collated from these strains (Table 2). First, it is evident that introduction of the gerU operon with the gerVB gene (strain GC423) complements the defective PV361 germination phenotype by permitting germination, at least in a fraction of the population, in response to all of the main single-trigger compounds. Complementation with the gerU operon alone (strain GC421) permits germination in response to glucose and leucine but not to proline or KBr, suggesting that GerVB is required for germination in response to proline and KBr. However, while the germination response to all single-trigger compounds is improved when gerVB is present in tandem with gerU, GerVB cannot function independently of GerU, as evidenced by strain GC422 (pUCTV-gerVB), which does not germinate in response to any of the single-trigger compounds. The physiological reason why only a fraction of the population should germinate has not been established, although perhaps the relatively high copy number of the pUCTV2-based plasmids (expected to be approximately 20 per cell [41]) compared to the low copy number of the native 165-kb plasmid (1 per cell [18]) may exert a deleterious effect. Alternatively, a degree of plasmid instability, which is not detectable by plate count methods employed in this study, leading to loss of the germination phenotype in a fraction of the spore population may account for incomplete germination.
TABLE 2.
Rates of germination of spores complemented with gerU and/or gerVBa
| Strain (receptor[s] present) | Rate of spore germination with the indicated germinantb
|
|||||
|---|---|---|---|---|---|---|
| Buffer | GPLK | Glucose | l-proline | l-leucine | KBr | |
| QM B1551 (wild type) | 22 | 100 | 100 | 99 | 99 | 90 |
| PV361 (plasmidless) | 1 | 9 | <1 | 1 | 1 | 9c |
| GC423 (GerU, GerVB) | 6 | 72 | 58 | 52 | 48 | 53 |
| GC421 (GerU) | 2 | 71 | 41 | 1 | 34 | 8 |
| GC422 (GerVB) | <1 | 9 | 1 | 1 | 1 | 7 |
Spores were germinated in Tris-HCl (5 mM, pH 7.8) for 40 min with 10 mM germinant (50 mM KBr). Spore germination was measured, and germination rates were calculated as described in Materials and Methods. Values are the means of at least duplicate experiments; standard deviation was <5% of the mean.
Rates of spore germination are given relative to the value for QM B1551 spores in 10 mM glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation.
KBr can cause aggregation of PV361 and derivative strains, which may affect the OD600 readings. Where appropriate, germination was also monitored as described in Materials and Methods by measuring DPA release, which showed similar results to OD600 analyses.
Further evidence that gerVB provides the proline recognition determinant was provided by experiments conducted to assess additive or synergistic germination responses triggered by combinations of germinants. Incubation of GC421 (pUCTV-gerU) spores in a mixture of glucose, proline, leucine, and KBr (GPLK) induces a germination response similar in magnitude to that observed in strain GC423 [pUCTV-(gerU-gerVB)], which is appreciably stronger than that induced by individual germinants (Table 2). Additive and synergistic effects are observed when GC421 spores are incubated in glucose with leucine or KBr, respectively (Table 3), but not when they are incubated with glucose and proline. The combination of leucine with KBr also shows a strong synergistic effect, increasing the proportion of germinated spores to approximately 61% compared to 34% and 8% for leucine and KBr as single germinants, respectively. Considered together, data presented to this point provide evidence that GerU forms a receptor for glucose and leucine, while KBr can be utilized as a cogerminant to synergistically enhance the germination response. Added to this, GerVB can interact with GerU to confer or unmask a cognate binding site for proline, while permitting KBr to act as a sole germinant.
TABLE 3.
Synergistic effects of germinant combinations on germination of PV361 complemented with the gerU operon (GC421)a
| Germinant | Rate of spore germinationb |
|---|---|
| GPLK | 69 |
| Glu and Glu | 38 |
| Glu and Pro | 39 |
| Glu and Leu | 53 |
| Glu and KBr | 57 |
| Leu and KBr | 61 |
Spores were germinated in Tris-HCl (5 mM, pH 7.8) supplemented with a 10 mM concentration of the respective germinant (KBr, 50 mM). Spore germination was measured, and germination were rates calculated as described in Materials and Methods. Values are the means of at least duplicate experiments; standard deviation was <5% of the mean.
Rates of spore germination are given relative to the value for QM B1551 spores in 10 mM glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation.
B-receptor proteins are interchangeable and confer germinant specificity.
To investigate the hypothesis that GerVB interacts with GerU proteins to create a proline receptor, we created a chimeric operon, gerU*, in which the B cistron of the gerU operon is dispensed with and replaced by gerVB. Spores of strain GC428, which carries a plasmid-borne copy of the gerU* operon, exhibit a strong germination response to proline, KBr, and glucose, suggesting that the GerVB protein is interacting with GerUA and GerUC to form a functional receptor complex (Table 4). The observation that both GerUB and GerVB can interact with GerUA and GerUC to form receptors with different specificities—GerUB confers the stronger leucine response while being nonresponsive to proline, where as GerVB confers strong responses to proline and KBr, with only a weak response to leucine—is of particular significance as this provides evidence for largely hitherto unrecognized receptor interchangeability, while suggesting that, at least for this system, it is the B protein of the receptor that determines germinant specificity.
TABLE 4.
Rates of germination of PV361 strains complemented with gerU, gerU* and gerUA-UCa
| Strain (receptor[s] present) | Rate of spore germination with the indicated germinantb
|
|||||
|---|---|---|---|---|---|---|
| Buffer | GPLK | Glucose | l-proline | l-leucine | KBr | |
| GC421 (GerU) | 2 | 71 | 41 | 1 | 34 | 8 |
| GC428 (GerU*) | 2 | 75 | 60 | 59 | 17 | 60 |
| GC424 (GerUA-UC) | 1 | 26 | 2 | <1 | 1 | 7 |
Spores were germinated in Tris-HCl (5 mM, pH 7.8) for 40 min with 10-mM germinant (50 mM KBr). Spore germination was measured, and germination rates were calculated as described in Materials and Methods. Values are the means of at least duplicate experiments; standard deviation was <5% of the mean.
Rates of spore germination are given relative to the value for QM B1551 spores in glucose, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation.
Interestingly, strain GC424, which carries only the gerUA and gerUC components of the gerU operon, exhibits a weak germination response to a combination of germinants, suggesting that the gerUA and gerUC proteins can physically interact with a B component from another receptor to confer a degree of functionality. Evidence for interchangeability between receptor proteins from different loci, while being suggested as a means of extending the range of germinant recognition in some species (8), has previously been presented for a strain of B. subtilis overexpressing an individual receptor component (13), although whether such interchangeability occurs under physiological conditions is not clear.
Evidence for other germinant receptors.
Data presented in this communication suggest that GerU and GerVB function cooperatively to trigger the spore germination response to glucose, proline, leucine, or KBr. These data do not preclude the possibility that other spore germinant receptors are required in addition to GerU/GerVB to facilitate this response. We have identified a novel GerA-type receptor operon using a degenerate PCR approach (unpublished data), while a region of DNA containing a receptor operon, GerA, was deposited in the GenBank database (accession number U61380) as far back as 1996. As far as we can establish, details regarding the functionality of GerA, which is chromosomally encoded (data not shown), have not been presented in the scientific literature. We decided to investigate the role of this receptor in spore germination by employing a temperature-dependent plasmid insertion mutagenesis approach to disrupt the gerAA gene, as described in Materials and Methods. A similar approach for the disruption of germination loci has been reported previously (8).
Strain GC413 (gerAA::pUCTV2) was observed to grow and sporulate with similar efficiency to the QM B1551 parent strain. Germination assays revealed, however, that disruption of this operon has little effect on the germination response to any of the single-trigger compounds (Fig. 3). In addition, strain GC417, which lacks both gerU/gerVB and has a disrupted gerA gene, germinates reasonably efficiently on complex rich medium (SNB) (Fig. 3). Considered together, these data suggest that GerA is functionally redundant, while at least one functional, chromosomally encoded receptor remains to be identified.
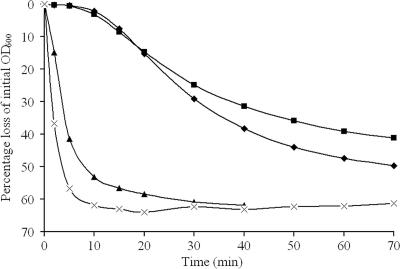
FIG. 3.
Germination of QM B1551 gerA (▴) in 5 mM Tris-HCl (pH 7.8) plus 10 mM glucose. Germination responses to other germinants were similar. Germination of PV361 (gerU/gerVB) (⧫) and PV361 (gerA gerU/gerVB) (▪) spores in supplemented nutrient broth plus 100 μg/ml chloramphenicol is shown. Germination of QM B1551 spores (×) in SNB is shown for comparison. Results are averages of two independent spore preparations (standard deviation is <7% of the mean).
GerU and GerVB confer recognition of novel germinants in B. megaterium KM.
B. megaterium KM, like B. megaterium QM B1551, has been employed extensively in biochemically based approaches to the study of spore germination (9, 17, 34). Unlike strain QM B1551, B. megaterium KM responds to a very narrow range of germinants, with l-alanine eliciting the strongest germination response. Similar amino acids, including l-leucine, can induce weak germination responses. PCR analyses conducted as part of this study revealed that while B. megaterium KM carries a chromosomal copy of the gerA operon, it appears to lack both the gerU-gerVB gene cluster and the grmA plasmid-borne antiporter gene. A mutant strain (GC425) carrying an insertion-deletion in the gerAA locus germinated normally in response to l-alanine (and leucine), indicating that gerA is not the l-alanine receptor in B. megaterium. However, when we transformed wild-type KM with plasmid pUCTV-(gerU-gerVB) to make strain GC426, we found that the spores of this transformant germinated in response to glucose, proline, and KBr in addition to alanine and leucine (Table 5). The strongest germination response was observed in a mixture of all four germinants, although as with PV361 transformants, the proportion of spores to germinate in response to novel germinants did not attain the level of the mutant nor wild-type response to l-alanine. These data also suggest that as yet unidentified proteins involved in signal transduction of the GPLK system are present and functional in B. megaterium KM.
TABLE 5.
Rates of germination of B. megaterium KM strainsa
| Strain (receptor[s] present) | Rate of spore germination with indicated germinantb
|
|||||
|---|---|---|---|---|---|---|
| l-alanine | l-leucine | GPLK | Glucose | l-proline | KBr | |
| KM (wild-type) | 100 | 71 | 64 | <1 | <1 | 12 |
| GC426 (GerU, GerVB) | 100 | 68 | 82 | 70 | 59 | 47 |
| GC425 (ΔgerAA::kan) | 96 | 69 | ND | ND | ND | ND |
Spores were germinated in Tris-HCl (5 mM, pH 7.8) supplemented with KCl (10 mM) and 10 mM germinant (50 mM KBr) for 40 min at 37°C. Spore germination was measured, and germination rates were calculated as described in Materials and Methods. Values are the means of at least duplicate experiments; standard deviation was <8% of the mean.
Rates of spore germination are given relative to the value for B. megaterium KM spores in l-alanine, which was set at 100. This value is equivalent to 100% germination after a 40-min incubation. ND, not determined.
DISCUSSION
Data presented in this communication comprise the first genetic analyses of receptor genes involved in spore germinant recognition in B. megaterium QM B1551. A previous study reported the observation that excision of the 165-kb plasmid leads to loss of the ability to germinate in response to any of the single-trigger germinants, suggesting that essential receptor genes reside on this plasmid (33). The tricistronic operon gerU and a monocistronic gene, gerVB, were identified as 165-kb plasmid-borne receptor homologues by sequence analysis, and while neither could be disrupted successfully by insertional mutagenesis or allelic exchange, complementation analyses conducted in this study provide strong evidence that these are the key plasmid-borne loci required for germination.
The monocistronic gerVB gene is of particular interest, since data presented in this study demonstrate for the first time that a monocistronically encoded receptor component can contribute to germinant specificity. Examination of the genomes of other species of Bacillus suggests that monocistronic germinant receptor genes are rare; virtually all are arranged as tricistronic operons, although Bacillus clausii KSM-K16 has a putative monocistronic gerAA component (GenBank accession number BAD62874). Whether this gene and other monocistronic genes that have yet to be identified contribute to germinant receptor function remains to be determined.
Complementation experiments involving fragments of the gerU/gerVB gene cluster and germination analyses conducted on the chimera receptor operon gerU* permit the delineation of receptor specificity with reasonable confidence. GerU proteins appear to form a receptor for glucose and leucine, while evidence is presented that GerVB interacts with GerUA and GerUC to form a functional receptor for glucose, proline, and KBr. The fact that both GerU and GerVB confer glucose recognition is perhaps not surprising, since both (the B cistrons in particular) appear to be orthologues of the B. subtilis GerK receptor, which has been proposed to be a glucose receptor (1, 14).
The functionality of the GerU* chimera also demonstrates an interchangeability between receptor components that, until recently (13), was thought unlikely to occur. It seems likely that the high degree of shared amino acid sequence identity between GerUB and GerVB (80%) permits this interchangeability, whereas homologous receptor components in other species are often highly divergent. Presumably, in wild-type spores the receptor complex comprises both GerUB and GerVB components, conferring germinability to both leucine and proline, although as yet we have no information on the stoichiometry or topology of the proteins that form the receptor.
In addition, the observation that GerU triggers germination in response to leucine but not proline while GerU* triggers germination in response to proline but exhibits only a weak response to leucine suggests that the B component of the receptor confers specificity, since only the B component differs in these systems. Indeed, the B proteins of the receptor are the only components of the GerA-type receptor family that show any similarity to other bacterial proteins, showing limited homology to single-component amino acid transporters, leading to their classification in evolutionary terms as a subgroup of the APC superfamily of transporters (16). Previous studies employing B. subtilis strains carrying point mutations in the gerAB gene of the alanine receptor, which lead to a requirement for higher concentrations of germinant, have also provided evidence that the B component of the receptor comprises the amino acid binding site (22, 31).
Spore germination in response to inorganic salts, often referred to as ionic germination, has previously been suspected to be mediated by a germinant receptor (5), and this study provides the first genetic evidence for this hypothesis. Potassium and other metal ions have long since been recognized as cogerminants (in the sense that while being essential cofactors, they cannot independently trigger germination) in a number of spore germination systems, including the asparagine, glucose, fructose, and potassium ion response of B. subtilis. The germination response to inorganic salts in B. megaterium QM B1551 differs however, in that both the cation (preferably an alkali metal) and the anion (either Cl−, Br− I−, or NO3−) have been demonstrated to influence the strength of the germination response (28). Data presented in this communication demonstrate that while GerU can utilize KBr as a cogerminant (evidenced by synergy observed when spores are incubated with both KBr and either glucose or leucine), GerVB interacting with GerUA and GerUC can efficiently utilize KBr as a single-trigger germinant.
It should be possible to further test these receptor descriptions by sequence analysis of QM B1551 mutants that display conditional germination responses (33, 39). Strain JV137, for example, germinates strongly in response to glucose but fails to germinate in response to proline (30), suggesting that the gerVB gene may have been subject to mutation. The GerU receptor also represents an interesting system for further study, since if GerVB is a paralogue of the GerUB protein, then mutations have accumulated following gene duplication that has led to altered germinant recognition capability. The high degree of shared identity between these proteins should therefore make this receptor system amenable to a site-directed mutagenesis approach to provide new insights to the residues involved in germinant recognition.
A second receptor operon, the chromosomally encoded GerA, is shown to be redundant, even when spores are germinated in complex medium in the absence of GerU/GerVB. The apparent nonfunctionality of this receptor, which is a close homologue of GerU, is most likely explained by the fact that the gerAB gene appears to encode a 329-residue protein that is C-terminally truncated. Hydropathy profiling suggests that this protein has only nine membrane-spanning helices as opposed to the 10 TMS normally observed for B-receptor components (16). Nonfunctional germinant receptors have also been identified in B. anthracis, where the structural genes for both GerA and GerY carry frameshift mutations (8). However, at least one functional germinant receptor remains to be identified in B. megaterium QM B1551, since spores lacking GerU/GerVB and GerA germinate reasonably efficiently on complex rich medium.
The observation that gerU and gerVB confer germinability to glucose, proline, and KBr with respect to B. megaterium KM, which germinates in response only to l-alanine and related amino acids, provides further evidence as to the functionality of these gene products. A similar experiment has been reported previously (36) in which a library of cloned QM B1551 genomic inserts was used to transform B. megaterium ATCC 19213, which germinates in response only to l-alanine. A transformant was described that germinates in response to glucose, leucine, and proline but not to K+ salts. Unfortunately, the fragment of DNA responsible for conferring the novel germination phenotype was not characterized at the molecular level although, intriguingly, the cloned insert was approximately only 2-kb in size, considerably smaller than the entire gerU/gerVB region. Thus, it may be that diverse strains of B. megaterium can be complemented by different fragments of the QM B1551 genome to confer recognition to novel germinants.
The observation that strain KM can form a functional GPLK-responsive receptor system when complemented with gerU/gerVB also provides further evidence as to the redundancy of the GrmA antiporter in B. megaterium spore germination, since a close homologue of this gene does not appear to be encoded within the KM genome. It is perhaps this aspect of this study that is the most difficult to explain, since GrmA homologues have previously been implicated in the germination response of both B. megaterium QM B1551 and B. cereus (37, 38). We can offer no good explanation for this observation—perhaps variations in strains or spore preparation procedures are important—and further studies focusing on grmA expression and protein localization may provide new insights to the potential role of this protein in spore germination.
Acknowledgments
We thank P. S. Vary (Northern Illinois University, De Kalb, IL) for the generous provision of strains and unpublished sequence information. We also acknowledge the gift of plasmid pUCTV2 from F. Meinhardt (Münster, Germany), and we thank K. Johnstone (Plant Sciences, University of Cambridge) for Bacillus megaterium KM.
Work in our laboratory is funded by grants awarded to C.R.L. by the BBSRC and the Home Office (CBRN).
Footnotes
Published ahead of print on 13 April 2007.
REFERENCES
- 1.Atluri, S., K. Ragkousi, D. E. Cortezzo, and P. Setlow. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J. Bacteriol. 188:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlass, P. J., C. W. Houston, M. O. Clements, and A. Moir. 2002. Germination of Bacillus cereus spores in response to l-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 148:2089-2095. [DOI] [PubMed] [Google Scholar]
- 3.Cabrera-Martinez, R.-M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corfe, B. M., R. L. Sammons, D. A. Smith, and A. Moir. 1994. The gerB region of the Bacillus subtilis 168 chromosome encodes a homologue of the gerA spore germination operon. Microbiology 140:471-478. [DOI] [PubMed] [Google Scholar]
- 5.Cortezzo, D. E., B. P. Setlow, and P. Setlow. 2004. Analysis of the action of compounds that inhibit the germination of spores of Bacillus species. J. Appl. Microbiol. 96:725-741. [DOI] [PubMed] [Google Scholar]
- 6.English, J. D., and P. S. Vary. 1986. Isolation of recombination defective and UV sensitive mutants of Bacillus megaterium. J. Bacteriol. 163:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feavers, I. M., J. Foulkes, B. Setlow, D. Sun, W. L. Nicholson, P. Setlow, and A. Moir. 1990. The regulation of transcription of the gerA spore germination operon of Bacillus subtilis. Mol. Microbiol. 4:275-282. [DOI] [PubMed] [Google Scholar]
- 8.Fisher, N. A., and P. C. Hanna. 2005. Characterization of Bacillus anthracis germinant receptors in vitro. J. Bacteriol. 187:8055-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, S. J., and K. Johnstone. 1986. The use of inhibitors to identify early events during Bacillus megaterium KM spore germination. Biochem. J. 237:865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornstra, L. M., Y. P. de Vries, W. M. de Vos, and T. Abee. 2006. Influence of sporulation medium composition on transcription of ger operons and the germination response of spores of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 72:3746-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 183:4317-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyatt, M. T., and H. S. Levinson. 1964. Effect of sugars and other carbon compounds on germination and postgerminative development of Bacillus megaterium spores. J. Bacteriol. 88:1403-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Igarashi, T., and P. Setlow. 2005. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J. Bacteriol. 187:2513-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irie, R., T. Okamoto, and Y. Fujita. 1982. A germination mutant of Bacillus subtilis deficient in response to glucose. J. Gen. Appl. Microbiol. 28:345-354. [Google Scholar]
- 15.Irie, R., Y. Fujita, and M. Kobayashi. 1996. Nucleotide sequence and gene organisation of the gerK spore germination locus of Bacillus subtilis 168. J. Gen. Appl. Microbiol. 42:141-153. [Google Scholar]
- 16.Jack, D. L., I. T. Paulsen, and M. H. Saier. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797-1814. [DOI] [PubMed] [Google Scholar]
- 17.Johnstone, K., G. S. Stewart, I. R. Scott, and D. J. Ellar. 1982. Zinc release and the sequence of biochemical events during triggering of Bacillus megaterium KM spore germination. Biochem. J. 208:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieselburg, M. K., M. Weickert, and P. S. Vary. 1984. Analysis of resident and transformant plasmids in Bacillus megaterium. Bio/Technology 2:254-259. [Google Scholar]
- 19.Levinson, H. S., and M. T. Hyatt. 1962. Nitrogenous compounds in germination and postgerminative development of Bacillus megaterium spores. J. Bacteriol. 83:1224-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCool, G. J., and M. C. Cannon. 2001. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol. 183:4235-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 22.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moir, A., E. H. Kemp, C. Robinson, and B. M. Corfe. The genetic analysis of bacterial spore germination. J. Appl. Bacteriol. 76:9S-16S. [PubMed]
- 24.Paidhungat, M., and P. Setlow. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J. Bacteriol. 181:3341-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paidhungat, M., and P. Setlow. 2000. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racine, F. M., S. S. Dills, and J. C. Vary. 1979. Glucose-triggered germination of Bacillus megaterium spores. J. Bacteriol. 138:442-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rode, L. J., and J. W. Foster. 1962. Ionic germination of spores of Bacillus megaterium QM B1551. Arch. Mikrobiol. 43:183-200. [DOI] [PubMed] [Google Scholar]
- 29.Rossignol, D. P., and J. C. Vary. 1979. Biochemistry of l-proline-triggered germination of Bacillus megaterium spores. J. Bacteriol. 138:431-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossignol, D. P., and J. C. Vary. 1979. l-Proline site for triggering Bacillus megaterium spore germination. Biochem. Biophys. Res. Commun. 89:547-551. [DOI] [PubMed] [Google Scholar]
- 31.Sammons, R. L., A. Moir, and D. A. Smith. 1981. Isolation and properties of spore germination mutants of Bacillus subtilis-168 deficient in the initiation of germination. J. Gen. Microbiol. 124:229-241. [Google Scholar]
- 32.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 33.Stevenson, D. M., D. Lach, and P. S. Vary. 1993. A gene required for germination in Bacillus megaterium is plasmid-borne, p. 197-207. In E. Balla and G. Berencsie (ed.), DNA transfer and gene expression in microorganisms. Intercept, Budapest, Hungary.
- 34.Stewart, G. S., K. Johnstone, F. Hagelberg, and D. J. Ellar. 1981. Commitment of bacterial spores to germinate. A measure of the trigger reaction. Biochem. J. 198:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swerdlow, B. M., B. Setlow, and P. Setlow. 1981. Levels of H+ and other monovalent cations in dormant and germinating spores of Bacillus megaterium. J. Bacteriol. 148:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tani, K., M. Kawanishi, J. Nishikawa, M. Sasaki, Y. Takubo, T. Nishihara, and M. Kondo. 1990. Identification of germination gene of Bacillus megaterium. Biochem. Biophys. Res. Commun. 167:402-406. [DOI] [PubMed] [Google Scholar]
- 37.Tani, K., T. Watanabe, H. Matsuda, M. Nasu, and M. Kondo. 1996. Cloning and sequencing of the spore germination gene of Bacillus megaterium ATCC 12872: similarities to the NaH-antiporter gene of Enterococcus hirae. Microbiol. Immunol. 40:99-105. [DOI] [PubMed] [Google Scholar]
- 38.Thackray, P. D., J. Behravan, T. W. Southworth, and A. Moir. 2001. GerN, an antiporter homologue important in germination of Bacillus cereus endospores. J. Bacteriol. 183:476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vary, J. C., and A. Kornberg. 1970. Biochemical studies of bacterial sporulation and germination XXI. Temperature-sensitive mutants for initiation of germination. J. Bacteriol. 101:327-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vary, P. S., and Y.-P. Tao. 1988. Development of genetic methods in Bacillus megaterium, p. 403-407. In A. T. Ganesan and J. A. Hoch (ed.), Genetics and biotechnology of bacilli, vol. 2. Academic Press, New York, NY. [Google Scholar]
- 41.Weisblum, B., M. Y. Graham, T. Gryczan, and D. Dubnau. 1979. Plasmid copy number control: isolation and characterization of high copy number mutants of plasmid pE194. J. Bacteriol. 137:635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittchen, K. D., and F. Meinhardt. 1995. Inactivation of the major extracellular protease from Bacillus megaterium DSM319 by gene replacement. Appl. Microbiol. Biotechnol. 42:871-877. [DOI] [PubMed] [Google Scholar]