Abstract
We describe mycobacterial phospholipase A activity (MPLA) and, using reverse genetics, have associated this activity with putative mycobacterial cutinase. PLAs, which hydrolyze fatty acids on phospholipids, play a significant role in human inflammatory states and disease pathogenesis. In prokaryotes, the recognition of their role in virulence is more recent. Cutinases are serine esterases whose primary substrate is cutin, the waxy exterior layer of plants. Mycobacterium tuberculosis has maintained seven putative cutinases, though it should not encounter cutin; we demonstrate that known cutinases and MPLA cleave phospholipids in a PLA-type manner and also hydrolyze Tween. We analyzed cutinase motifs in mycobacteria and found the motif very prevalent. All mycobacteria tested had MPLA activity. These studies suggest an alternative use for putative cutinases by the M. tuberculosis group that is likely related to MPLA activity and lipid metabolism.
Mycobacterium tuberculosis infects one-third of the world, makes over 8 million ill each year, and kills 1.8 million, 450,000 of whom are children (8). Though M. tuberculosis is one of the oldest known human pathogens, our ability to combat the spread of the disease remains insufficient, and the global health burden of tuberculosis is increasing (42). Keys to the success of the tubercle bacillus include its ability to persist within its host for decades, evading the immune system, and its uniquely complex lipid-rich cell wall; both are felt to be target areas for drug development (1, 3, 19, 25, 26). Though scientific advances have increased our understanding of the cell wall and persistence, much more remains to be learned. Lipid metabolism and cell wall remodeling are important components of this understanding. Our investigations describe a novel enzymatic activity in mycobacteria, mycobacterial phospholipase A (MPLA), which likely contributes to these processes and is associated with putative mycobacterial cutinases.
PLAs cleave phospholipids into lysophosphatidylcholine (LPC) and fatty acid (FA) (13). They are classified as PLA1 or PLA2 based on whether they cleave the sn-1 or sn-2 FA, respectively, at the ester linkage. They are further divided into groups based on cellular location, calcium dependence, and active site residues, among other attributes (11, 34). Some PLAs are part of the alpha-beta hydrolase fold superfamily (32). PLAs are essential components of bee and snake venom. They are well described for humans, in whom their dysregulation contributes to inflammation and disease (12, 44). In bacteria, PLAs as virulence factors are described for only a few pathogens (17). For mycobacteria, MPLA activity is alluded to in a few articles, though the protein imparting the activity has not been purified nor the gene encoding the active protein elucidated (2, 6, 24, 35, 38-41). The annotated M. tuberculosis amino acid sequence identifies no putative MPLAs, and similarity and motif searches are also unrevealing.
Cutinases, also members of the alpha-beta hydrolase fold superfamily, are serine esterases whose primary substrate is cutin, a component of the waxy, exterior layer of plants (5, 16, 20). Cutinases cleave the cutin polymer into monomers, facilitating plant invasion by cutinase-producing fungi. In addition to cutin, cutinase substrates include biodegradable plastics, tri- and diacylglycerols, and certain nitrophenyl esters. Phospholipase activity has not been previously associated with cutinases. Cutinase research primarily focuses on phytopathogenic fungi, on the use of cutinase in laundry agents to remove plant stains, and on the use of cutinase to degrade biodegradable plastics (4, 5, 10). Fusarium solani cutinase is perhaps the most well studied cutinase (14). The cutinase catalytic triad is a serine, an asparagine, and a histidine in an oxyanion hole. Unlike phospholipases, they do not exhibit interfacial activation, though they are stabilized by detergents (28). Though mycobacteria have open reading frames annotated as putative cutinases based on motif searches, there is no published research on them. M. tuberculosis has seven such annotations in its proteome, even though it does not encounter plants (and thus cutin) in its environment. This suggests an alternative use for this family of enzymes by M. tuberculosis.
In this study, we describe secreted MPLA activity in mycobacteria. Purification of this activity demonstrates that it is associated with mycobacterial cutinase. As exogenous active protein expression has not yet been possible, we present the data supporting the idea that mycobacterial cutinase is indeed a PLA. We analyze the putative cutinases in mycobacteria and explore the prevalence and cellular location of PLA and cutinase activity. We report phospholipids and polysorbates (Tween) as novel substrates for cutinases. We theorize that this novel PLA/cutinase activity has been conserved in mycobacteria because it plays an important role in mycobacterial lipid manipulation (cell wall or nutritional) and thus pathogenesis.
Throughout this paper we will use the term MPLA to designate the enzyme that we purified for its PLA activity from Mycobacterium smegmatis, which was subsequently found by mass spectrometry to be annotated as a cutinase (based on motif, not activity) but which has PLA, general lipase, and esterase activities.
MATERIALS AND METHODS
Materials.
The synthetic substrates p-nitrophenylbutyrate (NPB) and 2-nitrophenylpalmitate (NPP) were purchased from Sigma (N9876 and N2627, respectively). Polysorbates (Tween) 20, 40, 60, and 80 were purchased from Sigma (P7949, P1504, P1629, and P8074, respectively). Radiolabel phospholipids purchased from NEN Life Science Products included phosphatidylcholine (PC) l-a-dipalmitoyl (dipalmitoyl-1-14C) (NC682, 110 mCi/mM), bovine sphingomyelin (choline-methyl-14C) (NEC663, 52 mCi/mM), and phosphatidylethanolamine l-a-palmitoyl-2-arachidonyl (arachidonyl-1-14C) (NEC783, 48 mCi/mM). 3-Phosphatidyl-[3-14C]serine 1,2-dioleoyl (CFA757, 54 mCi/mM) was purchased from Amersham Biosciences. Unlabeled phospholipids were purchased from Sigma, and stocks were kept in chloroform at −20°C. Snake venom PLA2 from Naja mossambica mossambica (P-7778) and Thermomyces lanuginosus (L-0777) lipase were purchased from Sigma. Cutinase from F. solani was a kind gift from Unilever Research and Development, Vlaardingen, The Netherlands. All chemicals unless otherwise noted were purchased from Sigma.
Strains and growth conditions.
Gamma-irradiated whole cells of M. tuberculosis H37Rv, CSU 93, and Erdman as well as Mycobacterium bovis BCG were obtained from Colorado State University. M. smegmatis ATCC 14468, M. smegmatis mc2155 (ATCC 700084), and Mycobacterium marinum H1726 (ATCC 25039) were purchased from the ATCC. M. smegmatis was grown at 37°C in Luria-Bertani broth with 0.05% Tween 80, and M. marinum was grown at 32°C in Middlebrook 7H9 agar with oleic acid-albumin-dextrose-catalase enrichment and 0.05% Tween 80. Mycobacterial fractions obtained from Colorado State University included crude culture filtrate, crude culture filtrate proteins, cell wall, cell membrane, and cytosol from M. tuberculosis H37Rv.
Purification of M. smegmatis MPLA activity.
A 7.185-liter amount of M. smegmatis 14468 culture supernatant, grown for 24 h at 37°C, was ammonium sulfate precipitated in batches at 60%. After dialysis, the preparation was filtered with a 0.45-μm filter to remove particulate and cation exchange was performed on SP Sepharose columns (Amersham) with 50 mM Tris, pH 8.1. Activity remained in the flowthrough. All anion-exchange column assays were done with a NaCl gradient. Batches were applied to a UNO Q6 anion-exchange column (Bio-Rad) with 50 mM Tris-HCl at pH 8.1. Active fractions were applied to a UNO Q1 anion-exchange column (Bio-Rad) with 50 mM Tris-HCl at pH 8.6. Active fractions from all batches were then pooled and applied to the UNO Q1 column again at pH 9.2. Fraction activity was assessed with PLA activity on [14C]PC, and the efficiency of the purification step was assessed with run-to-run comparative quantitative FA release from PLA activity in trace units (measured with Quantity One software) per milligram of protein and later repeated with all steps simultaneously. The protein concentration was followed with the bicinchoninic acid (BCA) assay and was later repeated with a Nanodrop spectrophotometer. The final active fractions were spin concentrated 25-fold (Vivaspin 15, VS1512), loaded on a nondenaturing gel, and stained with colloidal blue. Protein bands were eluted, and the PLA activity of each band was assessed with a 14C radiolabel thin-layer chromatography (TLC) assay.
Gel electrophoresis conditions.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis was carried out according to the method of Laemmli (19a). Nondenaturing gel conditions for resolution of the liquid chromatography-purified M. smegmatis MPLA activity included upper chamber running buffer (40 mM Tris-HCl, pH 8.89, 40 mM glycine), lower chamber running buffer (60 mM Tris-HCl, pH 7.47), a 7.5% separating gel (237 mM Tris-HCl, pH 8.48), and a stack gel (40 mM Tris-HCl, pH 6.9). After separation proteins were visualized by staining with a colloidal blue staining kit (Invitrogen; LC6025) or a SilverQuest silver staining kit (Invitrogen; LC6070).
Protein concentration determination.
Protein concentrations were determined by either the BCA method (BCA assay kit; Pierce 23225) per protocol or a Nanodrop ND-1000 spectrophotometer per protocol.
Enzyme solutions for activity assays. (i) Control enzymes.
Six units of snake venom PLA2 was used per reaction for a PLA activity control. Lyophilized F. solani cutinase was resuspended at 3 mg/ml in H2O and then further diluted depending on the assay. T. lanuginosus lipase was diluted from stock depending on the assay.
(ii) Mycobacterial supernatants.
To assay M. smegmatis and M. marinum culture supernatants, 50 μl unprocessed supernatant was used. For M. tuberculosis H37Rv, crude filtered culture supernatant (50 μl) was used. In addition, 200 ml of crude filtered culture supernatant was precipitated with 80% ammonium sulfate and resuspended with 5 ml of 25 mM Tris, pH 7.4, with and without 0.5% Triton, and half of this was dialyzed to remove the ammonium sulfate. Fifty microliters of these solutions was then assayed. In addition, 160 μg of lyophilized culture filtrate protein from Colorado State University in 50 μl of 25 mM Tris, pH 7.4, with 0.5% Triton was assayed.
(iii) Mycobacterial gross cell wall/membrane fraction.
Mycobacterial pelleted cells were resuspended in 300 μl per 0.1 g (wet weight) of cells and then disrupted three times, on ice, with a tissue tearer (Biospec Products, model 985-370) at 30,000 rpm for 30 s. The cell debris was pelleted (15 min, 16,000 × g, 4°C), and 50 μl of this supernatant was used for assay.
(iv) M. tuberculosis H37Rv, fractionated.
One hundred sixty micrograms of cell wall, cytosol, and membrane was used per radiolabel assay, resuspended in 50 μl of 25 mM Tris, pH 7.4, with 0.5% Triton X-100. For the NPB assay, the final concentration of each M. tuberculosis H37Rv fraction in the 200-μl assay mixture was as follows: whole cells, 0.5 mg/ml; culture filtrate protein, 0.31 mg/ml; cell wall, 0.1 mg/ml; membrane, 0.25 mg/ml; and cytoplasm, 0.18 mg/ml, as assessed with a Nanodrop spectrophotometer. For substrate specificity and turnover rate assays, partially purified MPLA after the liquid chromatography steps was used. Purified MPLA after the native gel electrophoresis purification step was also checked for MPLA activity on [14C]PC and for cutinase activity on NPB and NPP.
PLA activity assays.
Unlabeled dipalmitoyl PC was resuspended at 23 mM in chloroform. Cold PC (0.25 μl) and 0.1 μl of 14C-radiolabeled phospholipid per reaction were dried down and then resuspended with 50 μl of 50 mM Tris, pH 7.4, buffer with 0.5% Triton per reaction mixture to make mixed micelles. This solution was sonicated three times for 30 s each, with vortexing in between. Fifty microliters of this substrate was then added to 50 μl of enzyme solution and incubated at 37°C for 2 to 3 hours. The reaction was quenched with 100 μl of chloroform-methanol-HCl (50:50:0.3, vol/vol/vol), vortexed, and centrifuged at 13,000 rpm for 2 minutes. The organic phase was then plated on 60-angstrom silica plates (Whatman 4807-425) that were prerun in chloroform-methanol (1:1, vol/vol) and then placed in hexane-diethyl ether-acetic acid (70:30:1, vol/vol/vol) solvent. Lipids were visualized by being charred with 10% cupric sulfate (wt/vol)-8% phosphoric acid (vol/vol).
Polysorbate (Tween) activity assays.
Polysorbate was added to 50 mM Bis-Tris, pH 7.0, at a concentration of 2% (wt/vol). Fifty microliters of this was added to 50 μl of enzyme solution and incubated for 4 hours at 37°C. The reaction mixture was quenched with 100 μl of HCl-methanol-HCl (50:50:0.3, vol/vol/vol), vortexed, and centrifuged at 13,000 rpm for 2 minutes. The organic phase was then plated on 60-angstrom silica plates (Whatman 4861-820) and placed in chloroform-methanol-acetic acid (13:3:1, vol/vol/vol) solvent. Lipids were visualized by being charred with 10% cupric sulfate (wt/vol)-8% phosphoric acid (vol/vol).
Cutinase activity assays.
Cleavage of NPB and that of NPP were used as surrogates for cutinase activity. One hundred fifty microliters of 10 mM NPB resuspended in 50 mM Bis-Tris, pH 7.0, with 5% glycerol was added to 50 μl of enzyme solutions and control buffer in a 96-well plate and incubated at 25°C. For NPP, 20 mg was resuspended in 2.65 ml acetonitrile and isopropanol (1:4, vol/vol); 10 μl of this was added to 140 μl of 50 mM Bis-Tris, pH 7.0, with 5% glycerol; and this was added to 50 μl of enzyme solution. Absorbance was read with a plate spectrophotometer at 415 nm at various time points.
Turnover rate calculations.
For calculation of turnover rate, an extinction coefficient for p-nitrophenyl of 15,250 mol−1 cm−1 and a path length of 0.8 cm for the 96-well plate were used in Beer's law equation (18). Masses used were as follows: M. smegmatis putative cutinase, 29.175 kDa; F. solani cutinase, 22.362 kDa; and T. lanuginosus, 29.923 kDa.
Mass spectrometry of purified MPLA.
Proteins from partially purified samples were resolved by native and SDS-polyacrylamide gel electrophoresis, bands were excised and digested in-gel with trypsin, and mass spectrometry was performed as described previously (36). Data were analyzed using SEQUEST to compare the theoretical tandem mass spectrometry spectra of an annotated M. smegmatis library and compare data against the annotated M. tuberculosis H37Rv genome (7).
RESULTS
Identification and purification of MPLA activity in mycobacteria.
While we were investigating phospholipase C activity in M. tuberculosis, products consistent with MPLA activity (FA and LPC) were seen on TLC, and mass spectrometry confirmed their presence (data not shown). Because a putative gene encoding an MPLA could not be identified by BLAST and motif searching, purification for reverse genetics was undertaken. The purification is summarized in Table 1. MPLA activity was purified from M. smegmatis 14468 instead of M. tuberculosis H37Rv because M. smegmatis, unlike H37Rv, secretes the MPLA activity into the culture supernatant and is not a biosafety level 3 pathogen. Purification was followed by TLC analysis using [14C]PC as a substrate. Purification included a 60% ammonium sulfate precipitation of 7.2 liters of culture supernatant, followed by a cation-exchange step. In cation exchange, MPLA activity was in the flowthrough, and though this step did not add to the purification, it removed particulate not removed by the 0.45-μm filter that would otherwise clog the UNO Q columns. The flowthrough from cation exchange was then applied to anion exchange on an UNO Q6 column, pH 8.1, followed by an UNO Q1 column at pH 8.6. Active fractions were then applied to the UNO Q1 column again at pH 9.2. At this point in purification, specific activity had increased from 0.2 to 38,062 trace units of FA per milligram, a purification of 222,585-fold. This product was designated “partially purified MPLA” and is referred to as such in this paper. These active fractions were then spin concentrated 10-fold, and native gel electrophoresis was performed. Four bands were visible with Coomassie colloidal blue (Fig. 1A); these bands, as well as eight other areas of the gel without visible protein, were eluted, and MPLA activity was assessed by TLC (Fig. 1B). Activity was associated with only one band eluted from the native gel, and this fraction was also active on NPB and NPP (see below). This product was designated “purified MPLA.” This preparation produced only one band on a silver-stained SDS electrophoretic gel, at approximately 30 kDa (Fig. 1C).
TABLE 1.
Purification of MPLA activity from M. smegmatis
| Purification step for MPLA activity from M. smegmatis 14468 | Vol (ml) | Total proteina (mg) | Total activityb (trace units of FA) | Sp act (trace units of FA mg−1) | % Yield | Purification factor (fold) |
|---|---|---|---|---|---|---|
| Culture supernatant | 7,185 | 11,783 | 23,719,984 | 0.171 | 100 | 1 |
| 60% ammonium sulfate | 63.6 | 91.6 | 5,169,812 | 616 | 22 | 3,602 |
| precipitation | ||||||
| Cation-exchange flowthrough | 108.8 | 97.0 | 4,382,557 | 466 | 18.5 | 2,725 |
| UNO Q6, pH 8.1 | 90.0 | 13.5 | 552,123c | 3,029 | 2.3 | 17,713 |
| UNO Q1, pH 8.6 | 50 | 1.5 | 51,327c | 22,812 | 0.22 | 133,403 |
| UNO Q1, pH 9.2 | 16 | 0.88 | 29,476c | 38,062 | 0.12 | 222,585 |
| Native gel electrophoresis | 0.3 | 0.03 | —d |
Total protein was measured using the BCA assay.
Total activity was measured using a [14C]PC assay and Quantity One software to measure trace units of released FA.
Total activity in these specimens may be inaccurate because all of the substrate was consumed.
These calculations were not made because purified preparation was conserved for mass spectrometry and other assays.
FIG. 1.
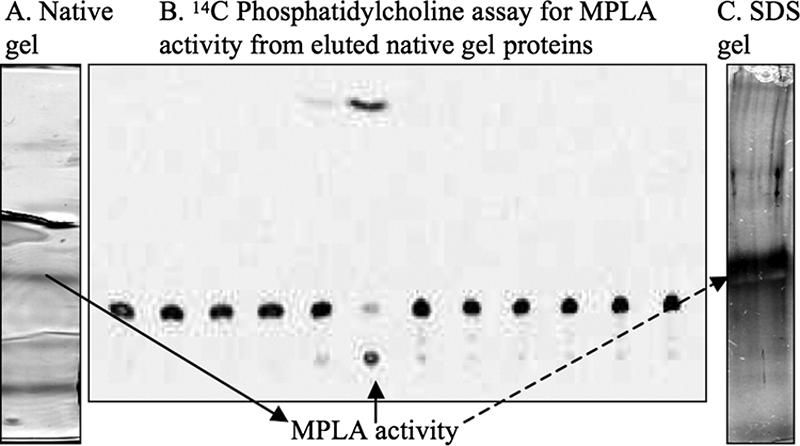
(A) Native gel after column purification of M. smegmatis MPLA activity. (B) The gel was cut into 12 fractions, protein was eluted, and radiolabel 14C TLC was performed for MPLA activity; activity is in the sixth fraction from the indicated band (solid arrow) on the native gel. Mass spectrometry was done on this fraction. (C) An SDS gel of the active fraction showed one band at 30 kDa (dashed arrow).
Identification of the gene encoding MPLA activity in M. smegmatis.
Activity eluted from the native gel during purification was digested with trypsin and subjected to mass spectrometry. Mass densities were then compared to the annotated proteome of M. smegmatis mc2155. Four trypsin fragments were identified, two in the M. smegmatis homologue of M. tuberculosis Rv1926c, annotated as an immunoprotective extracellular protein, and two in Rv3452, annotated as cutinase 4.
Analysis of Rv1926c did not show any lipase motifs, while analysis of Rv3452 revealed it as a serine esterase in the same superfamily (alpha-beta hydrolase fold) as some PLAs, and thus this family of enzymes was pursued.
Analysis of cutinases in mycobacteria.
Searches in NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), Tuberculist (http://genolist.pasteur.fr/TubercuList/), Pfam (http://www.sanger.ac.uk/Software/Pfam/), and TIGR (http://www.tigr.org/) databases found that all mycobacteria in these databases have 1 to 11 open reading frames annotated as cutinases (Table 2). M. tuberculosis H37Rv has eight such annotations (Table 2). Six of these indeed have an identifiable cutinase motif, P-x-[STA]-x-[LIV]-[IVT]-x-[GS]-G-Y-S-[QL]-G and C-x-(3)-D-x-[iv]-c-x-G-[gst]-x(2)-[LIVM]-x(2,3)-H (http://ca.expasy.org/cgi-bin/prosite-search-ac?PDOC00140), with all three catalytic sites (S, D, and H [shown by bold]); one has a partial motif; and one does not contain the motif and appears to be a partial protein adjacent to another probable cutinase. M. tuberculosis CSU 93 and M. bovis each have seven putative cutinases. The M. smegmatis cutinase that we identified by mass spectrometry is most similar to M. tuberculosis H37Rv 3452 (42%). H37Rv 3452 has 37% similarity with F. solani cut12 and 57 to 72% similarity with the other M. tuberculosis putative cutinases (Fig. 2). The mycobacterial cutinases are designated as such based on the presence of the motif, not on activity.
TABLE 2.
Database search of available annotated mycobacterial open reading frames for putative cutinases and MPLA activity determined by [14C]dipalmitoyl PC assaya
| Organism with putative cutinases | No. of cutinases | Gene designation(s) | MPLA activity by TLCb |
|---|---|---|---|
| M. tuberculosis H37Rv | 7 | Rv1758, 1984c, 2301, 3451, 3452, 3724, 3802c | Yes |
| M. tuberculosis CSU 93 | 7 | MT3827, 3909, 3557, 1805, 2037, 3559, 2358 | Yes |
| M. tuberculosis Erdman | ? | Yes | |
| M. bovis subsp. bovis AF2122/97 | 7 | Mb3832c, 2006c, 3751, 1788, 3482, 3481, 2323 | Yes |
| M. bovis BCG strain Pasteur 1173P2 | 6 | BCG 1789, 2317, 3517, 3518, 3784, 3864c | Yes |
| M. avium subsp. paratuberculosis | 11 | MAP 4236c, 1476c, 4237c, 3495c, 0218, 0333, 3428c, 2020, 1662c, 1680c, 2304 | NT |
| M. avium 104 | 10 | MAV 0216, 0369, 1682, 2169, 2741, 2759, 2961, 4283, 4394, 4396 | NT |
| M. smegmatis mc2 155 | 11 | Msmeg 0194, 1184, 1403, 1526, 1528, 1529, 2095, 2474, 4465, 5878, 6354 | Yes |
| M. smegmatis 14468 | ? | Yes | |
| M. leprae TN | 1 | ML 0099 | NT |
| M. marinum 25039 | ? | Yes | |
| M. marinum 11566 | ? | Yes | |
| M. vanbaalenii PYR-1 | 10 | Mvan 0371, 1283, 1439, 1440, 1441, 1924, 3105, 3344, 5793, 5804 | NT |
The data were compiled from Tuberculist, Pfam, TIGR, and NCBI. The search was done using the search term “cutinase” for annotation and with the protein sequence from M. tuberculosis H37Rv 3452 in a Blast search. ?, proteome not available.
See Fig. 3. NT, not tested.
FIG. 2.
Alignment of a known cutinase, F. solani cut12, with M. smegmatis cutinase that we found through reverse genetics of MPLA activity and the eight putative H37Rv cutinases. Active sites (S, D, and H) are in bold; the cutinase motif is underlined. All putative cutinases except 3724a have a cutinase motif, and all except 1758 and 3724b appear to have secretion signals.
Activity of M. smegmatis purified MPLA on the cutinase substrates NPB and NPP.
Described cutinase substrates include cutin, tri- and diacyclglycerols, nitrophenyl esters, and biodegradable plastics. Though techniques for making cutin powder have been described, our attempts to make a workable cutin substrate were unsuccessful. This difficulty is well recognized, and many authors recommend using NPB as a surrogate for cutinase activity (33). Purified MPLA, as well as control F. solani cutinase and T. lanuginosus lipase, was active in this assay (Tables 3 and 4). We present gross activity and a calculated turnover rate (molecules per minute [Table 4]); the experiment was done in triplicate, on partially purified MPLA after all liquid chromatography steps. The turnover rate for our purified protein is very low compared to those of known cutinases under these conditions, though it was steady over time out to 165 min. Snake venom PLA2 was not active on NPB (data not shown). While NPB reflects esterase activity, NPP reflects lipase activity (29). Purified MPLA, as well as control F. solani cutinase and T. lanuginosus lipase, was active in this assay while snake venom PLA2 was not (Table 3).
TABLE 3.
Substrate specificities of known PLA2, known cutinases, and our purified MPLA
| Enzyme | Specificity for substratea:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PC | PE | PS | SM | NPB | NPP | T20 | T40 | T60 | T80 | |
| Snake venom PLA2 | + | + | + | − | − | − | − | − | − | − |
| T. lanuginosus lipase | + | + | + | − | + | + | + | + | + | + |
| F. solani cutinase | + | + | + | − | + | + | + | + | + | + |
| M. smegmatis purified MPLA | + | + | + | + | + | + | + | + | + | + |
PE, phosphatidylethanolamine; PS, phosphatidylserine; SM, sphingomyelin; T20 to T80, Tween 20 to Tween 80, respectively; −, not specific; +, specific.
TABLE 4.
Total activity and turnover ratea
| Enzyme | Total NPB activity (A410) | Turnover rate (min−1) |
|---|---|---|
| F. solani cutinase | 3.1 | 5,024 ± 77 |
| T. lanuginosus lipase | 0.43 | 916 ± 74 |
| M. smegmatis purified MPLA | 0.84 | 0.8 ± 0.1 |
Total absorbance and turnover rate data are averages of three independent determinations on NPB. Data are for the 60-minute time point. Protein concentrations were determined prior to dilution with a Nanodrop spectrophotometer; after dilution concentrations should be 75 ng/ml for F. solani cutinase and T. lanuginosus lipase.
MPLA activity in mycobacteria.
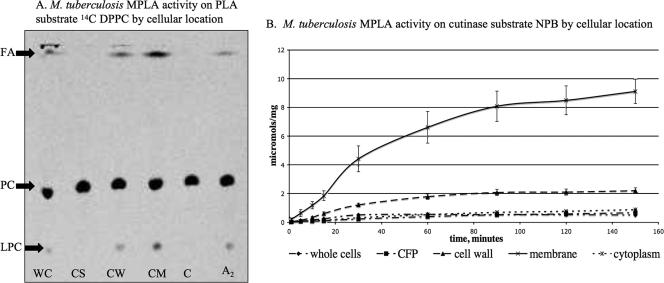
All mycobacteria tested, including M. tuberculosis H37Rv, CSU 93, and Erdman; M. bovis and M. bovis BCG (Pasteur); M. marinum 25039 and 11566; and M. smegmatis 14468 and mc2155, have MPLA activity by [14C]PC TLC assay (Fig. 3). An interesting difference did exist, however. M. marinum 25039 and 11566 and M. smegmatis 14468 and mc2155 all had MPLA activity in their culture supernatant, while activity was not detected in the supernatant of M. tuberculosis H37Rv. Because some of the cutinases are described in culture filtrate protein from H37Rv (Rv1984c, Rv3452, and Rv2301), there was concern that our result was due to specimen processing (37). We thus tested culture filtrate protein, irradiated cell-depleted supernatant, and cell-depleted supernatant with and without ammonium sulfate precipitation and still found no culture supernatant MPLA activity in M. tuberculosis H37Rv. Activity in M. tuberculosis H37Rv was associated with the cell pellet, and upon fractionation the activity was in the cell membrane and cell wall fractions in both a radiolabeled PC assay and an NPB assay (Fig. 4), by both total protein and μmol mg−1. The average activity (μmol mg−1) at 60 minutes was as follows: whole cells, 0.57; culture filtrate protein, 0.40; cell wall, 1.80; cell membrane, 6.62; and cytoplasm, 0.54.
FIG. 3.
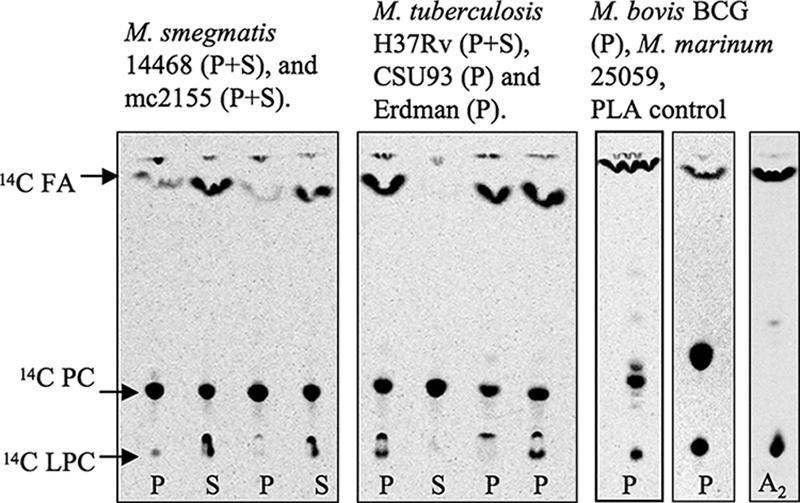
MPLA activity determined by TLC of various mycobacterial strains. Cell pellets (P) and culture supernatants (S) were assayed using [14C]dipalmitoyl PC labeled on both palmitic acids. The presence of [14C]FA and [14C]LPC indicates hydrolysis of [14C]dipalmitoyl PC from MPLA activity. A2, snake venom PLA2. Culture supernatant from H37Rv is lyophilized culture filtrate (shown). Nonlyophilized H37Rv culture supernatant and nonlyophilized ammonium sulfate-precipitated H37Rv culture supernatant were also tested and were inactive; M. marinum 11566 (P+S) and M. marinum 25059 (P) were tested and were active (data not shown).
FIG. 4.
(A) TLC of M. tuberculosis H37Rv MPLA activity by cellular location using [14C]dipalmitoyl PC (DPPC) labeled on both palmitic acids. The presence of [14C]FA and [14C]LPC indicates hydrolysis of [14C]dipalmitoyl PC from PLA activity. WC, whole cells; CS, culture supernatant; CW, cell wall; CM, cell membrane; C, cytosol; A2, snake venom PLA2. Results were consistent with those in panel B. (B) MPLA activity of M. tuberculosis H37Rv by cellular location on cutinase substrate NPB in μmol per mg protein. Activity is predominantly in cell membrane and cell wall in both the PLA and the cutinase assays. CFP, culture filtrate protein.
PLA activity in M. smegmatis partially purified MPLA and known cutinases.
Because it is difficult to see why mammalian pathogens such as those in the M. tuberculosis complex would retain seven cutin-degrading enzymes, we sought to explore possible alternative substrates. Radiolabeled phospholipids in a TLC assay were used to investigate PLA activity of purified MPLA and known cutinases. F. solani cutinase, which contains a full cutinase motif, and T. lanuginosus lipase, which does not contain the full cutinase motif but is known to have cutinase activity as well as extended-spectrum lipase activity, were investigated along with snake venom PLA2 and partially purified MPLA. F. solani, T. lanuginosus, snake venom PLA2, and partially purified MPLA were all active on PC, phosphatidylethanolamine, and phosphatidylserine. Partially purified MPLA alone was active on sphingomyelin (Fig. 5).
FIG. 5.
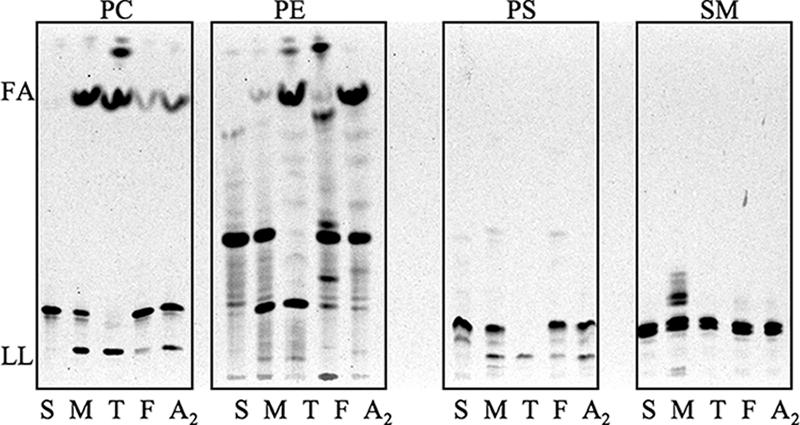
PLA activity by TLC of known cutinases and M. smegmatis partially purified MPLA on various substrates. PC, [14C]PC labeled on both palmitic acids; PE, [14C]phosphatidylethanolamine sn-2 arachidonic acid; PS, [14C]phosphatidylserine labeled on the head group; SM, [14C]sphingomyelin labeled on the head group. The presence of [14C]FA and/or [14C]lysophospholipid (LL) indicates hydrolysis of [14C]phospholipid from PLA activity. Lanes: S, substrate alone; M, M. smegmatis partially purified MPLA; T, T. lanuginosus lipase; F, F. solani cutinase; A2, snake venom PLA2.
Tween-hydrolyzing activity of M. smegmatis partially purified MPLA and known cutinases.
While we were investigating optimal conditions for supernatant MPLA activity in M. smegmatis, it was noted that the addition of Tween 80 to the culture medium enhanced MPLA activity. As Tween compounds are FA esters of sorbitan polyethoxylates, we investigated the ability of partially purified MPLA and known cutinases to release FA from Tween in a TLC assay. All of these enzymes, but not snake venom PLA2, cleave Tween 20, 40, 60, and 80 (Fig. 6), which are predominantly composed of lauric, palmitic, stearic, and oleic acids, respectively.
FIG. 6.
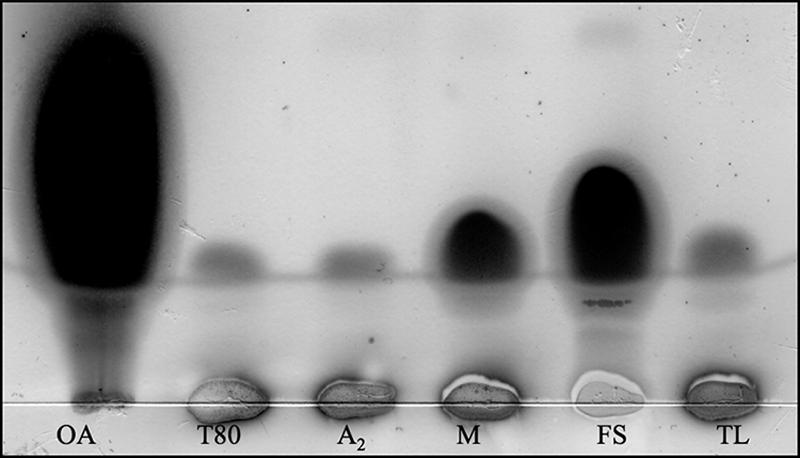
Tween hydrolysis by known cutinases and M. smegmatis partially purified MPLA assessed by TLC. The presence of oleic acid indicates hydrolysis. Lanes: OA, oleic acid; T80, Tween 80 alone; A2, snake venom PLA2; M, M. smegmatis partially purified MPLA; FS, F. solani cutinase (7.5 μg/ml); TL, T. lanuginosus lipase (7.5 μg/ml), all with Tween 80. M. smegmatis partially purified MPLA and F. solani and T. lanuginosis cutinases, but not snake venom PLA2, also hydrolyzed Tween 20, Tween 40, and Tween 60 (data not shown).
DISCUSSION
Mycobacteria have MPLA activity that is associated with putative cutinase. Though MPLA activity has been alluded to in a few publications, this is the first paper to definitively confirm this activity using radiolabeled 14C and the first paper to associate this activity with a specific mycobacterial protein.
We present the evidence that associates mycobacterial MPLA activity with mycobacterial cutinase. First, the presence of a cutinase motif correlates with MPLA activity, as the motif and the activity are found in all mycobacteria tested. Second, the cellular localization of M. tuberculosis MPLA activity correlates with the localization of cutinase activity, as both are found in the cell membrane and cell wall. Third, purification of MPLA activity from M. smegmatis yielded a protein identified as a putative cutinase by mass spectrometry, and this purified MPLA has cutinase activity by NPB and NPP assays, while known snake venom PLA does not. Fourth, we report that known cutinases have PLA activity, and thus it is reasonable that a protein purified for its MPLA activity is a putative cutinase. Fifth, partially purified MPLA and known cutinases, but not snake venom PLA, share another previously undescribed group of substrates, the Tweens.
Furthermore, it is logical that cutinases have PLA activity. The alpha-beta hydrolase fold superfamily encompasses many enzymes. Cutinases are the smallest members of this family, and in our analysis they bear particular resemblance to the group VII PLA2s, and to the other PLA2s using a catalytic serine, though without the C2 domain or a cap over their catalytic domain. However, for cutinases to be established as PLAs, certain criteria put forth in the literature must be fulfilled (34). We will pursue this once we have exogenously expressed protein. Though PLA activity may not be the primary activity of all cutinases, PLA activity is clearly a part of the cutinase repertoire. In pathogens that do not encounter cutin, the presence of these enzymes likely represents an evolutionary divergence, and activity on other substrates is likely to be more important. Our data add a broad range of phospholipids and the Tweens to the list of cutinase substrates. Of particular interest is partially purified MPLA's activity on sphingomyelin, a phospholipid not known to exist in mycobacteria (27).
The activity of these enzymes on Tween is also intriguing, as Tween is widely added to mycobacterial culture medium to prevent cell clumping. A longtime debate has been the concern over Tween as a viable carbon source and the influence of Tween on growth (21, 31), antimicrobial susceptibility (45), and the mycobacterial cell wall (22, 23). The ability of cutinases to cleave these compounds, releasing FAs, provides insight into possible uses of Tween by mycobacteria and the possibility that Tween could alter experimental results, especially in studies of mycobacterial lipids (43). We are pursuing further investigations in this regard.
Though the MPLA activity is not specific to pathogenic mycobacteria, the difference in the locations of the activity—in the cell membrane and wall in M. tuberculosis H37Rv and additionally in the culture supernatant in strains of M. marinum and M. smegmatis—is intriguing and may suggest an environmental adaptation. Extracellular secretion may provide an advantage for an environmental organism that degrades plant matter, while for an intracellular pathogen this may be a disadvantage due to toxicity. We cannot explain why the cutinase proteins published as being in the culture supernatant are not active under our MPLA assay conditions. Either these enzymes were inactivated in processing, are inhibited, or need a certain cofactor, or not all proteins with a cutinase motif have PLA activity. Notably, it has long been known that esterase activity is absent from the culture supernatant of M. tuberculosis, in contrast to other mycobacteria, as this property was used as a tool to differentiate them; our results are consistent with this observation (9).
The low turnover rate for NPB by partially purified MPLA compared to that of known cutinases may reflect damage to MPLA during the purification process, conditions which are not optimized, or a difference in substrate specificity. Formal kinetic studies will be undertaken when purified recombinant protein is available, as well as kinetic studies on other substrates.
While a few other bacteria have cutinases, the majority of organisms found with the motif are fungi. No proteins with a cutinase motif have been identified in humans. Though cutinases are important in phytopathogens, which can occasionally cause opportunistic infections, this is the first publication on cutinases in a strictly mammalian pathogen. The cutinase motif is prevalent in environmental as well as pathogenic strains of mycobacteria. Though M. tuberculosis complex organisms have retained seven open reading frames annotated as cutinases, it is likely that evolutionarily the activities of these enzymes have diverged. Though individually the M. tuberculosis cutinase genes are not essential, according to Himar1-based transposon mutagenesis in H37Rv (15, 30), their redundancy implies an important role. As there is no rationale for cutinases in mammalian hosts, we propose that these enzymes act on other FA-containing substrates.
Conclusion.
Mycobacteria including the M. tuberculosis complex have MPLA activity, and this activity is associated with proteins annotated as mycobacterial cutinases. Known cutinases from F. solani and T. lanuginosus have PLA activity as well, supporting this association. The redundancy of cutinases in the M. tuberculosis genome suggests an important role, and as this pathogen should not encounter cutin, their activity must be directed at alternative substrates, and possibilities include an array of phospholipids and Tweens. The activity of these enzymes may contribute to virulence through their likely modification of mycobacterial and environmental lipids, either for cell wall remodeling or for carbon scavenging.
Acknowledgments
This work was supported by a grant (K08 AI050646) from the National Institute of Allergy and Infectious Diseases to Sarah K. Parker and by a grant (HL62608) from the National Heart, Lung and Blood Institute to Michael L. Vasil.
Cutinase from F. solani was a kind gift from Unilever Research and Development, Vlaardingen, The Netherlands. We thank the Colorado State University of the NIH, NIAID contract NO1 AI75320 entitled “Tuberculosis Research Materials and Vaccine Testing,” for the gamma-irradiated M. tuberculosis H37RV products, Karen Dobos for mass spectrometry work, and Dennis Knudson and Richard Slayden for providing an annotated M. smegmatis genome in FASTA format.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Barry, C. E., III. 2001. Interpreting cell wall ‘virulence factors’ of Mycobacterium tuberculosis. Trends Microbiol. 9:237-241. [DOI] [PubMed] [Google Scholar]
- 2.Bopape, M. C., H. C. Steel, R. Cockeran, N. M. Matlola, P. B. Fourie, and R. Anderson. 2004. Antimicrobial activity of clofazimine is not dependent on mycobacterial C-type phospholipases. J. Antimicrob. Chemother. 53:971-974. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, P. J. 2003. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 83:91-97. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho, C. M., M. R. Aires-Barros, and J. M. Cabral. 1998. Cutinase structure, function and biocatalytic applications. Electron. J. Biotechnol. 1:160-173. [Google Scholar]
- 5.Carvalho, C. M., M. R. Aires-Barros, and J. M. Cabral. 1999. Cutinase: from molecular level to bioprocess development. Biotechnol. Bioeng. 66:17-34. [DOI] [PubMed] [Google Scholar]
- 6.Cholo, M. C., H. I. Boshoff, H. C. Steel, R. Cockeran, N. M. Matlola, K. J. Downing, V. Mizrahi, and R. Anderson. 2006. Effects of clofazimine on potassium uptake by a Trk-deletion mutant of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 57:79-84. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T. 2002. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology 148:2919-2928. [DOI] [PubMed] [Google Scholar]
- 8.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 9.Cox, F. R., C. E. Slack, M. E. Cox, E. L. Pruden, and J. R. Martin. 1978. Rapid Tween 80 hydrolysis test for mycobacteria. J. Clin. Microbiol. 7:104-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degani, O., S. Gepstein, and C. G. Dosoretz. 2002. Potential use of cutinase in enzymatic scouring of cotton fiber cuticle. Appl. Biochem. Biotechnol. 102-103:277-289. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, E. A. 1994. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 269:13057-13060. [PubMed] [Google Scholar]
- 12.Dennis, E. A. 1995. Potential phospholipase A2s involved in inflammatory diseases. Agents Actions Suppl. 46:35-39. [DOI] [PubMed] [Google Scholar]
- 13.Dessen, A. 2000. Structure and mechanism of human cytosolic phospholipase A(2). Biochim. Biophys. Acta 1488:40-47. [DOI] [PubMed] [Google Scholar]
- 14.Egmond, M. R., and J. de Vlieg. 2000. Fusarium solani pisi cutinase. Biochimie 82:1015-1021. [DOI] [PubMed] [Google Scholar]
- 15.Ho, T. B., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotelier, T., L. Renault, X. Cousin, V. Negre, P. Marchot, and A. Chatonnet. 2004. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins. Nucleic Acids Res. 32:D145-D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Istivan, T. S., and P. J. Coloe. 2006. Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology 152:1263-1274. [DOI] [PubMed] [Google Scholar]
- 18.Kurioka, S., and M. Matsuda. 1976. Phospholipase C assay using p-nitrophenylphosphoryl-choline together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal. Biochem. 75:281-289. [DOI] [PubMed] [Google Scholar]
- 19.Kusner, D. J. 2005. Mechanisms of mycobacterial persistence in tuberculosis. Clin. Immunol. 114:239-247. [DOI] [PubMed] [Google Scholar]
- 19a.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Longhi, S., and C. Cambillau. 1999. Structure-activity of cutinase, a small lipolytic enzyme. Biochim. Biophys. Acta 1441:185-196. [DOI] [PubMed] [Google Scholar]
- 21.Lyon, R. H., H. C. Lichstein, and W. H. Hall. 1963. Effect of Tween 80 on the growth of tubercle bacilli in aerated cultures. J. Bacteriol. 86:280-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masaki, S., G. Sugimori, A. Okamoto, J. Imose, and Y. Hayashi. 1991. Effect of Tween 80 on formation of the superficial L1 layer of the Mycobacterium avium-Mycobacterium intracellulare complex. J. Clin. Microbiol. 29:1453-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masaki, S., G. Sugimori, A. Okamoto, J. Imose, and Y. Hayashi. 1990. Effect of Tween 80 on the growth of Mycobacterium avium complex. Microbiol. Immunol. 34:653-663. [DOI] [PubMed] [Google Scholar]
- 24.Matlola, N. M., H. C. Steel, and R. Anderson. 2001. Antimycobacterial action of B4128, a novel tetramethylpiperidyl-substituted phenazine. J. Antimicrob. Chemother. 47:199-202. [DOI] [PubMed] [Google Scholar]
- 25.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 26.Munoz-Elias, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortalo-Magne, A., A. Lemassu, M. A. Laneelle, F. Bardou, G. Silve, P. Gounon, G. Marchal, and M. Daffe. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 178:456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prompers, J. J., A. Groenewegen, C. W. Hilbers, and H. A. Pepermans. 1999. Backbone dynamics of Fusarium solani pisi cutinase probed by nuclear magnetic resonance: the lack of interfacial activation revisited. Biochemistry 38:5315-5327. [DOI] [PubMed] [Google Scholar]
- 29.Rhee, J. K., D. G. Ahn, Y. G. Kim, and J. W. Oh. 2005. New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl. Environ Microbiol. 71:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 31.Sattler, T. H., and G. P. Youmans. 1948. The effect of “Tween 80,” bovine albumin, glycerol, and glucose on the growth of Mycobacterium tuberculosis var. hominis (H37Rv). J. Bacteriol. 56:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaloske, R. H., and E. A. Dennis. 2006. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761:1246-1259. [DOI] [PubMed] [Google Scholar]
- 33.Schomburg, I., A. Chang, and D. Schomburg. 2002. BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 30:47-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Six, D. A., and E. A. Dennis. 2000. The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta 1488:1-19. [DOI] [PubMed] [Google Scholar]
- 35.Steel, H. C., N. M. Matlola, and R. Anderson. 1999. Inhibition of potassium transport and growth of mycobacteria exposed to clofazimine and B669 is associated with a calcium-independent increase in microbial phospholipase A2 activity. J. Antimicrob. Chemother. 44:209-216. [DOI] [PubMed] [Google Scholar]
- 36.Turner, J., K. M. Dobos, M. A. Keen, A. A. Frank, S. Ehlers, I. M. Orme, J. T. Belisle, and A. M. Cooper. 2004. A limited antigen-specific cellular response is sufficient for the early control of Mycobacterium tuberculosis in the lung but is insufficient for long-term survival. Infect. Immun. 72:3759-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weldingh, K., I. Rosenkrands, S. Jacobsen, P. B. Rasmussen, M. J. Elhay, and P. Andersen. 1998. Two-dimensional electrophoresis for analysis of Mycobacterium tuberculosis culture filtrate and purification and characterization of six novel proteins. Infect. Immun. 66:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler, P. R., K. Bulmer, and C. Ratledge. 1990. Enzymes for biosynthesis de novo and elongation of fatty acids in mycobacteria grown in host cells: is Mycobacterium leprae competent in fatty acid biosynthesis? J. Gen. Microbiol. 136:211-217. [DOI] [PubMed] [Google Scholar]
- 39.Wheeler, P. R., K. Bulmer, and C. Ratledge. 1991. Fatty acid oxidation and the beta-oxidation complex in Mycobacterium leprae and two axenically cultivable mycobacteria that are pathogens. J. Gen. Microbiol. 137:885-893. [DOI] [PubMed] [Google Scholar]
- 40.Wheeler, P. R., and C. Ratledge. 1992. Control and location of acyl-hydrolysing phospholipase activity in pathogenic mycobacteria. J. Gen. Microbiol. 138:825-830. [DOI] [PubMed] [Google Scholar]
- 41.Wheeler, P. R., and C. Ratledge. 1988. Use of carbon sources for lipid biosynthesis in Mycobacterium leprae: a comparison with other pathogenic mycobacteria. J. Gen. Microbiol. 134:2111-2121. [DOI] [PubMed] [Google Scholar]
- 42.World Health Organization. March 2006, posting date. Tuberculosis. Fact sheet 104. World Health Organization, Geneva, Switzerland. www.who.int/mediacentre/factsheets/fs104/en/.
- 43.Wright, E. L., M. Pourshafie, and W. W. Barrow. 1992. Mycobacterium avium rough-to-smooth colony conversion resulting from growth in Tween 80 without presence of type-specific glycopeptidolipid antigens. FEMS Microbiol. Lett. 77:209-216. [DOI] [PubMed] [Google Scholar]
- 44.Yedgar, S., D. Lichtenberg, and E. Schnitzer. 2000. Inhibition of phospholipase A(2) as a therapeutic target. Biochim. Biophys. Acta 1488:182-187. [DOI] [PubMed] [Google Scholar]
- 45.Youmans, A. S., and G. P. Youmans. 1948. The effect of “Tween 80” in vitro on the bacteriostatic activity of twenty compounds for Mycobacterium tuberculosis. J. Bacteriol. 56:245-252. [PMC free article] [PubMed] [Google Scholar]