Abstract
In Pseudomonas aeruginosa, the narK1K2GHJI operon encodes two nitrate/nitrite transporters and the dissimilatory nitrate reductase. The narK1 promoter is anaerobically induced in the presence of nitrate by the dual activity of the oxygen regulator Anr and the N-oxide regulator Dnr in cooperation with the nitrate-responsive two-component regulatory system NarXL. The DNA bending protein IHF is essential for this process. Similarly, narXL gene transcription is enhanced under anaerobic conditions by Anr and Dnr. Furthermore, Anr and NarXL induce expression of the N-oxide regulator gene dnr. Finally, NarXL in cooperation with Dnr is required for anaerobic nitrite reductase regulatory gene nirQ transcription. A cascade regulatory model for the fine-tuned genetic response of P. aeruginosa to anaerobic growth conditions in the presence of nitrate was deduced.
The most efficient way for the gram-negative bacterium Pseudomonas aeruginosa to generate energy in the absence of oxygen is through denitrification. During this process, molecular oxygen is replaced by nitrate as the terminal electron acceptor. Nitrate (NO3−) is reduced in four consecutive steps, via nitrite (NO2−), nitric oxide (NO), and nitrous oxide (N2O) to dinitrogen (N2). This process is vital for growth and survival under microaerobic and anaerobic conditions as found in biofilms and microcolonies of infectious P. aeruginosa (1, 25a). The majority of earlier investigations focused on the enzymology and regulation of nitrite (NO2−)-to-dinitrogen (N2) conversion (27). Here, the regulatory network for the onset of nitrate respiration under oxygen-limiting conditions was elucidated using reporter gene fusions, strains carrying mutated regulatory genes, and site-directed mutagenesis of potential regulator binding sites.
Importance of narGHJI, narXL, anr, and dnr for anaerobic growth of P. aeruginosa.
In order to confirm the importance of the nitrate reductase genes narGHJI and the regulatory genes anr, dnr, and narXL for the anaerobic growth of P. aeruginosa, knockout mutants were characterized concerning their growth behavior. P. aeruginosa Anr is the oxygen-sensing regulatory protein homologue to Escherichia coli Fnr (19, 26). Dnr of P. aeruginosa belongs to the Crp-Fnr superfamily of transcriptional regulators and was reported to activate transcription of the genes nir, nor, and nos (6, 9). In Pseudomonas stutzeri, DnrD was shown to detect NO (13, 22). NarXL is a nitrate-responding two-component regulatory system (14). All investigated P. aeruginosa mutant strains failed to grow under anaerobic nitrate respiratory conditions (data not shown). They did not reveal any growth phenotype when tested under aerobic conditions (data not shown). These experiments identify narL, anr, dnr, and narG as key players in the anaerobic growth of P. aeruginosa.
Transcriptional control of the nar locus is mediated by the narXL-narK1 intergenic region.
In E. coli and P. stutzeri, Fnr- and NarXL-dependent transcription of the narGHJI operon is mediated by the narG upstream region (8, 15, 23, 24). In contrast to these observations, inspection of the 200-bp 5′ upstream region of narG in P. aeruginosa revealed no obvious binding motifs for the Fnr homologue Anr or the nitrate response regulator NarL. Using the two PnarG1 and PnarG2 reporter gene fusions, containing 100 bp and 411 bp of the upstream region of the narG gene fused to the reporter gene lacZ in the pQF50 plasmid, respectively, we showed that these DNA fragments did not mediate transcriptional activation of PnarG-lacZ under any of the tested aerobic and anaerobic growth conditions (Table 1). Information regarding primer sequences, construction of all reporter gene fusions, mutated promoter constructs, and various strains can be provided upon request. We failed to detect a transcriptional start site using primer extension experiments upstream of narG (data not shown). Additionally, the intergenic region between narK2 and narG was successfully amplified from cDNA synthesized from mRNA extracted from anaerobically grown P. aeruginosa PAO1 cells, confirming that narG is cotranscribed with the narK1K2 genes located upstream. Consequently, the 173-bp DNA fragment localized between narX and narK1 harbors two divergently oriented promoters (Fig. 1A). This was also recently detected using a promoter predictor program and mutational analysis (19a). The 5′ end of narK1K2GHJI mRNA was localized at 29 bp upstream of the translational start codon of narK1, and the 5′ end of the narX mRNA was localized at 45 bp upstream of the translational start codon of narX, by using primer extension analysis on an ALF model DNA sequencer (Pharmacia) (16).
TABLE 1.
Regulation of the narK1K2GHJI transcription by Anr, Dnr, IHF, and NarL
| P. aeruginosa strain (genotype) | Promoter constructc | Gene in transb | β-Galactosidase activity (Miller units)a
|
|||||
|---|---|---|---|---|---|---|---|---|
| + O2 + nitrate | + O2 + nitrite | + O2 | − O2 + nitrate | − O2 + nitrite | − O2 fermentative | |||
| PAO1 | PnarG1 | — | 3 | 4 | 5 | 11 | 10 | 7 |
| PAO1 | PnarG2 | — | 9 | 8 | 6 | 10 | 10 | 8 |
| PAO1 | PnarK1 | — | 11 | 10 | 12 | 720 | 390 | 159 |
| PAO6261 (Δanr) | PnarK1 | — | 6 | 6 | 5 | 8 | 12 | 7 |
| PAO6261 (Δanr) | PnarK1 | dnr | — | — | — | 134 | 60 | 53 |
| PAO1 | PnarK1 | dnr | — | — | — | 761 | 399 | 329 |
| RM536 (Δdnr) | PnarK1 | — | 11 | 10 | 9 | 256 | 195 | 104 |
| RM536 (Δdnr) | PnarK1 | anr | — | — | — | 678 | 161 | 268 |
| PAO1 | PnarK1 | anr | — | — | — | 846 | 200 | 441 |
| PAO1 | PnarK1ΔAnr | — | 8 | 6 | 5 | 19 | 16 | 21 |
| CHA-A2 (ΔihfA)d | PnarK1 | — | 4 | 5 | 5 | 72 | 53 | 37 |
| PAO1 | PnarK1ΔIHF | — | 8 | 7 | 6 | 299 | 152 | 183 |
| PAO9104 (ΔnarL) | PnarK1 | — | 4 | 4 | 5 | 14 | 10 | 22 |
| PAO9104 (ΔnarL) | PnarK1 | narL | 9 | 8 | 10 | 630 | 378 | 126 |
| PAO1 | PnarK1ΔNarL1 | — | 10 | 9 | 10 | 32 | 33 | 30 |
| PAO1 | PnarK1ΔNarL2 | — | 11 | 8 | 11 | 147 | 107 | 91 |
| PAO1 | PnarK1ΔNarL3 | — | 2 | — | — | 138 | — | — |
All values are results of three independent experiments performed in triplicate. Bacterial strains were grown aerobically and anaerobically in LB medium supplemented with 10 mM sodium nitrate or 1 mM sodium nitrite as described previously (7). Arginine fermentation conditions were achieved as outlined previously (21). The β-galactosidase activities are given in Miller units and are calculated as described previously (18). Standard deviations were between 3 and 13% of given values. —, not measured.
The plasmids pHA411 (anr) and pHA541Ω (dnr) provided the anr and dnr genes in trans as described previously (4, 5). Vector without an insert served as background control. —, no gene in trans.
Details for the construction of tested reporter gene fusions and P. aeruginosa mutants will be provided upon request.
CHA-A2 is not isogenic to PAO1. Due to the clear cut results obtained with this strain, the data obtained are presented.
FIG. 1.
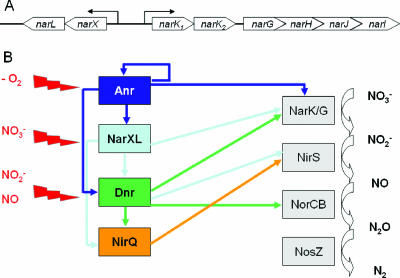
(A) Schematic representation of the nar locus in Pseudomonas aeruginosa. The narXL and narK1K2GHJI operons share a divergently oriented promoter region covering 173 bp. The 5′ end of the narXL mRNA was localized at 45 bp upstream of the translational start of narX. The narK1K2GHJI genes are transcribed as an operon starting at 29 bp upstream of the narK1 translational start. (B) The currently elucidated regulatory network for the onset of denitrification in P. aeruginosa. The major initial signal to turn on the denitrification pathway in P. aeruginosa is low-oxygen tension. This signal is measured by the Fe-S clusters attached to Anr (27). Anr increases the transcription of the narXL operon encoding a two-component regulatory system responding to the presence of nitrate. Anr and NarL cooperatively activate the dnr gene for the third involved regulatory system, Dnr, responding to NO (5). The fourth regulatory system, NirQ, in turn requires Dnr and NarL for its formation (4). Under low-oxygen tension conditions and in the presence of nitrate, Anr and NarL activate in concert with the DNA bending protein IHF, the narK1K2GHJI operon encoding nitrate/nitrite transporters, and the structural genes for the respiratory nitrate reductase. The enzyme converts nitrate into nitrite. Now, both Dnr and NirQ, most likely responding to N-oxides, are essential for the gene regulatory scenario required for the formation of the three enzyme complexes that catalyze the conversion of nitrite into N2.
Regulation of the narK1K2GHJI promoter.
A reporter gene fusion, PnarK1-lacZ containing 435 bp upstream of the translational start codon of narK1 was constructed. The reporter gene activity was found to be induced 65-fold when P. aeruginosa was grown under anaerobic conditions in the presence of nitrate compared to that under identical but aerobic conditions. A 39-fold induction was detected when nitrate was replaced with nitrite, and still a 13.3-fold induction was present during arginine fermentative conditions (Table 1). Subsequently, the PnarK1 construct was introduced into the two P. aeruginosa regulatory mutant strains PAO6261 (Δanr) and RM536 (dnr::Tc). Aerobic narK1 promoter activity in the wild-type and in the two mutant strains was comparably low (Table 1). No obvious narK1 promoter activity was detected in the anr mutant strain under any of the tested anaerobic conditions. Deletion of the dnr gene resulted in 2.8-fold lower promoter activity levels than that in the wild-type strain under anaerobic growth conditions in the presence of nitrate. A 2-fold and 1.5-fold reduction of wild-type reporter gene activity was observed for anaerobic growth in the presence of nitrite and under fermentative growth conditions, respectively. Thus, Anr is essential, and Dnr enhances narK1 activation (Fig. 1B). A regulatory cascade was proposed where Anr activates the expression of dnr, which in turn activates target promoters (5). To determine whether both regulators act in a coordinated manner, gene complementation assays were performed. The anr mutant strain carrying the PnarK1-lacZ promoter reporter gene fusion was transformed with the dnr gene on an expression plasmid (pHA541Ω) in trans. Reporter gene expression increased significantly compared to that of the anr mutant without dnr in trans. However, the values obtained were below those of reporter gene expression in the wild-type strain or the wild-type strain carrying a dnr expression plasmid (Table 1). A minor but significant role for Dnr in the anaerobic activation of the narK1 promoter was indicated, in which Dnr function was independent of Anr. Introducing an anr expression plasmid (pHA411) into the dnr mutant led to narK1 promoter activity levels that were almost the same as those of the wild type (Table 1). Consequently, the narK1 promoter represents another example of the dual actions of Anr and Dnr, besides those of the P. aeruginosa hemF and hemN promoters (Fig. 1B) (18). A highly conserved potential Anr/Dnr binding site (5′-TTGATTCCTATCAA-3′; conserved nucleotides in the Anr binding site are underlined) centered at −40.5 bp upstream of the 5′ end of the narK1 mRNA, was detected using Virtual Footprint of PRODORIC software (www.prodoric.de). Three potential NarL binding sites, NarL1 (5′-TACCTCT-3′) at −108 bp, NarL2 (5′-TACGGCT-3′) at −113 bp, and NarL3 (5′-TACCTCC-3′) at −208 bp with respect to the narK1 mRNA 5′ end, were found. One putative binding site for the DNA bending protein IHF (5′-CAATAATTTCAGCCG-3′) was proposed at −119 bp upstream of the narK1 mRNA 5′ end.
Next, nucleotide exchanges (5′-TTGATTCCTATCAA-3′ to 5′-TCGATTCCTACTTA-3′) were introduced into the Anr box consensus motif (PnarK1ΔAnr). Putative regulator binding sites were mutated using a QuikChange mutagenesis kit (Stratagene, Amsterdam, The Netherlands) or via crossover PCR (12) (details can be provided on request). No obvious promoter activity was detected under any of the tested conditions (Table 1). These results confirm the importance of the putative Anr binding site centered at −40.5 bp upstream of the transcriptional start for anaerobic induction of narK1. The PnarK1-lacZ reporter gene fusion was next introduced into the IHF (CHA-A2) and narL (PAO9104) mutant strains. The β-galactosidase activities were found to be significantly reduced (10-fold and 51-fold, respectively) for both mutants, indicating the involvement of both proteins in narK1 transcription (Table 1 and Fig. 1B). Complementation of the narL mutant with a narL expression plasmid (pRK-LM) nearly restored wild-type-level promoter activity (Table 1). To verify the importance of a predicted IHF binding motif within the narK1 promoter, the motif was mutated from 5′-CAATAATTTCAGCCG-3′ to 5′-GGGGAATTTCAGCCG-3′ (PnarK1ΔIHF). The 2.4-fold decrease observed for the promoter activity of wild-type P. aeruginosa strain PAO1 carrying a PnarK1ΔIHF-lacZ fusion indicated the importance of the predicted IHF binding motif in narK1 promoter activation (Table 1). The failure to completely eliminate narK1 promoter activity with the introduced mutations might be due to residual IHF binding capacity of the mutated site or to an additional as-yet-unknown second IHF binding site in the narK1 promoter. Additionally, the IHF protein can be replaced by less binding sequence-specific HU proteins (11, 17). The potential NarL binding sites NarL1 and NarL3 were correctly oriented for narK1 activation; NarL2 was oriented in the opposite direction. Mutagenesis of the NarL1 binding site (PnarK1ΔNarL1) resulted in a nearly total loss of reporter gene activities under denitrifying conditions (Table 1). Mutagenesis of NarL2 (PnarK1ΔNarL2) decreased narK1 promoter activity down to a level which was in the range of that of the IHF mutant. Since the NarL2 and the IHF binding motifs overlap, secondary effects of the NarL2 mutagenesis on the IHF binding site cannot be excluded. Mutagenesis of the third NarL binding site, NarL3 (PnarK1ΔNarL3), abolished reporter gene activities, as shown in Table 1. Consequently, NarL1 and NarL3 are at least important for the anaerobic induction of the narK1 promoter.
P. aeruginosa possesses a second dissimilatory nitrate-reducing system localized in the periplasm, encoded by the napEFDABC operon. No obvious influence of tested regulators, oxygen tension and nitrate or nitrite availability on napEFDABC promoter activity was detected using a napE promoter reporter gene fusion (Table 1). Clearly, napEFDABC expression is not coregulated with the onset of denitrification.
Regulation of the narXL promoter.
To study the regulation of the narXL promoter, a PnarX-lacZ fusion carrying 206 bp of the narX promoter region (PnarX) was constructed. A moderate 1.7- to 2.0-fold promoter induction was observed during anaerobic nitrate and nitrite respiratory and fermentative conditions (Table 2). The aerobic constitutive narXL transcription was independent of all regulators tested (Table 2). Moreover, P. aeruginosa NarXL did not autoregulate its own gene expression and did not require IHF for expression (8, 20). However, P. aeruginosa narX promoter activity was slightly reduced under any of the tested anaerobic conditions in the case of a missing Anr or Dnr regulator (Table 2 and Fig. 1B). To distinguish between cascade regulation and dual activities by Anr and Dnr, the dnr gene was expressed in trans from a plasmid in the anr mutant PAO6261 carrying the PnarX-lacZ reporter gene fusion. As a control, anr was expressed in the dnr mutant RM536 harboring the same fusion. In both experiments, full anaerobic expression was not restored, indicating a dual function for both regulators at the narXL promoter. For weaker Anr-dependent promoters in P. aeruginosa, Anr half-site reactivity has been reported (14, 25). Therefore, both half sites of the potential Anr binding site centered at −60.5 bp upstream of the narX mRNA 5′ end were mutated independently, from 5′-TTGATTCCTATCAA-3′ to 5′-TGAATTCCTATCAA-3′ in PnarXΔAnr1 and to 5′-TTGATTCCTAAGAA-3′ in PnarXΔAnr2. Decreased reporter gene activities of both mutated narX reporter gene fusions were at the levels observed for the reporter gene fusions with the intact narXL promoter tested with the regulatory mutants PAO6261 (Δanr) and RM536 (dnr::Tc), respectively (Table 2). These results demonstrated that both half sites of the Anr/Dnr binding sequence are involved in anaerobic narX transcriptional activation. The location of the potential Anr/Dnr binding site at −60.5 bp might provide an explanation for the moderate 1.7-fold induction of the narX promoter under anaerobic conditions.
TABLE 2.
Anaerobic induction of the napE and narXL promoters by NarL, IHF, Anr, and Dnr and NarL influence on the expression of Pdnr-lacZ, PnirQ-lacZ, Panr-lacZ, PnirS-lacZ, and PnorC-lacZ
| P. aeruginosa strain | Promoter constructc | Gene in transb | β-Galactosidase activity (Miller units)a
|
|||||
|---|---|---|---|---|---|---|---|---|
| + O2 + nitrate | + O2 + nitrite | + O2 | − O2 + nitrate | − O2 + nitrite | − O2 fermentative | |||
| PAO1 | PnapE | — | 352 | 410 | 361 | 291 | 305 | 310 |
| PAO6261 (Δanr) | PnapE | — | 402 | 576 | 408 | 350 | 361 | 338 |
| RM536 (Δdnr) | PnapE | — | 238 | 253 | 262 | 286 | 284 | 239 |
| PAO9104 (ΔnarL) | PnapE | — | 322 | 313 | 292 | 400 | 396 | 320 |
| PAO1 | PnarX | — | 293 | 269 | 251 | 504 | 539 | 519 |
| PAO9104 (ΔnarL) | PnarX | — | 220 | 225 | 216 | 523 | 426 | 580 |
| CHA-A2 (ΔihfA)d | PnarX | — | 300 | 310 | 285 | 526 | 549 | 530 |
| PAO6261 (Δanr) | PnarX | — | 246 | 259 | 241 | 367 | 239 | 336 |
| PAO6261 (Δanr) | PnarX | anr | 272 | — | — | 622 | — | — |
| PAO6261 (Δanr) | PnarX | dnr | 272 | — | — | 372 | — | — |
| RM536 (Δdnr) | PnarX | — | 259 | 280 | 257 | 259 | 291 | 282 |
| RM536 (Δdnr) | PnarX | dnr | 257 | — | — | 582 | — | — |
| RM536 (Δdnr) | PnarX | anr | 298 | — | — | 398 | — | — |
| PAO1 | PnarXΔAnr1 | — | 194 | 216 | 200 | 280 | 320 | 316 |
| PAO1 | PnarXΔAnr2 | — | 241 | 269 | 228 | 331 | 393 | 365 |
| PAO1 | Pdnr | — | 7 | 8 | 5 | 210 | 132 | 122 |
| PAO9104 (ΔnarL) | Pdnr | — | 8 | 6 | 7 | 12 | 14 | 15 |
| PAO1 | PnirQ | — | 17 | 22 | 12 | 327 | 427 | 60 |
| PAO9104 (ΔnarL) | PnirQ | — | 8 | 6 | 8 | 32 | 53 | 24 |
| PAO9104 (ΔnarL) | PnirQ | dnr | 11 | 12 | 10 | 5 | 12 | 7 |
| PAO1 | Panr | — | 103 | 114 | 105 | 233 | 293 | 322 |
| PAO9104 (ΔnarL) | Panr | — | 81 | 93 | 92 | 191 | 206 | 268 |
| PAO1 | PnirS | — | 12 | 23 | 29 | 470 | 852 | 268 |
| PAO9104 (ΔnarL) | PnirS | — | 15 | 14 | 15 | 321 | 627 | 331 |
| PAO9104 (ΔnarL) | PnirS | dnr | 13 | — | — | 521 | — | — |
| PAO1 | PnorC | — | 18 | 28 | 25 | 1053 | 1642 | 493 |
| PAO9104 (ΔnarL) | PnorC | — | 22 | 17 | 18 | 480 | 1627 | 307 |
| PAO9104 (ΔnarL) | PnorC | dnr | 25 | — | — | 880 | — | — |
All values are results of three independent experiments performed in triplicate. Bacterial strains were grown aerobically and anaerobically in LB medium supplemented with 10 mM sodium nitrate or 1 mM sodium nitrite as described previously (7). Arginine fermentation conditions were achieved as outlined previously (21). The β-galactosidase activities are given in Miller units and are calculated as described previously (18). Standard deviations for the experiment shown were between 3 and 12%. —, no gene in trans.
The anr and dnr genes were cloned in a vector compatible with the lacZ fusion vector pQF50 and cotranscribed where indicated. Vector without an insert served as background control. —, not measured.
Details for the construction of tested reporter gene fusions and P. aeruginosa mutants will be provided upon request.
CHA-A2 is not isogenic to PAO1. Due to the clear cut results obtained with this strain, the data obtained are presented.
The NarL regulator and dnr, nirQ, anr, nirS, and norC transcription.
Transcriptional lacZ fusions of the regulatory genes dnr, nirQ, and anr and the structural genes nirS (encoding nitrite reductase) and norC (encoding nitric-oxide reductase cytochrome c subunit) were introduced into P. aeruginosa PAO1 and the narL mutant strain PAO9104. In agreement with previous findings, the Pdnr-lacZ fusion was found to be induced by Anr under anaerobic conditions (5). Moreover, dnr transcription was highly dependent on NarL (Table 2 and Fig. 1B). In agreement, highly conserved NarL binding sites were localized, centered at 189 bp and 114 bp upstream of the translational start point. The ATP-binding protein NirQ from P. aeruginosa activates respiratory nitrite reductase activity (10). The expression of the PnirQ-lacZ fusion was found to be strongly induced under anaerobic conditions. Like Pdnr-lacZ expression, PnirQ-lacZ transcription was found to be dependent on NarL (Table 2 and Fig. 1B). Arai et al. described Anr- and Dnr-dependent nirQ expression (4). Since our experiments showed that expression of dnr in turn is also dependent on NarL, we investigated whether the observed NarL dependence of the nirQ promoter was caused by an indirect effect via a NarL-dependent decrease of Dnr levels. The nirQ promoter activities were still found to be low even in the presence of a high Dnr concentration, which confirmed a direct effect of NarL on nirQ expression (Fig. 1B). In agreement with these observations, the nirQ promoter harbored potential NarL binding sites centered at positions 97 bp and 4 bp upstream of the translational start site. Clearly, Dnr- and NarL-dependent NirQ formation provides the regulatory link between nitrate and nitrite respiration. We did not observe NarL dependence of anr expression (Table 2). The nitrite reductase nirS gene expression occurred under anaerobic conditions and was induced by nitrate and nitrite (3). Anr mediates this transcriptional activation indirectly via the induction of dnr expression in the regulatory cascade, as outlined previously (2, 4). Anaerobic PnirS-lacZ induction in the narL mutant in the presence of nitrate or nitrite was only about 1.4-fold lower than that in wild-type PAO1 (Table 2). Therefore, the effect of NarL on nirS transcription seemed to be of an indirect nature, most likely via the missing induction of dnr (Fig. 1B). Consequently, dnr expression in trans complemented the observed expression phenotype. Additionally, the narL mutant, expressing the structural genes for the nitrate reductase at a low level, failed to efficiently convert nitrate into nitrite. However, high nitrite levels are important for nirS induction as well. Almost identical results were obtained for the analysis of a PnorC-lacZ fusion in a narL mutant. Transcription of norC is also dependent on Anr and Dnr and the presence of nitrite (2). The observed strong nitrite-dependent induction is not completely understood. Either dnr transcription is activated via formed N-oxides, or an additional as-yet-unknown system is involved. Obtained results clearly showed that norC transcription is not directly regulated by NarL (Table 2 and Fig. 1B). Again, an indirect effect via NarL-dependent dnr expression was concluded.
Acknowledgments
We thank Elisabeth Härtig for critically reading the manuscript. We are indebted to Dieter Haas (Université de Lausanne, Switzerland) for providing the P. aeruginosa anr mutant.
This study was sponsored by grants from the Deutsche Forschungsgemeinschaft (DFG), the Bundesministerium für Bildung und Forschung (BMBF), the DFG-European Graduate College Pseudomonas: Pathogenicity and Biotechnology program 653, and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Arai, H., M. Hayashi, A. Kuroi, M. Ishii, and Y. Igarashi. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 187:3960-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai, H., Y. Igarashi, and T. Kodama. 1995. Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 371:73-76. [DOI] [PubMed] [Google Scholar]
- 3.Arai, H., Y. Igarashi, and T. Kodama. 1991. Nitrite activates the transcription of the Pseudomonas aeruginosa nitrite reductase and cytochrome c-551 operon under anaerobic conditions. FEBS Lett. 288:227-228. [DOI] [PubMed] [Google Scholar]
- 4.Arai, H., Y. Igarashi, and T. Kodama. 1994. Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 58:1286-1291. [DOI] [PubMed] [Google Scholar]
- 5.Arai, H., T. Kodama, and Y. Igarashi. 1997. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol. Microbiol. 25:1141-1148. [DOI] [PubMed] [Google Scholar]
- 6.Arai, H., M. Mizutani, and Y. Igarashi. 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149:29-36. [DOI] [PubMed] [Google Scholar]
- 7.Eschbach, M., K. Schreiber, K. Trunk, J. Buer, D. Jahn, and M. Schobert. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186:4596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Härtig, E., U. Schiek, K. U. Vollack, and W. G. Zumft. 1999. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J. Bacteriol. 181:3658-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa, N., H. Arai, and Y. Igarashi. 1998. Activation of a consensus FNR-dependent promoter by DNR of Pseudomonas aeruginosa in response to nitrite. FEMS Microbiol. Lett. 166:213-217. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi, N. R., H. Arai, T. Kodama, and Y. Igarashi. 1998. The nirQ gene, which is required for denitrification of Pseudomonas aeruginosa, can activate the RubisCO from Pseudomonas hydrogenothermophila. Biochim. Biophys. Acta 1381:347-350. [DOI] [PubMed] [Google Scholar]
- 11.Herrera, M. C., and J. L. Ramos. 2007. Catabolism of phenylalanine by Pseudomonas putida: the NtrC-family PhhR regulator binds to two sites upstream from the phhA gene and stimulates transcription with sigma70. J. Mol. Biol. 366:1374-1386. [DOI] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 14.Krieger, R., A. Rompf, M. Schobert, and D. Jahn. 2002. The Pseudomonas aeruginosa hemA promoter is regulated by Anr, Dnr, NarL and Integration Host Factor. Mol. Genet. Genomics 267:409-417. [DOI] [PubMed] [Google Scholar]
- 15.Li, S. F., and J. A. DeMoss. 1988. Location of sequences in the nar promoter of Escherichia coli required for regulation by Fnr and NarL. J. Biol. Chem. 263:13700-13705. [PubMed] [Google Scholar]
- 16.Marino, M., H. C. Ramos, T. Hoffmann, P. Glaser, and D. Jahn. 2001. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD). J. Bacteriol. 183:6815-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Martín, J., and V. De Lorenzo. 1997. Coactivation in vitro of the σ54-dependent promoter pu of the TOL plasmid of Pseudomonas putida by HU and the mammalian HMG-1 protein. J. Bacteriol. 179:2757-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rompf, A., C. Hungerer, T. Hoffmann, M. Lindenmeyer, U. Romling, U. Gross, M. O. Doss, H. Arai, Y. Igarashi, and D. Jahn. 1998. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 29:985-997. [DOI] [PubMed] [Google Scholar]
- 19.Sawers, R. G. 1991. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol. Microbiol. 5:1469-1481. [DOI] [PubMed] [Google Scholar]
- 19a.Sharma, V., C. E. Noriega, and J. J. Rowe. 2006. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart, V., and J. Parales, Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vander Wauven, C., A. Piérard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vollack, K. U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker, M. S., and J. A. DeMoss. 1994. NarL-phosphate must bind to multiple upstream sites to activate transcription from the narG promoter of Escherichia coli. Mol. Microbiol. 14:633-641. [DOI] [PubMed] [Google Scholar]
- 24.Walker, M. S., and J. A. DeMoss. 1991. Promoter sequence requirements for Fnr-dependent activation of transcription of the narGHJI operon. Mol. Microbiol. 5:353-360. [DOI] [PubMed] [Google Scholar]
- 25.Yamano, Y., T. Nishikawa, and Y. Komatsu. 1993. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1. Mol. Microbiol. 8:993-1004. [DOI] [PubMed] [Google Scholar]
- 25a.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann, A., C. Reimmann, M. Galimand, and D. Haas. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5:1483-1490. [DOI] [PubMed] [Google Scholar]
- 27.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]