Abstract
The chemolithotrophic ammonia-oxidizing bacterium Nitrosomonas europaea is known to be highly resistant to starvation conditions. The transcriptional response of N. europaea to ammonia addition following short- and long-term starvation was examined by primer extension and S1 nuclease protection analyses of genes encoding enzymes for ammonia oxidation (amoCAB operons) and CO2 fixation (cbbLS), a third, lone copy of amoC (amoC3), and two representative housekeeping genes (glyA and rpsJ). Primer extension analysis of RNA isolated from growing, starved, and recovering cells revealed two differentially regulated promoters upstream of the two amoCAB operons. The distal σ70 type amoCAB promoter was constitutively active in the presence of ammonia, but the proximal promoter was only active when cells were recovering from ammonia starvation. The lone, divergent copy of amoC (amoC3) was expressed only during recovery. Both the proximal amoC1,2 promoter and the amoC3 promoter are similar to gram-negative σE promoters, thus implicating σE in the regulation of the recovery response. Although modeling of subunit interactions suggested that a nonconservative proline substitution in AmoC3 may modify the activity of the holoenzyme, characterization of a ΔamoC3 strain showed no significant difference in starvation recovery under conditions evaluated. In contrast to the amo transcripts, a delayed appearance of transcripts for a gene required for CO2 fixation (cbbL) suggested that its transcription is retarded until sufficient energy is available. Overall, these data revealed a programmed exit from starvation likely involving regulation by σE and the coordinated regulation of catabolic and anabolic genes.
Ammonia-oxidizing bacteria fulfill an important biological role because they carry out the first reaction in the oxidative pathway of the nitrogen cycle. These bacteria obtain energy for growth from the oxidation of ammonia and acquire the majority of their carbon through the fixation of CO2 via the Calvin cycle. Since these microorganisms must compete with plants and other microorganisms that assimilate ammonia for biosynthesis (47, 48), it is likely that they have developed adaptive strategies to cope with periods of ammonia starvation. Both Nitrosomonas cryotolerans and Nitrosomonas europaea have been shown to be particularly resilient to energy source deprivation. Following 10 weeks of starvation, N. cryotolerans retained 85% of initial viability, showed no apparent degradation of DNA, protein, or RNA, and maintained an active electron transport system (21). Studies of N. europaea demonstrated persistence of the AmoB protein and immediate oxidation of added ammonia following 1 year of starvation for ammonia (38, 51). This remarkable resistance to starvation likely reflects unique physiological facets of these highly specialized microorganisms. In this study, we examined transcription of genes for key energy-generating (ammonia oxidation) and energy-consuming (CO2 fixation) reactions during recovery of N. europaea from ammonia starvation.
Ammonia-oxidizing bacteria convert ammonia to nitrite (NO2−) in two enzymatic reactions. Ammonia is first oxidized to hydroxylamine (NH2OH) in an energy-consuming step by the membrane-bound ammonia monooxygenase (AMO), a three-subunit holoenzyme encoded by the amoCAB operon. Electrons for energy generation and biosynthesis are derived from the oxidation of hydroxylamine to nitrite by the periplasmic hydroxylamine oxidoreductase (52). All betaproteobacterial ammonia oxidizers characterized to date have multiple copies of amoCAB and an additional copy of amoC (4, 25, 33, 37, 43). In N. europaea, the sequence of the protein encoded by the monocistronic amoC (AmoC3) diverges from the two nearly identical copies of AmoC (AmoC1,2) encoded by duplicate amoCAB operons (67.5% identity, 81.4% similarity). Although cotranscribed with amoA and amoB (43), an abundant monocistronic transcript of amoC has been shown to be remarkably stable in N. europaea during starvation (43).
Very little is known about the function of AmoC, other than a likely localization to the cell membrane as a subunit of ammonia monooxygenase (25), nor is there direct information about the possible role for the divergent copy, even though the conservation of amoC3 among ammonia-oxidizing bacteria suggests that it serves an important function. Our current analyses demonstrate that transcription of amoC3 is specific to recovery from ammonia starvation, thus implicating this subunit of the holoenzyme in the recovery of ammonia-oxidizing bacteria from starvation. Comparative promoter analysis showed that amoC3 transcription is correlated with elevated transcription of the operon copies of amoC as part of the poststarvation recovery response. Additional transcript analyses revealed a programmed exit from starvation in which transcription of genes encoding energy demanding anabolic functions are delayed relative to the synthesis/repair of central energy-generating systems.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotide primers.
Strains of N. europaea and Escherichia coli are described in Table 1. N. europaea was grown in liquid medium containing 5 mM (NH4)2SO4 and 20 mM HEPES (pH 7.5) or 12.5 mM (NH4)2SO4 and 50 mM HEPES (pH 7.5); 150 μM CaCl2, 150 μM MgSO4, 5 μM FeNa EDTA, 250 μM KH2PO4, 1.5 mM NaHCO3, 41 nm NaMoO4, 120 nm MnCl2, 0.84 nm CoCl2, 35 nm ZnSo4, 8 nm CuSO4, and 0.0003% phenol red as a pH indicator. Solid medium for growth of N. europaea was prepared by supplementing the liquid medium with 1% phytagel (Sigma) and adjusting the final concentrations of selected media components to 25 mM (NH4)2SO4, 100 mM HEPES (pH 7.5), 750 μM CaCl2, and 750 μM MgSO4. Ammonia-free medium was prepared by omitting (NH4)2SO4. E. coli was grown in LB medium as previously described (39). Plasmids and oligonucleotide primers are described in Tables 1 and 2, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| N. europaea | ||
| ATCC 19718 | Sequenced strain | ATCC |
| NCIMB 11850 | ATCC 25978 (strain C-31) | NCIMB |
| PMB1 | ΔamoC3::aacC1 (Gmr) N. europaea ATCC 19718 | This study |
| Plasmids | ||
| pCR4 | TA PCR cloning vector (Kanr AmprlacZα-ccdB ColE1 ori) | Invitrogen |
| pCRII | Dual promoter TA PCR cloning vector (Kanr AmprlacZα f1 ori ColE1 ori) | Invitrogen |
| pPMB8 | Upstream region of amoC1 cloned into pCR4 | This study |
| pPMB10 | Upstream region of amoC3 cloned into pCRII | This study |
| pPMB11 | Upstream region of cbbL cloned into pCRII | This study |
| pCM351 | Allelic exchange vector with aacC1 flanked by loxP sites (Ampr Genr Tetr) | 31 |
| pPMB25 | 340-bp upstream flanking region of amoC3 cloned upstream of aacC1 cassette in pCM351 | This study |
| pPMB34 | 450-bp downstream flanking region of amoC3 cloned downstream of aacC1 cassette in pPMB25 | This study |
TABLE 2.
Oligonucleotide primers and probes
| Name | Sequence | Reference |
|---|---|---|
| amoC1-140r | GGCAAAGCTCCAATACCTC | This study |
| S1amoC1-140r | GGCAAAGCTCCAATACCTCCGCCAGATACATCTGAAAGGCTAGTCACCTTGACCTTTTTAGTCAAGCCCTTTTTTATCCA | This study |
| amoC1-66r | CACTATCGCAAAGAAACCAC | This study |
| S1amoC1-66r | CACTATCGCAAAGAAACCACAATAACATCATGGTCTCCGCTTGATTTTCGACTCCGTATTTCTCGTTTTGTTCCGGCAAA | This study |
| C23 | ATAGCCTCTCGACGAGACTGATGA | 16 |
| NEUamoC3_54r | CGGTTTATCAGTGACTTGCTG | This study |
| NEUcbbL40r | GGTATTCTTTAACACCAGCG | This study |
| NEUglyA32r | ACCTGCTCAATCGTCAAGC | This study |
| NEUrpsJ95r | GTACGCTTGGCTGTTTCTAC | This study |
| C3upEcoR1F1 | CGGAATTCCGTGACGATGATGGCGATGACCTC | This study |
| C3upNco1R1 | CATGCCATGGCATGCGTATTCCTGGCTCAGAACTGC | This study |
| C3downApa1F2 | TGCAATGGGCCCATGGTTGGAACAGTCTGAATACCATCC | This study |
| C3downSac1R2 | TGCATCCGAGCTCGAATCGGCGAAAAAGCGGAG | This study |
| C3Gf1 | TCATCATTTGCGGGGGAAC | This study |
| C3Gr1 | TGCTGCTGCGTAACATCGTTG | This study |
| C3Gf2 | AAGTTGGGCATACGGGAAGAAG | This study |
| C3Gr2 | GCAGGCACAAACACCACAGATG | This study |
| C3f233 | TCGTTATTGCCTTGGCAGCATTC | This study |
| C3r822 | CGGAGTTGTTACGACATCGTC | This study |
Starvation conditions and cell sampling procedure.
Medium (25.4 liters) containing 5 mM (NH4)2SO4 and 20 mM HEPES (pH 7.5) was inoculated with 650 ml of a mid-exponential culture of N. europaea ATCC 19718. Nitrite levels in the culture were measured colorimetrically as previously described (23). After 48 h of growth (after approximately 3.75 mM NH3 was consumed), 1.3 liters of the exponential phase culture was harvested by filtration, resuspended in 750 μl RNA extraction buffer (50 mM sodium acetate, 10 mM EDTA, pH 5.1), and frozen in liquid nitrogen. Following an additional 14 h of growth, the remaining 24.7 liters of culture was harvested by filtration and resuspended in 290 ml ammonia-free medium containing 50 mM HEPES (pH 7.5) in a 500-ml flask. The concentrated (85-fold) N. europaea cells were starved of ammonia with gentle agitation for a period of up to 500 h. Starved cells were sampled at 24, 144, and 500 h. Cells were prepared for RNA extraction by centrifugation of 10 ml of culture (3,200 × g, 10 min, 0°C), resuspension in 1.5 ml of RNA extraction buffer, and being frozen in liquid nitrogen. At 24, 144, and 500 h, 50 ml of culture was removed and spiked with 12.5 mM (NH4)2SO4 to initiate recovery. During recovery, 10 ml of recovering cells was sampled at 20, 60, 180, 360, and 720 min and prepared for RNA extraction as described above. As a control, 20 ml of starved cells following 24, 144, and 500 h of starvation was spiked with 12.5 mM Na2SO4 and sampled at 20 and 720 min (10 ml per time point). After centrifugation, supernatants were filtered and retained for analysis of nitrite concentration. RNA was prepared by standard hot phenol-chloroform extraction under acidic conditions (39). In an alternate experiment, N. europaea NCIMB 11850 was starved for a total of 144 h. Samples for RNA extraction were obtained from exponential-phase cells, cells starved for 144 h, and recovering cells 3 and 13 h following the addition of 12.5 mM (NH4)2SO4. Cell samples were collected and processed as described above.
Primer extensions.
Plasmids are described in Table 1, and oligonucleotide primers are described in Table 2. Primer extensions were performed as previously described (39). Optikinase (USB) was used to end-label primers C23, NEUamoC3_54r, NEUcbbL40r, NEUglyA32r, and NEUrpsJ95r with [γ-32P]dATP (3,000 Ci/mmol; PerkinElmer). For primer extensions using RNA from the 500-h starvation of N. europaea ATCC 19718, 2.5 μg total RNA was used with primer C23 and 4.5 μg total RNA was used with primers NEUamoC3_54r and NEUcbbL40r. For primer extensions using RNA from the 144-h starvation of N. europaea NCIMB 11850, 3 μg of RNA was used in reactions with all of the above primers. Superscript III reverse transcriptase (Invitrogen) was used with N. europaea ATCC 19718 RNA, and Superscript II reverse transcriptase (Invitrogen) was used with NCIMB 11850 RNA. Sequencing ladders were prepared by cycle sequencing using the SequiTherm EXCEL II DNA sequencing kit (Epicenter). Plasmids pPMB8, pPMB10, and pPMB11 were used as templates with the respective labeled primers C23, NEUamoC3_54r, and NEUcbbL40r. Primer extension reactions were fractionated on a 6% polyacrylamide gel and visualized by autoradiography of dried gels.
S1 nuclease digestion.
S1 analysis was preformed as previously described (39) using RNA from N. europaea ATCC 19718 starved for 2 weeks in spent medium before the addition of 12.5 mM (NH4)2SO4. Each 80-mer probe (amoC1-140r and amoC1-66r) was purified by polyacrylamide gel electrophoresis. Optikinase (USB) was used to end-label amoC1-140r (targeting amoCp1) and amoC1-66r (targeting amoCp2) with [γ-32P]dATP (3,000 Ci/mmol; PerkinElmer). Each probe was hybridized with 5 μg of total RNA before digestion with 50, 250, or 500 U/ml S1 nuclease (Invitrogen). The digestion products were fractionated on a 6% polyacrylamide gel and visualized as described above.
mRNA secondary structure prediction.
The web interface for the mfold version 3.0 algorithm (http://www.bioinfo.rpi.edu/applications/mfold/) was used for all RNA structure predictions (49). As this version allows predictions at different temperatures, initial foldings were run using temperatures between 65°C and 85°C to predict the most stable secondary structures for each RNA molecule. To obtain ΔG values for each structure, mfold version 3.1 was employed at a fixed temperature of 37°C (32, 54).
Construction of a ΔamoC3 strain.
The allelic exchange vector, pCM351, was used to replace the amoC3 gene with the gentamicin resistance determinant, aacC1, in the same orientation. The upstream flanking region of amoC3 was amplified by PCR using primers C3upEcoR1F1 and C3upNco1R1. The resulting PCR product was digested with EcoRI and NcoI and cloned upstream of aacC1 in pCM351 to create pPMB25. The downstream flanking region of amoC3 was amplified using primers C3downApa1F2 and C3downSac1R2. The resulting PCR product was digested with ApaI and SacI and cloned downstream of aacC1 to create pPMB34. pPMB34 was linearized using NheI prior to subsequent electroporation of N. europaea ATCC 19718, as previously described (15). Recombinant clones were selected by plating on solid media containing 5 μg/ml gentamicin (USB). Replacement of amoC3 was confirmed by PCR using primers specific to amoC3, the gentamicin cassette, and upstream and downstream of the recombination sites. In addition, primer extension analysis using RNA from recovering cells was used to confirm that the mutant strain did not express amoC3. The mutant was designated N. europaea PMB1.
Assay of the recovery phenotype for the ΔamoC3 strain.
N. europaea ATCC 19718 and N. europaea PMB1 were grown in triplicate 500-ml batch cultures. At late-exponential phase, cells were harvested by filtration and resuspended in ammonia-free medium. Cell densities were normalized using optical density at 600 nm and starved for up to 4.5 months. Periodically during the starvation, an aliquot of cells was spiked with 0.020 mM or 25 mM NH3. In a separate pulse-feed experiment, 7-day stationary-phase cultures of N. europaea ATCC 19718 and N. europaea PMB1 were transferred in triplicate to ammonia-free medium. NH3 was periodically added to the cultures to induce several cycles of starvation and recovery. The recovery response in all experiments was monitored by measuring nitrite production as previously described (23).
AMO structure modeling.
Structural modeling of AMO was performed with the comparative modeling protocol on the PROTINFO structure prediction server (http://protinfo.compbio.washington.edu) using the particulate methane monooxygenase crystal structure as a template (28). This comparative modeling protocol has been shown to work well in the CASP protein structure prediction experiments (18, 19). Initial models were constructed using a minimum perturbation approach that aims to preserve as much information as possible from the template structure solved by X-ray crystallography. Variable side chains and main chains were then built using a graph theory clique-finding approach that explores a variety of possible conformations for the respective side chains and main chains and finds the optimal combination using an all-atom scoring function (42). These approaches are described in further detail in references 40 and 41. PDB files of all AMO structure models are available from the Computational Biology Research Group in the Department of Microbiology at the University of Washington (http://data.compbio.washington.edu/misc/downloads/amoc/).
RESULTS
Transcription of amoC1,2.
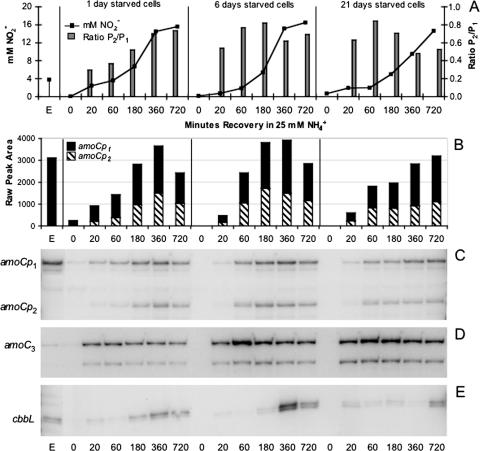
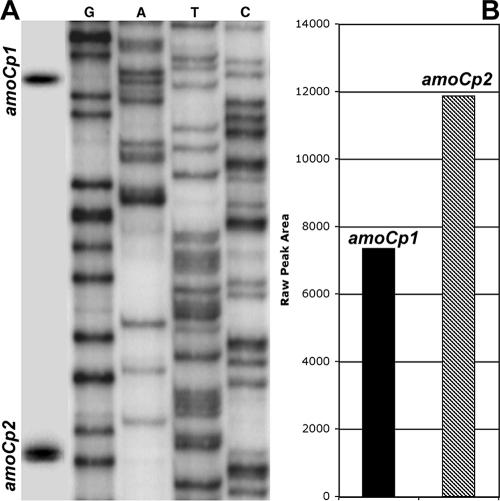
Primer extension analysis was used to monitor transcription of amoC1,2 in actively growing, starved, and recovering cells of N. europaea ATCC 19718 and N. europaea NCIMB 11850. Our data demonstrated two differentially regulated promoters upstream of amoC1,2. Only the distal (amoCp1) promoter is active during exponential growth, while both amoCp1 and the proximal (amoCp2) promoter are utilized during recovery (Fig. 1). Promoter activity of amoCp1 relative to amoCp2 during recovery was shown to vary with the length of starvation. Although our study did not determine absolute transcript abundance, the relative activities of the amoCp1 and amoCp2 promoters could be determined by using densitometry to evaluate the signal intensity of each alternative transcript within the same lane of the primer extension gel. Since the same amount of sample was loaded for each time point in the sample series, changes in transcript abundance could be estimated over time given that the reactions were prepared using the same labeled primer and loaded on the same gel. Initial RNA concentrations were measured in triplicate using the Agilent Bioanalyzer 2100 to ensure greatest accuracy in RNA quantification. Relative abundance of different transcripts (i.e., amoC1,2 versus amoC3) could not be accurately determined due to differences in primer labeling efficiency and exposure times. The activity of amoCp2 peaked at 3 to 6 h in cells recovering from 1 or 6 days of starvation but remained below the activity of amoCp1 (Fig. 1). In contrast, analysis of N. europaea NCIMB 11850 recovering from 1 month of starvation in spent medium showed that the amoCp2 promoter was nearly twice as active as amoCp1 (Fig. 2).
FIG. 1.
Primer extension analysis of amoC1,2, amoC3, and cbbL expression during exponential growth and recovery of N. europaea ATCC 19718 from 1, 6, and 21 days of starvation. (A) Nitrite accumulation and ratios of amoCp2 transcripts to amoCp1 transcripts (determined by densitometry) during recovery. (B) amoC1,2 expression levels, as determined by densitometry, showing the contribution of each promoter to total amoC1,2 abundance. (C, D, E) Primer extension analysis of amoC1,2, amoC3, and cbbL, respectively. Transcripts derived from each amoC1,2 promoter are indicated at left.
FIG. 2.
(A) Primer extension analysis of amoC1,2 following a 3-h recovery of a 1-month stationary-phase culture of N. europaea NCIMB 11850. Transcription start sites associated with each promoter are indicated at left. (B) amoC1,2 expression levels, as determined by densitometry.
Transcription of amoC3.
Transcription of the divergent, monocistronic amoC3 gene was found to be differentially regulated with respect to growth phase. The amoC3 transcript could barely be detected by primer extension analysis in exponentially growing cells of N. europaea ATCC 19718 (Fig. 1) and only at low levels in N. europaea NCIMB 11850 (data not shown). Expression of amoC3 was also absent during the entire starvation period. During recovery of starved cells, expression of amoC3 was significantly up-regulated in both N. europaea ATCC 19718 (Fig. 1) and N. europaea NCIMB 11850 (data not shown). Expression levels of amoC3 followed a general trend related to the length of the starvation period. Longer periods of starvation resulted in higher expression levels of amoC3 during the subsequent recovery period (Fig. 1).
Transcription of cbbL, glyA, and rpsJ.
As a control, the expression of several additional genes of various functional role categories was also examined. The expression of cbbL (large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase), glyA (glycine hydroxymethyltransferase), and rpsJ (S10 ribosomal protein) were monitored in actively growing, starved, and recovering cells of N. europaea. Since cbbL encodes a subunit of an anabolic enzyme involved in CO2 assimilation (50), it was hypothesized that its transcription would be regulated differently than the genes encoding the catabolic ammonia monooxygenase. The cbbL transcript was detected in exponential-phase cells but not in starved cells (Fig. 1). Expression of cbbL during the recovery period was delayed several hours with respect to the expression of amoC1,2 and amoC3 (Fig. 1). Longer periods of starvation resulted in a longer lag between the initiation of recovery and the expression of cbbL (Fig. 1). The “housekeeping” genes, glyA and rpsJ, exhibited similar expression profiles in N. europaea NCIMB 11850. Both transcripts were detected in actively growing cells but not during the starvation period. Expression of glyA and rpsJ was induced within the first 3 h of recovery following 6 days of starvation at levels similar to that of exponential phase cells (data not shown).
Identification of transcription start sites and promoter consensus sequences.
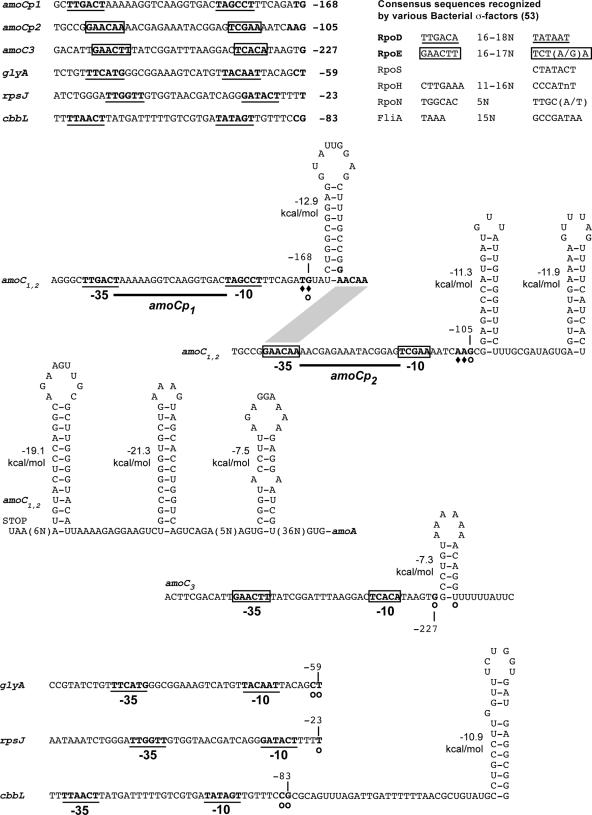
The transcription start sites for amoC1,2, amoC3, cbbL, glyA, and rpsJ were first determined by primer extension. S1 nuclease protection was then used to confirm the transcription start sites for amoC1,2. The −35 and −10 promoter sequences were identified by visual inspection of sequences upstream of the identified transcription start sites (53). The start sites for the promoters amoCp1 and amoCp2 are, respectively, 168 to 169 and 105 to 107 nucleotides upstream of the start codon for amoC1,2 (Fig. 3). These positions differ by approximately 2 nucleotides (nt) from those reported by Hommes et al. for amoC1,2 (16), likely reflecting the use of a heterologous sequence for positional identification in the prior study. As shown previously, the amoCp1 promoter most closely matches the eubacterial σ70 consensus sequence (16). Although the amoCp2 promoter also shows homology to the eubacterial σ70 consensus sequence, as reported by Hommes et al. (16), further inspection of this region revealed features highly similar to the consensus sequence for the eubacterial extracytoplasmic function (ECF) σ factor, encoded by the rpoE gene in N. europaea (Fig. 3). This promoter assignment also places the putative promoter closer to the −35 and −10 regions. The putative amoC3 promoter for the major primer extension product, 227 nt upstream of the start codon, shows some homology to σ70 and σ32 consensus sequences, but also most closely matches the consensus sequence for ECF σ factors (Fig. 3). No obvious promoter sequence could be found upstream of the putative start site corresponding to the minor primer extension product for amoC3 (209 nt upstream of the amoC3 start codon). The transcription start sites for cbbL, glyA, and rpsJ are 83 to 84, 59 to 60, and 23 nt upstream of their respective start codons. These genes, all primarily up-regulated during exponential growth, appear to be controlled by σ70 promoters (Fig. 3).
FIG. 3.
Transcription start sites, promoters, and predicted 5′ untranslated region secondary structures associated with amoC1,2, amoC3, glyA, rpsJ, and cbbL. Transcription start sites determined by primer extension and S1 analysis are indicated by open circles and diamonds, respectively. Putative σ70 promoters are underlined and putative σECF promoters are boxed. ΔG values for each structure were determined by mfold version 3.1 at a fixed temperature of 37°C. Start codons (AUG) and stop codons (UAA) are indicated as needed. The positions of the transcription start sites relative to the start codons are indicated next to each start site. Bacterial promoter consensus sequences were obtained from the review by Wösten (53).
Transcript secondary structures.
The stability of amoC transcripts may in part be determined by stable stem-loop structures identified near the 5′ and 3′ termini. These types of structures have been shown to protect mRNA from degradation (7, 9, 11, 45). The mfold algorithm predicted stable stem-loop structures in the 5′ untranslated regions of amoC1,2, amoC3, and cbbL, as well as the region immediately downstream of amoC1,2 (Fig. 3). The start sites for amoCp1 and amoCp2 are located at the base of highly stable stem-loop structures, having ΔG values of −12.9 and −11.3 kcal/mol, respectively (Fig. 3). In addition, a highly stable (ΔG = −19.1 kcal/mol) stem-loop is predicted immediately downstream of the stop codon for amoC1,2 (Fig. 3). The 5′ termini for both primer extension products for amoC3 are located at the base of a hairpin structure with a ΔG value of −7.3 kcal/mol (Fig. 3). In contrast, the cbbL transcript lacks a stable stem-loop structure associated with its transcription start site but does have a predicted structure located approximately 32 nt downstream of the start site with a ΔG value of −10.9 kcal/mol (Fig. 3).
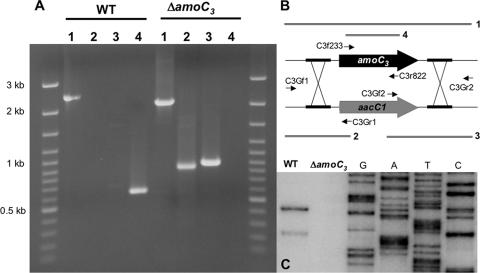
Characterization of a ΔamoC3 mutant.
An amoC3 deletion mutant of N. europaea ATCC 19718 (ΔamoC3::aacC1 strain PMB1) was constructed to further evaluate possible differences in the function of AmoC1,2 and AmoC3. The deletion was confirmed by PCR and the absence of an amoC3 transcript during recovery from starvation (Fig. 4). The deletion mutant was initially characterized by examining its recovery from starvation relative to the wild type. No significant differences in nitrite production between the wild type and mutant were observed over a 10-day recovery period following the addition of 25 mM NH3 to cultures starved of ammonia for 4.5 months (data not shown). The recovery phenotype was also assayed after addition of 20 μM NH3 to a culture starved for 47 days, a concentration below the estimated KS of 50 μM NH3 for N. europaea (26) (data not shown). As with the higher concentration of NH3, no significant differences in nitrite production were observed. Pulse-feeding of ammonia to 7-day-old stationary-phase cultures of wild-type and mutant strains was then examined, since our transcription data suggest that this feeding regimen would selectively enrich for AmoC3 (Fig. 3). A slight but replicable difference (standard deviation) in ammonia oxidation activity was observed when the cultures were initially pulse-fed 400 μM ammonia. However, nitrite production converged with subsequent additions of ammonia (data not shown).
FIG. 4.
Verification of the amoC3 deletion. (A) PCR analysis of the recombination site. (B) Primers used (Table 1) and PCR products corresponding to each lane. (C) Primer extension analysis of amoC3 expression in wild-type (WT) and ΔamoC3 cells following 20 min of recovery in 25 mM NH3 after 24 h of starvation.
AmoC1,2, AmoC3, and PmoC structure comparisons.
The recently published crystal structure of the evolutionarily related particulate methane monooxygenase from the methane-oxidizing bacterium, Methylococcus capsulatus (Bath), provides an additional comparative framework to examine possible functional differences between the divergent AmoC subunits (28). Like amoCAB, the pmoCAB operon exists in multiple copies and there is an additional copy of pmoC (44). AmoC1 from N. europaea ATCC 19718 and PmoC1 from M. capsulatus (Bath) are 41.0% identical and 62.3% similar, while AmoC3 and PmoC3 are 42.3% identical and 57.7% similar. Unlike AmoC3 which is 67.5% identical and 81.4% similar to AmoC1,2, PmoC3 is more similar to the operon copies of PmoC (89.6% identity and 91.5% similarity), with all differences located at the termini of the protein in contrast to a more uniform distribution in AmoC3.
A comparative alignment of PmoC and AmoC sequences revealed an amino acid substitution in AmoC3 near a conserved metal binding center. As revealed by the crystal structure of PmoC, this tetrahedral metal binding center involves 3 residues (D156, H160, and H173) from PmoC and 1 residue (E195) from PmoA. All 3 residues from PmoC are conserved in both AmoC1,2 and AmoC3 (Fig. 5). However, there is a nonconservative proline substitution at position 157 in AmoC3 (V155 in AmoC1 and I146 in PmoC1) (Fig. 5). PmoC3 does not have a substitution at this position compared to the operon copies of PmoC (Fig. 5). The structural consequences of this mutation were then examined using a knowledge-based all-atom scoring function developed for protein structure prediction. The all-atom function computes a stability score by summing up the individual scores of all the atomic interactions in a protein structure. The individual scores are derived from atomic preferences in a database of experimentally determined structures (42).
FIG. 5.
ClustalW alignment of AmoC and PmoC amino acid sequences. Identities are highlighted in black, and similarities are highlighted in gray. Conserved residues involved in coordination of the tetrahedral metal binding site are indicated by asterisks. The proline substitutions in the AmoC3 proteins from N. europaea ATCC 19718 and Nitrosomonas eutropha C71 are boxed.
The stability of the AMO proteins were evaluated for both the wild-type AmoC sequences as well as Ile, Pro, and Val variants, both individually and in the context of AmoA and AmoB. When the AmoC1 or AmoC3 proteins are scored as monomers, the structure with the Pro variant is the least stable structure. The AmoC1 structure containing a V155P mutation has a higher all-atom stability score than the wild-type AmoC1 structure, and the wild-type AmoC3 structure has a higher all-atom score than AmoC3 structures containing P157I and P157V mutations (lower scores indicate greater stability). This difference in stability was expected, since the Pro change occurs toward the end of an alpha helix and disrupts the hydrogen-bonding pattern necessary for helical conformation. However, this relationship is reversed when the AmoA, AmoB, and AmoC structures are scored as multimers. In this case, the holoenzyme containing AmoC1 or AmoC3 with a Pro residue at this position is more stable than the respective Val or Ile variants. Since the Pro/Val residues are located at sites that are not directly in contact with the AmoA and AmoB chains, it is not immediately clear how the holoenzyme structure containing the Pro variant has increased stability (or conversely, why the Ile and Val structures have decreased stability only in the context of the multimer).
DISCUSSION
The starvation physiology of ammonia-oxidizing bacteria is characterized by a high tolerance to energy source deprivation while retaining the ability to quickly respond to the presence of ammonia, even after starvation periods of nearly a year (51). Ammonia-oxidizing bacteria do not exhibit the well-characterized starvation response strategies described for other bacteria. For instance, they do not undergo cellular differentiation, such as reductive cell division or sporulation, and in contrast to many heterotrophs, few proteins are induced in response to starvation conditions (6, 30, 34). This is consistent with the observation that N. europaea does not have the alternative sigma factor RpoS which controls the stress response during stationary phase (8, 53). Yet, ammonia-oxidizing bacteria maintain a state of cellular readiness during periods of starvation that is manifested during the subsequent recovery response. Therefore, available data suggest that a more complete appreciation of the adaptive physiology of this organism must include understanding the genetic and physiological processes associated with recovery from starvation.
Our studies demonstrate a major role of the AmoC subunit in recovery from starvation. The long-lived amoC1,2 message presumably contributes to recovery following relatively short-term starvation periods of several days. Recovery from longer-term starvation is associated with a high level of transcription from both promoters for the amoC1,2 operons and a recovery-specific activation of transcription of amoC3. While these analyses confirmed the presence of two promoters for amoC1,2, as previously reported by Hommes et al. (16), the earlier study showed, in contrast to our results, that transcripts originating from both promoters are present in starved cells and that only the distal promoter is active during outgrowth from starvation (16). Differences in starvation and recovery conditions may account for some differences in results between these two studies. In the Hommes et al. study, N. europaea ATCC 19718 was grown for 3 days to late exponential phase, harvested by centrifugation, washed three times with buffer (0.2 mM MgCl2 and 50 mM NaH2PO4, pH 7.5), and stored as a pellet at 4°C for at least 24 h to allow the endogenous mRNA to decay before transcription was induced by the addition of 50 mM NH3 (16). In contrast, our protocol limited the manipulation of the cells to more physiologically relevant conditions; cells were starved as a suspension in fresh growth medium lacking ammonia at room temperature (approximately 21°C). To address these differences, the effects of starvation time and temperature on transcription of amoC1,2 were investigated. When we used our protocol to examine cells starved for either 1 or 6 days at 4°C or 20°C in ammonia-free medium, the addition of ammonia induced transcription from both promoters (data not shown). These studies also showed that mRNA was more stable when cells were stored at 4°C; the longer transcript could still be detected after 6 days of starvation at 4°C but not after 6 days of starvation at 20°C (data not shown). Thus, variation in conditions antecedent to harvest may have influenced the results earlier reported (16).
Control of amoC1,2 and amoC3 transcription by an ECF sigma factor implies a role of AmoC in stress response. ECF sigma factors are often responsive to environmental stimuli and membrane stress conditions that result in misfolded proteins in the periplasm (1, 53). The fact that AMO is a membrane protein is consistent with this function of ECF sigma factors. Inactivation of the membrane-bound AMO during starvation may serve as one signal for increased transcription of the amoCAB operons and recovery-specific transcription of amoC3 during the subsequent recovery period.
As initiation is typically the rate-limiting step of transcription (17), our data indicate that the tandem promoters upstream of the amoCAB operon are utilized to enhance transcription during the recovery period. There are a number of examples of tandem promoters in bacteria, many of which are regulated by different sigma factors (5, 10, 12, 13, 36). For instance, in Shewanella violacea, the tandem σ70 and σ54 promoters that control expression of glnA are differentially regulated with respect to hydrostatic pressure (20). Also, the tandem rrn promoters in E. coli are differentially regulated with respect to growth rate such that the proximal promoter is preferentially utilized during the outgrowth of cells from stationary phase (35). Utilization of amoCp2 presumably enables cells to increase expression levels beyond that observed in actively growing cells (Fig. 1).
Transcript stability can determine protein stoichiometry in some systems and has also been suggested to conserve energy at the expense of tight regulatory control (14, 27). The stability of the bulk mRNA pool increases during nutrient limitation in many bacteria and is hypothesized to enhance survival by decreasing the energetic requirements for transcription (2, 3, 24). The proximity of a stable secondary structure to the 5′ end of a transcript (within 4 to 7 bp) has been shown to be the primary determinant of messenger stability (7, 9, 11, 45). The 5′ stem-loop structures associated with amoC1,2 transcripts are within 4 bp of the termini and should be sufficient to impart significant stability to the message (Fig. 3). In addition, there is a stable structure located downstream of the amoC1,2 stop codon that likely protects these transcripts from 3′-5′ degradation (9). Although this structure lacks a downstream poly(U) sequence characteristic of many intrinsic transcription terminators in E. coli, there is evidence that a poly(U) sequence is not necessary for efficient termination in some species (46). While further studies are warranted, these predicted secondary structures could explain the high abundance of the 1.1-kb monocistronic amoC message present in N. europaea cells (16, 43).
In contrast to the elevated expression of genes encoding enzymes responsible for catabolic processes and energy generation, strict control over CO2 fixation is supported by a delay in the expression of cbbL encoding the large subunit of RuBisCO (Fig. 1). These data are consistent with a previous study by Johnstone and Jones which demonstrated that CO2 uptake was delayed approximately 4 h before it began to steadily increase in recovering cells of N. cryotolerans that had been starved for 5 weeks (22). Unlike glyA and rpsJ which also have putative σ70 promoters, cbbL is not expressed immediately upon addition of ammonia to starved cells. This result indicates that another regulatory factor is involved in the control of cbbL expression during recovery. Since there is no apparent stable stem-loop structure associated with the 5′ terminus of the cbbL transcript, a lower stability compared to amoC may impart greater transcriptional control of cbbL messenger abundance.
The third, divergent, lone copy of amoC (amoC3) apparently has a specific role in the recovery of cells from starvation. Similar to the operon copies of amoC, the transcription start site of amoC3 is associated with a short stem-loop structure that may influence the stability of the amoC3 message (Fig. 3). Since no promoter could be assigned to the minor primer extension product, it is most likely an artifact caused by premature termination of the reverse transcriptase at this stem-loop. A poly(U) sequence immediately downstream of this stem-loop structure implicates another regulatory control, since probable transcription termination in this region would need to be suppressed during recovery. However, the amoC3 deletion mutant showed only modest differences from the wild type in its recovery phenotype under the conditions so far evaluated, suggesting a subtle role in recovery physiology that may in part be masked by the operon variants.
The conservation of the metal binding site in PmoC and AmoC implies a function beyond that of a chaperone or membrane anchor as suggested previously (25). Lieberman and Rosenzweig have hypothesized that the metal binding site of PmoC may function in catalysis or as an electron transfer center due to the proximity of a putative quinone binding site (29). The close proximity of the proline substitution to the metal center in AmoC3 could affect these possible functions by altering the local structure of this metal binding site. In contrast, our observations of the increased stability of the AMO holoenzyme containing AmoC3 and that an ECF sigma factor appears to control the increased transcription of all three amoC copies during recovery indicate that AmoC3 may serve a role as a specialized chaperone as suggested previously. As such, AmoC3 may facilitate association of AMO subunits in the membrane during recovery but not function optimally, as illustrated by the lack of amoC3 transcription during active growth (Fig. 1) and the likely disruptive effects of a proline substitution near the metal binding site of AmoC3 (Fig. 5). We are now examining the stoichiometry of the AMO subunits under different starvation and recovery scenarios to identify conditions that may enhance phenotypic differences between the wild type and the amoC3 deletion mutant. Many proteins involved in ammonia oxidation are stable and highly abundant (38, 51). It may not be possible to detect subtle differences between the wild type and the amoC3 deletion mutant if the level of AmoC1,2 is significantly greater that the quantities of AmoC3 produced during the recovery of wild-type cells.
Our results demonstrate that cellular resources are primarily directed toward the regeneration of ammonia oxidation activity during the recovery of ammonia-oxidizing bacteria from starvation. In particular, the high abundance and putative stability of amoC transcripts suggest that the physiological importance of AmoC has been underestimated, during both active growth and recovery from starvation. The presence of a conserved, divergent copy of amoC that is primarily expressed during recovery underscores this point and presents a unique opportunity to make further advances in understanding the structure and function of AMO, which has been recalcitrant to purification and in vitro enzymatic studies. Although technically challenging, the development of strains in which the stoichiometry of amoC copies is controlled would facilitate in vivo studies aimed at understanding the functional nature of AMO. In addition, our results indicate that the RpoE stress response regulon has a role in the recovery physiology of N. europaea. Characterizing other members of this regulon is likely to further our understanding of the unique starvation and recovery response of ammonia-oxidizing bacteria.
Acknowledgments
This work was partially supported by the Department of Energy's Office of Biological and Environmental Sciences under the GTL-Genomics Program via the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov).
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Gene Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertson, N. H., and T. Nystrom. 1994. Effects of starvation for exogenous carbon on functional messenger RNA stability and rate of peptide chain elongation in Escherichia coli. FEMS Microbiol. Lett. 117:181-188. [DOI] [PubMed] [Google Scholar]
- 3.Albertson, N. H., T. Nystrom, and S. Kjelleberg. 1990. Functional messenger RNA half-lives in the marine Vibrio sp S14 during starvation and recovery. J. Gen. Microbiol. 136:2195-2199. [Google Scholar]
- 4.Arp, D. J., L. A. Sayavedra-Soto, and N. G. Hommes. 2002. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178:250-255. [DOI] [PubMed] [Google Scholar]
- 5.Barrios, H., H. M. Fischer, H. Hennecke, and E. Morett. 1995. Overlapping promoters for two different RNA polymerase holoenzymes control Bradyrhizobium japonicum nifA expression. J. Bacteriol. 177:1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollmann, A., I. Schmidt, A. M. Saunders, and M. H. Nicolaisen. 2005. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvet, P., and J. G. Belasco. 1992. Control of Rnase E mediated RNA degradation by 5′-terminal base pairing in Escherichia coli. Nature 360:488-491. [DOI] [PubMed] [Google Scholar]
- 8.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: An old problem with some new twists. Prog. Nucleic Acid Res. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 10.de Zamaroczy, M., F. Delorme, and C. Elmerich. 1990. Characterization of three different nitrogen-regulated promoter regions for the expression of glnB and glnA in Azospirillum brasilense. Mol. Gen. Genet. 224:421-430. [DOI] [PubMed] [Google Scholar]
- 11.Emory, S. A., P. Bouvet, and J. G. Belasco. 1992. A 5′-terminal stem loop structure can stabilize messenger RNA in Escherichia coli. Gene Dev. 6:135-148. [DOI] [PubMed] [Google Scholar]
- 12.Govantes, F., J. A. Albrecht, and R. P. Gunsalus. 2000. Oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon: roles of multiple promoters and the Fnr-1 and Fnr-2 binding sites. Mol. Microbiol. 37:1456-1469. [DOI] [PubMed] [Google Scholar]
- 13.Grafe, S., T. Ellinger, and H. Malke. 1996. Structural dissection and functional analysis of the complex promoter of the streptokinase gene from Streptococcus equisimilis H46A. Med. Microbiol. Immunol. 185:11-17. [DOI] [PubMed] [Google Scholar]
- 14.Heck, C., R. Rothfuchs, A. Jager, R. Rauhut, and G. Klug. 1996. Effect of the pufQ-pufB intercistronic region on puf mRNA stability in Rhodobacter capsulatus. Mol. Microbiol. 20:1165-1178. [DOI] [PubMed] [Google Scholar]
- 15.Hommes, N. G., L. A. Sayavedra-Soto, and D. J. Arp. 1998. Mutagenesis and expression of amo, which codes for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 180:3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hommes, N. G., L. A. Sayavedra-Soto, and D. J. Arp. 2001. Transcript analysis of multiple copies of amo (encoding ammonia monooxygenase) and hao (encoding hydroxylamine oxidoreductase) in Nitrosomonas europaea. J. Bacteriol. 183:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu, L. M. 2002. Promoter clearance and escape in prokaryotes. Biochim. Biophys. Acta 1577:191-207. [DOI] [PubMed] [Google Scholar]
- 18.Hung, L. H., S. C. Ngan, T. Y. Liu, and R. Samudrala. 2005. PROTINFO: new algorithms for enhanced protein structure predictions. Nucleic Acids Res. 33:W77-W80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung, L. H., and R. Samudrala. 2003. PROTINFO: secondary and tertiary protein structure prediction. Nucleic Acids Res. 31:3296-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikegami, A., K. Nakasone, C. Kato, Y. Nakamura, I. Yoshikawa, R. Usami, and K. Horikoshi. 2000. Glutamine synthetase gene expression at elevated hydrostatic pressure in a deep-sea piezophilic Shewanella violacea. FEMS Microbiol. Lett. 192:91-95. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone, B. H., and R. D. Jones. 1988. Physiological-effects of long-term energy-source deprivation on the survival of a marine chemolithotrophic ammonium-oxidizing bacterium. Mar. Ecol. Prog. Ser. 49:295-303. [Google Scholar]
- 22.Johnstone, B. H., and R. D. Jones. 1988. Recovery of a marine chemolithotrophic ammonium-oxidizing bacterium from long-term energy-source deprivation. Can. J. Microbiol. 34:1347-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenney, D. R., and D. W. Nelson. 1982. Nitrogen—inorganic forms, p. 643-693. In A. L. Page (ed.), Methods of soil analysis, part 2. American Society of Agronomy, Madison, WI.
- 24.Kjelleberg, S., N. Albertson, K. Flardh, L. Holmquist, A. Jouperjaan, R. Marouga, J. Ostling, B. Svenblad, and D. Weichart. 1993. How do nondifferentiating bacteria adapt to starvation. Antonie Leeuwenhoek 63:333-341. [DOI] [PubMed] [Google Scholar]
- 25.Klotz, M. G., J. Alzerreca, and J. M. Norton. 1997. A gene encoding a membrane protein exists upstream of the amoA/amoB genes in ammonia oxidizing bacteria: A third member of the amo operon? FEMS Microbiol. Lett. 150:65-73. [DOI] [PubMed] [Google Scholar]
- 26.Koops, H. P., and A. Pommerening-Roser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 27.Kuzj, A. E., P. S. Medberry, and J. L. Schottel. 1998. Stationary phase, amino acid limitation and recovery from stationary phase modulate the stability and translation of chloramphenicol acetyltransferase mRNA and total mRNA in Escherichia coli. Microbiology 144(Pt 3):739-750. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman, R. L., and A. C. Rosenzweig. 2005. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434:177-182. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman, R. L., and A. C. Rosenzweig. 2005. The quest for the particulate methane monooxygenase active site. Dalton Trans. 3390-3396. [DOI] [PubMed]
- 30.Loewen, P. C., and R. Henggearonis. 1994. The role of the sigma-factor sigma(S) (Katf) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 31.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 32.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 33.McTavish, H., J. A. Fuchs, and A. B. Hooper. 1993. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. Bacteriol. 175:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee, T. K., A. Raghavan, and D. Chatterji. 1998. Shortage of nutrients in bacteria: the stringent response. Curr. Sci. India 75:684-689. [Google Scholar]
- 35.Murray, H. D., and R. L. Gourse. 2004. Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol. Microbiol. 52:1375-1387. [DOI] [PubMed] [Google Scholar]
- 36.Nasser, W., M. Rochman, and G. Muskhelishvili. 2002. Transcriptional regulation of fis operon involves a module of multiple coupled promoters. EMBO J. 21:715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norton, J. M., J. J. Alzerreca, Y. Suwa, and M. G. Klotz. 2002. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 177:139-149. [DOI] [PubMed] [Google Scholar]
- 38.Pinck, C., C. Coeur, P. Potier, and E. Bock. 2001. Polyclonal antibodies recognizing the AmoB protein of ammonia oxidizers of the beta-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 67:118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Samudrala, R., and M. Levitt. 2002. A comprehensive analysis of 40 blind protein structure predictions. BMC Struct. Biol. 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samudrala, R., and J. Moult. 1998. A graph-theoretic algorithm for comparative modeling of protein structure. J. Mol. Biol. 279:287-302. [DOI] [PubMed] [Google Scholar]
- 42.Samudrala, R., and J. Moult. 1998. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 275:895-916. [DOI] [PubMed] [Google Scholar]
- 43.Sayavedra-Soto, L. A., N. G. Hommes, J. J. Alzerreca, D. J. Arp, J. M. Norton, and M. G. Klotz. 1998. Transcription of the amoC, amoA and amoB genes in Nitrosomonas europaea and Nitrosospira sp. NpAV. FEMS Microbiol. Lett. 167:81-88. [DOI] [PubMed] [Google Scholar]
- 44.Stolyar, S., A. M. Costello, T. L. Peeples, and M. E. Lidstrom. 1999. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology 145(Pt 5):1235-1244. [DOI] [PubMed] [Google Scholar]
- 45.Unniraman, S., M. Chatterji, and V. Nagaraja. 2002. A hairpin near the 5′ end stabilises the DNA gyrase mRNA in Mycobacterium smegmatis. Nucleic Acids Res. 30:5376-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unniraman, S., R. Prakash, and V. Nagaraja. 2001. Alternate paradigm for intrinsic transcription termination in eubacteria. J. Biol. Chem. 276:41850-41855. [DOI] [PubMed] [Google Scholar]
- 47.Verhagen, F. J., and H. J. Laanbroek. 1991. Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats. Appl. Environ. Microbiol. 57:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verhagen, F. J. M., H. J. Laanbroek, and J. W. Woldendorp. 1995. Competition for ammonium between plant roots and nitrifying and heterotrophic bacteria and the effects of protozoan grazing. Plant Soil 170:241-250. [Google Scholar]
- 49.Walter, A. E., D. H. Turner, J. Kim, M. H. Lyttle, P. Muller, D. H. Mathews, and M. Zuker. 1994. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. USA 91:9218-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, X. M., L. A. Sayavedra-Soto, and D. J. Arp. 2004. The transcription of the cbb operon in Nitrosomonas europaea. Microbiology 150:1869-1879. [DOI] [PubMed] [Google Scholar]
- 51.Wilhelm, R., A. Abeliovich, and A. Nejidat. 1998. Effect of long-term ammonia starvation on the oxidation of ammonia and hydroxylamine by Nitrosomonas europaea. J. Biochem. 124:811-815. [DOI] [PubMed] [Google Scholar]
- 52.Wood, P. M. 1986. Nitrification as a bacterial energy source, p. 39-62. In J. I. Prosser (ed.), Nitrification. IRL Press, Oxford, United Kingdom.
- 53.Wösten, M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 54.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Boston, MA.