Abstract
Bacterial translation initiation factor IF1 is an S1 domain protein that belongs to the oligomer binding (OB) fold proteins. Cold shock domain (CSD)-containing proteins such as CspA (the major cold shock protein of Escherichia coli) and its homologues also belong to the OB fold protein family. The striking structural similarity between IF1 and CspA homologues suggests a functional overlap between these proteins. Certain members of the CspA family of cold shock proteins act as nucleic acid chaperones: they melt secondary structures in nucleic acids and act as transcription antiterminators. This activity may help the cell to acclimatize to low temperatures, since cold-induced stabilization of secondary structures in nascent RNA can impede transcription elongation. Here we show that the E. coli translation initiation factor, IF1, also has RNA chaperone activity and acts as a transcription antiterminator in vivo and in vitro. We further show that the RNA chaperone activity of IF1, although critical for transcription antitermination, is not essential for its role in supporting cell growth, which presumably functions in translation. The results thus indicate that IF1 may participate in transcription regulation and that cross talk and/or functional overlap may exist between the Csp family proteins, known to be involved in transcription regulation at cold shock, and S1 domain proteins, known to function in translation.
Shifting exponentially growing Escherichia coli cells from 37°C to 15°C elicits a cold shock response, during which the synthesis of most cellular proteins is strongly decreased while the synthesis of several cold shock proteins is strongly increased. The most highly produced cold shock protein is the CspA protein. E. coli has nine CspA homologues, only some of which are cold shock inducible. CspA and its homologues destabilize secondary structures in both RNA and DNA and are therefore referred to as nucleic acid chaperones (13). In our previous studies, we showed that CspA and its homologues CspC and CspE act as transcription antiterminators (1). Stabilization of secondary structures in RNA upon a temperature downshift affects transcription elongation by RNA polymerase. Therefore, the RNA chaperoning activity of CspA and its cold-inducible homologues might facilitate transcription at low temperatures. Indeed, the nucleic acid melting activity of CspA family proteins is essential both for transcription antitermination and for cold acclimation of cells (27).
X-ray and nuclear magnetic resonance structures of E. coli CspA and Bacillus subtilis CspB reveal a barrel of five antiparallel β-strands with a surface-exposed patch of several aromatic amino acids (8, 23, 31, 32, 34). These amino acids are involved in nucleic acid binding activity (11, 35), and some are essential for nucleic acid chaperoning activity (27, 29).
The common fold of CspA homologues has been named the cold shock domain (CSD). The structure of the CSD is very similar to the S1 domain structure. On the basis of this similarity, the CSD and S1 domains are grouped into the OB (oligomer binding) fold (21). S1, a ribosomal protein, is essential for initiation of translation of several mRNAs, mainly those that contain long 5′ untranslated regions that would be expected to form secondary structures. It has been suggested that S1 may melt nucleic acid secondary structures (15), and S1 is believed to facilitate translation initiation by removing the secondary-structure elements from untranslated regions (38, 40). S1-like domains are also present in polynucleotide phosphorylase, translation initiation factors IF1 and IF2, NusA, RNase E, and other nucleic acid binding proteins (3). The functional similarity between the CSD and S1 domain proteins is suggested by observations that the cold-sensitive phenotype of an E. coli strain harboring a quadruple deletion of csp genes is complemented by expression of the polynucleotide phosphorylase S1 domain (43) and that heterologous expression of E. coli IF1 in B. subtilis suppresses growth defects of a cspB cspC double-deletion mutant (41). IF1 levels are induced two- to threefold upon cold shock (9), and some mutations in the infA gene coding for IF1 result in cold sensitivity (5), further strengthening the possible role of S1 domain proteins in cold acclimation. Together, these observations suggest that there exists cross talk and/or functional overlap between proteins known to be involved in transcription regulation upon cold shock (CspA family proteins) and proteins known to function in translation (S1 domain proteins).
Translational initiation factor IF1 is an essential 71-amino-acid S1 domain protein (7). While several functions, such as increasing the rate of 70S ribosome dissociation and subunit association (10) and involvement in the fidelity of translation initiation through stimulation of IF2 and IF3 activities (30, 42), have been attributed to IF1, its essential function(s) remains to be determined. The observations that the 3-dimensional structure of IF1 closely resembles that of CspA (36) and that IF1 can compensate for the absence of Csp's, proteins whose essential function is melting of nucleic acid secondary structures and transcription antitermination, suggest that IF1 may also possess these functions and that these functions may be essential. Here we test these conjectures directly. We show that IF1 indeed has an RNA chaperone activity and acts as a transcription antiterminator in vivo and in vitro. However, neither of these activities is essential, and they may not be required for the role of IF1 in translation.
MATERIALS AND METHODS
Bacterial strains.
The E. coli wild-type strain JM83 (44) and strain RL211 (17) were used in this study. Bacterial cultures were grown in Luria-Bertani broth (LB). Antibiotics such as ampicillin (50 μg ml−1) kanamycin (Km; 25 μg ml−1), or chloramphenicol (30 μg ml−1) were added as required. The infA deletion strain was constructed as described below.
Disruption of the infA gene.
The E. coli infA gene was replaced with a kanamycin cassette (kan) by recombineering (4, 39, 45). Recombineering is a novel technique of manipulating the DNA sequences in bacterial cells using the Red recombination functions of phage λ (4). It has been shown to be very efficient in generating gene knockouts in E. coli (2, 16, 45). Recombineering was carried out according to the methods described by Thomason et al. (39) with some modifications. Briefly, the recombinogenic E. coli DY330 cells [W3110 ΔlacU169 gal490 pglΔ8 λcI857 Δ(cro-bioA)] were induced for Red functions at an A600 of ∼0.5 and transformed by electroporation with 200 ng of a linear DNA kan cassette replacing the chromosomal infA gene. The cells were collected into 0.9 ml LB, incubated for 2 h at 32°C, and incubated on LB agar plates supplemented with Km (25 μg/ml) for 3 days at 32°C to reveal Kmr recombinants. The kan cassette for infA disruption was made by PCR amplification of the Tn5 kan gene with a Platinum Taq DNA polymerase high-fidelity kit (Invitrogen) using forward primer TATCTTGCCGGTTCAAATTACGGTAGTGATACCCCAGAGGATTAGTTGCCAGCTGGGGCGCCCTCTG and reverse primer ACCTTTTACTCGTTCTTTCTCTTCGC CCATCAGGCGGTAAAACAATCAGAAGAACTCGTCAAGAAG. Each primer contained a 5′ segment of 45-nucleotide (nt) sequence homologous to chromosomal regions flanking infA, followed by a 21-nt priming sequence (underlined) complementary to the 5′ and 3′ regions of the kan gene including its promoter region. Thus, the entire infA open reading frame was precisely replaced with the kan gene fragment containing its promoter region and the complete kan open reading frame.
Recombinant colonies were purified on LB-Km plates at 32°C and resuspended in 20 μl of sterile water, and 1 μl was used as a template for PCR with the following pair of 24-nt primers flanking the infA open reading frame: forward check primer, TTTCGGAGTAATGTGCC GAACCTG; reverse check primer, TCCAAAGTACTTCATACATATCAC. The resulting NB223 strain (DY330 infA<>kan/infA+) was used for further studies.
Site-directed mutagenesis.
The single-amino-acid point mutation (H34R) within the infA coding region was introduced through site-directed mutagenesis using PCR by the methods described by Lerner et al. and Spee et al. (18, 37). The resultant DNA fragment was cloned into the pINIII (12) and pET17b vectors. The plasmids (pINIIIH34RinfA and pET17binfAH32R) were sequenced to confirm the mutant sequences.
In vivo functional analysis of the infA deletion mutant.
The design of the assay for functional analysis of the infA mutant was based on the fact that IF1 is essential for bacterial growth (7). Cells include rare cells with large chromosomal duplications (merodiploids), in which one copy of an essential gene can be disrupted. Recombineering has recently been used to directly identify a number of essential genes in E. coli (2). The assay is simple and is based on the survival of essential gene knockouts only as cells containing duplications (19)—in this case, having both the wild-type allele and the infA<>kan allele, infA<>kan/infA+ (2, 14, 16). These duplications can be visualized on an agarose gel as two PCR fragments amplified from the duplicated regions of the replaced essential gene, where one fragment represents the wild-type infA+ while the other represents the replaced infA<>kan. Normally, such duplication is quite large and cannot be transduced by P1 into another strain. However, if the recipient strain contains a plasmid expressing the wild-type allele of the replaced essential gene, e.g., infA+, then infA<>kan can be transduced into the chromosome, showing one PCR fragment on the agarose gel (2). Thus, any plasmid carrying functionally active infA should allow efficient P1 transduction of infA<>kan, whereas a functionally inactive infA mutant cannot give rise to transductants.
To assess the functional activity of the infA mutant, the W3110 wild-type strain was transformed with pINIII plasmids harboring genes encoding wild-type or mutant [IF1(H34R)] IF1 protein. Phage P1 was grown on NB223 containing the infA<>kan/infA+ duplication and was transduced (20) into W3110 cells transformed with the plasmids described above. Cells were spread on LB-Km plates and grown at 37°C for 2 days to reveal transductants. Transductants were purified on LB-Km plates, and individual colonies were analyzed by PCR for the configuration of the chromosomal infA gene, as described above. If such recombinations occur, the mutant infA gene on a plasmid can support cell growth. The growth characteristics of the infA mutant were analyzed by growing the transductants with the respective plasmid-borne infA mutant on LB plates at 37°C.
Expression and purification of the proteins.
The method for purification of CspE was reported previously (1). IF1 and its mutant IF1(H34R) protein were cloned into the pET17b vector and were purified by Q-Sepharose, SP-Sepharose, and hydroxyapatite column chromatography. In short, IF1 proteins were purified using E. coli BL21(DE3) cells harboring the respective plasmids. Transformed cells were grown to early-logarithmic phase at 37°C in M9-Casamino Acids medium. Expression of IF1 proteins was induced by 1.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Cells were harvested by centrifugation at 4,500 × g for 30 min at 4°C and then suspended in 20 mM Tris-HCl, pH 7.5. A French press was used at 900 lb/in2 for cell lysis, followed by centrifugation at 10,000 × g for 10 min to remove debris. The supernatant was subjected to ultracentrifugation at 40,000 × g for 2 h. Protein was purified by 60% ammonium sulfate precipitation. The pellet was solubilized in 10 mM Tris-HCl, pH 7.5, and then dialyzed against the same buffer. The dialysate was subjected to Q-Sepharose (Pharmacia) fast-flow column chromatography using the same buffer. IF1 protein was eluted by washing the column with the same buffer, and the fractions were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and dialyzed against 10 mM potassium phosphate buffer, pH 7.0. This was followed by SP-Sepharose column chromatography (Pharmacia). The protein was eluted in a 100 to 1,000 mM sodium chloride gradient in the same buffer. The homogeneity of the protein was checked by Coomassie blue and silver staining. The protein concentration was determined by the Bradford method.
In vitro transcription.
In vitro transcription was carried out as described previously (1). A DNA template containing the T7A1 promoter fused to the tR2 terminator was used. The elongation complexes stalled at position +20 (EC20) were prepared in transcription buffer (40 mM KCl, 40 mM Tris-HCl [pH 7.9], 10 mM MgCl2) containing Ni2+-nitriloacetic acid agarose, 20 nM T7 A1 promoter-DNA fragments, 40 nM His-tagged RNA polymerase, and 0.5 mM ApU. Reaction mixtures were incubated at 37°C for 15 min to form the open complexes and were then transferred to room temperature. Then 50 μM ATP, 50 μM GTP, and 2.5 μM [α-32P]CTP (300 Ci/mmol) were added. After a 10-min incubation, the agarose beads were thoroughly washed with transcription buffer. Equal aliquots of purified EC20 were then supplemented with CspE or wild-type or mutant IF1 protein (1.5 μg) and nucleoside triphosphates (250 μM), and the reaction mixtures were incubated at room temperature for 10 min. After the transcription reactions, 20 mM EDTA and 10 mg/ml heparin were added to the reaction mixtures to avoid nonspecific retardation of RNA in the gel. The reactions were terminated by formamide-containing loading buffer. The products were analyzed by urea-PAGE (7 M urea-10% polyacrylamide) followed by autoradiography and phosphorimaging analysis.
KMnO4 probing.
The DNA template Invertbeacon sequence with a 4-nt arm and a 9-bp stem, used for studying the melting mechanism of CspE, has been described previously (28) and is shown in Fig. 3. The DNA was end labeled with [γ-32P]ATP using polynucleotide T4 kinase (New England Biolabs). The KMnO4 probing reactions were carried out as described elsewhere (26, 28). The reaction mixtures (18 μl) containing the labeled DNA template and CspE/IF1 proteins in 10 mM potassium phosphate buffer (pH 7.0) were incubated at room temperature for 5 min and then treated with KMnO4 (1 mM) for 15 s at 37°C. Reactions were terminated by the addition of β-mercaptoethanol to 330 mM, followed by extraction with phenol, precipitation in ethanol, and a 30-min treatment with 10% (vol/vol) piperidine at 90°C. The reaction products were analyzed by urea-6% polyacrylamide gels.
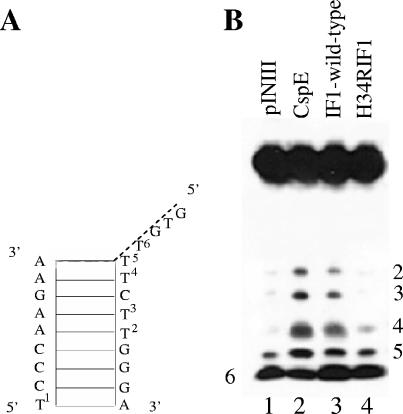
FIG. 3.
Mutation in IF1 affects its nucleic acid melting activity. (A) Schematic drawing of the DNA substrate. The numbering of thymines corresponds to that in panel B. (B) The melting of the stem structure of the DNA substrate (with a 4-nt 5′ overhang and a 9-bp stem) with addition of CspE or IF1 was followed with KMnO4 probing as described in Materials and Methods. Results of a control reaction without added protein (lane 1) and of reactions with added CspE (lane 2), wild-type IF1 (lane 3), or IF1(H34R) (lane 4) are shown. Thymines in the stem are numbered 1 to 5. A thymine residue that lies immediately outside the stem region is numbered 6. The gel is representative of an experiment carried out at least three times.
Nucleic acid binding assay.
IF1 and its mutant were tested for RNA binding by using a radiolabeled RNA template described previously (13). Filter binding assays were carried out as described previously (25). The binding assay was carried out in a 15-μl reaction mixture containing binding buffer (10 mM Tris-HCl buffer [pH 8.0] containing 1 mM EDTA, 10 mM KCl, and 7.4% glycerol) and 50 fmol of RNA. After incubation on ice for 20 min, the reaction mixtures were passed through nitrocellulose filters, which were washed thoroughly to remove unbound RNA. Radioactivity retained on the filter was measured by a liquid scintillation counter. About 1% of the input radioactivity was detected as background in the absence of any protein in the reaction mixture. This background was subtracted from the measured amounts to obtain specific binding values. Similar assays were carried out to test the DNA binding of IF1 and its mutant protein by using the DNA substrate, with a 4-nt 5′ overhang and a 9-bp stem structure (28), used for KMnO4 probing in this study.
RESULTS
Transcription antitermination by IF1 in vivo.
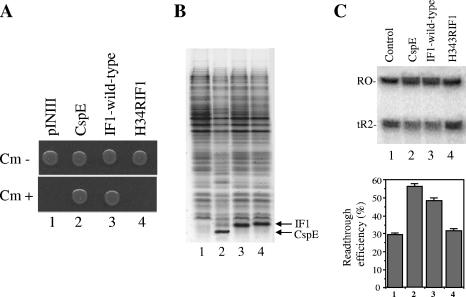
E. coli strain RL211, designed by Landick et al. (17), contains the cat gene preceded by a strong ρ-independent trpL terminator and is therefore sensitive to chloramphenicol. When transcription termination at trpL is reduced, the cat gene is expressed, and cells become resistant to chloramphenicol. We previously used this strain to demonstrate the transcription antitermination activity of CspA family proteins (1, 24, 27, 29). The strain was also used to demonstrate the transcription antitermination activity of the wheat Csp protein WCSP1 by Nakaminami et al. (22). RL211 cells transformed with the infA expression plasmid were spotted onto LB plates with or without chloramphenicol. Cells carrying a cspE expression plasmid were also spotted onto the same plate. Previously, we showed that CspE antiterminates transcription both in vivo and in vitro (1, 27). We used this protein as a positive control throughout this work. As a negative control, cells harboring a pINIII vector plasmid (12) were used. All cells grew equally well in the absence of chloramphenicol, while cells carrying the cspE expression plasmid but not the pINIII plasmid alone grew in the presence of chloramphenicol (Fig. 1A). Cells expressing infA also grew in the presence of chloramphenicol. We therefore conclude that overproduction of IF1 decreases transcription termination at the trpL terminator.
FIG. 1.
Transcription antitermination activity of IF1 in vivo and in vitro. (A) E. coli RL211 cells containing a cat gene cassette positioned downstream of the trpL terminator were transformed with the pINIII vector either alone or containing a cloned cspE or infA gene and were spotted onto LB plates containing 50 μg/ml ampicillin and 1 mM IPTG, with or without 30 μg/ml chloramphenicol (Cm). The results of cell growth after 1 and 2 nights for plates without and with chloramphenicol, respectively, are presented. (B) In vitro transcription assays were carried out as described in Materials and Methods. A DNA template containing the T7A1 promoter fused to the tR2 terminator was used, and CspE or IF1 was included in the reaction mixture. The products were analyzed by urea-PAGE (7 M urea-10% polyacrylamide). The readthrough efficiency was calculated as described in the text, and the mean values calculated from three independent experiments are shown for each lane in the bar graph on the right.
Transcription antitermination by IF1 in vitro.
The results presented above suggest that like CspA-like proteins, IF1 may act as a transcription antiterminator. To test this notion directly, the IF1 protein was overproduced, purified, and tested for its effect on transcription termination in vitro. Stalled E. coli RNA polymerase elongation complexes were prepared on a DNA template containing the T7 A1 promoter followed by the ρ-independent bacteriophage λ terminator tR2 (46). Transcription was resumed by the addition of nucleoside triphosphates in the presence or in the absence of CspE or IF1 (27 μM) (Fig. 1B). In the absence of IF1 or CspE, two transcription products, corresponding to the runoff (Fig. 1B, lane 1, upper band) and the shorter terminated transcripts (Fig. 1B, lane 1, lower band) were observed. Under the conditions used, readthrough efficiency, defined as the ratio of the amount of runoff transcript to the sum of runoff and terminated transcripts, was ∼30%. This value was highly reproducible in several independent experiments. Amounts of transcripts produced were measured for three independent experiments and are presented in Fig. 1B. As expected, CspE decreased transcription termination at tR2 (Fig. 1B, lane 2), and readthrough efficiency increased to 56%. IF1 had a similar effect (48% readthrough [Fig. 1B, lane 3]). We therefore conclude that IF1 can antiterminate transcription in vitro. This activity likely causes the expression of the cat gene observed in the in vivo experiments described above.
The His34 residue of IF1 is essential for transcription antitermination.
The transcription antitermination activity of Csp family proteins requires their ability to melt the secondary structure in nucleic acids (27). Our previous analysis showed that three residues of CspE—Phe17, Phe30, and His32—are involved in nucleic acid melting, most likely though stacking interactions with nucleic acid bases (27, 29). The His32 residue is conserved in IF1 (His34). We created a point substitution of this IF1 residue with arginine. In CspE, the H32R substitution abolishes the nucleic acid melting, transcription antitermination, and cold acclimation functions without affecting structural stability or the ability to interact with single-stranded nucleic acids (27).
We first tested the effect of the H34R substitution in IF1 on transcription antitermination activity in vivo using the RL211 cells described above. As can be seen from Fig. 2A, cells expressing mutant infA grew normally in the absence of chloramphenicol but did not grow in its presence. Cells expressing cspE or wild-type infA grew in the presence of chloramphenicol, as expected. The difference was not due to decreased levels of mutant IF1 production, as revealed by the SDS gel analysis shown in Fig. 2B. The result thus suggests that His34 is essential for the transcription antitermination activity of IF1.
FIG. 2.
Mutation in IF1 affects its transcription antitermination activity in vivo and in vitro. (A) The in vivo transcription antitermination assay using the RL211 cells was carried out as for the experiment for which results are presented in Fig. 1A. Cells were transformed with the pINIII vector either alone or carrying cspE, wild-type infA, or infA(H34R). Results of cell growth after 1 night (plates without chloramphenicol) and 2 nights (plates with chloramphenicol) are presented. Cm, chloramphenicol. (B) SDS-PAGE analysis of induction patterns of CspE, IF1, and its mutant. Lane 1, cells with pINIII vector alone; lane 2, pINIIIcspE; lane 3, pINIIIInfA; lane 4, pINIIIH34RinfA. Bands corresponding to overexpressed CspE or IF1 are indicated. (C) The in vitro transcription antitermination assay was carried out, using purified CspE or wild-type or mutant IF1, as described for the experiment for which results are presented in Fig. 1B. The products were analyzed by urea-PAGE (7 M urea-10% polyacrylamide). RO and tR2 indicate the runoff and tR2-terminated transcripts, respectively. Mean values calculated from three independent experiments are shown in the bar graph below the gel.
We confirmed this result in vitro using purified mutant IF1 protein. The transcription assay was carried out as described above using CspE and wild-type IF1 as controls. As can be seen from Fig. 2C, CspE (lane 2) and wild-type IF1 (lane 3) increased readthrough efficiency to 56% and 48%, respectively; however, for reaction mixtures containing the mutant IF1 protein (lane 4), the readthrough efficiency was ∼32%, very close to the 29% readthrough observed in the absence of added protein. This result is consistent with the in vivo result and shows that the H34R substitution abolishes the transcription antitermination function of IF1.
Nucleic acid melting activity of IF1.
We conducted further testing to determine if the loss of the transcription antitermination activity of IF1 is due to the loss of its nucleic acid melting activity. The melting of a nucleic acid substrate containing secondary structures can be followed by the appearance of KMnO4-sensitive thymines in regions where the double-stranded nucleic acid structure is melted. The DNA melting substrate used in this study has a 4-nt single-stranded 5′ overhang and a 9-bp double-stranded stem, as shown in Fig. 3A (see also reference 28). The stem contains three thymine residues that are not accessible to KMnO4 (Fig. 3B, lane 1). In the presence of either CspE or IF1, the three thymines become KMnO4 sensitive (Fig. 3B, lanes 2 and 3, respectively), indicating that the double-stranded stem is melted. In contrast, no KMnO4-sensitive bands are detected for reactions with added IF1(H34R) (Fig. 3B, lane 4), implying that the stem region remains double stranded in the presence of this protein. Thus, a substitution in IF1 that abolishes its transcription antitermination activity also abolishes IF1-induced nucleic acid melting. The result strongly suggests that, like Csp's, IF1 antiterminates transcription by disrupting the formation of a termination hairpin in the nascent RNA.
The H34R substitution does not affect the nucleic acid binding activity of IF1.
The result presented above could be a trivial consequence of the lack of nucleic acid binding by the IF1(H34R) mutant. The nucleic acid binding activity of the mutant protein was examined in filter binding assays using radioactively labeled RNA or DNA substrates as described in Materials and Methods. The IF1(H34R) protein bound both RNA and DNA substrates as efficiently as the wild-type protein (data not shown). Thus, the nucleic acid binding activity of IF1 is not affected by the H34R substitution.
The transcription antitermination and nucleic acid melting activities of IF1 are not essential for the cell.
Our results establish that IF1 exhibits activities similar to those of CspE, which may explain the previously reported observation that overexpression of IF1 complements certain defects observed in csp mutants. In order to test if these activities of IF1 are essential for cell growth, we prepared E. coli cells harboring a disruption of the chromosomal copy of the infA gene that codes for IF1 by replacing it with a kanamycin cassette. The infA gene is essential, and the cells carrying a chromosomal infA<>kan replacement are viable only if there is an additional wild-type copy of infA either in the form of a chromosomal duplication, infA<>kan/infA+, or expressed in trans from a complementing plasmid. Likewise, the infA knockout is also viable if complemented by a functionally active infA mutant. The pINIIIH34RinfA plasmid (see Materials and Methods for details), as well as the wild-type infA plasmid, complemented the growth of infA knockout cells (Fig. 4). From this experiment, we conclude that the H34R substitution does not affect the function of IF1 that is essential for supporting cell growth. It therefore follows that nucleic acid melting by IF1, and as a consequence its ability to antiterminate transcription, are not essential for the cell. Conversely, the essential function of IF1 (presumably in translation) is not related to nucleic acid melting. Thus, these two activities of IF1 are independent.
FIG. 4.
Complementation of infA deletion cells by wild-type and mutant IF1 protein. The P1 phage was grown on NB223 containing the infA<>kan/infA+ duplication and transduced into W3110 cells transformed with either plasmid pINIII alone, pINIIIinfA, or pINIIIH34RinfA. Cells were plated onto LB plates containing kanamycin and were incubated at 37°C for 2 days.
The data presented in Fig. 4 were obtained at 37°C; an identical result was also obtained at 15°C, a temperature at which cells undergo cold shock (data not shown). Since earlier data obtained with B. subtilis showed that overproduction of IF1 can compensate for the growth defect of a double csp mutant, we wondered if overproduction of E. coli IF1 might have a similar effect on a cold-sensitive E. coli quadruple csp deletion mutant. However, even overproduction of the wild-type, antitermination-capable IF1 did not suppress the cold sensitivity of the quadruple deletion strain (data not shown). The lack of suppression was not related to the lack of overproduction of IF1; therefore, IF1 and Csp's may have different specificities for the transcription terminators on which they act, at least in E. coli. Alternatively, products of promoter-distal genes whose transcription is increased due to IF1 transcription antitermination activity may somehow interfere with the cold shock response. These possibilities are currently being tested using DNA microarray technology.
DISCUSSION
Proteins belonging to the S1 family and the CSD family share a common fold and thus are included in the OB fold family (21). Recent observations suggest that some functions of the S1 domain and CSD proteins might overlap (41, 43). The goal of this work was to test if IF1 can function as a transcription antiterminator similarly to Csp proteins. Our results clearly demonstrate that IF1 indeed can melt secondary structures in nucleic acids and that this activity leads to transcription antitermination, both in vivo and in vitro. Our observation that IF1 is an RNA chaperone is consistent with a previous report which showed that IF1 alters the structures of various oligonucleotides and disrupts nucleic acid interactions in vitro (33). During the preparation of this article, an interesting report by Croitoru et al. was published (6). The authors showed that IF1 activates splicing of the group I intron of the thymidylate synthase gene from phage T4 and suggested that this activity requires the RNA chaperoning activity of IF1. None of the IF1 mutants obtained in that study were deficient in the RNA chaperoning activity, and thus these mutants did not allow assessment of the relevance of this activity to the presumably essential function of IF1 in translation.
The mechanism of the IF1 melting activity must be similar to that of CspA and its homologues, since substitution of a structurally homologous His residue abolishes the activity in both classes of proteins. This result opens the possibility that other S1 domain proteins may also affect transcription elongation and termination by RNA polymerase inside the cell. Indeed, Sukhodolets et al. recently showed that the prototypical S1 domain protein, ribosomal protein S1, promotes transcriptional cycling by enhancing the release of the transcript from RNA polymerase. However, these authors were not able to detect S1-dependent unwinding of DNA-RNA duplexes by S1 and suggested a passive role of S1 in stimulating transcription (38).
It is generally believed that the essential function of IF1 is that of the translation initiation factor. Our results suggest that the RNA chaperoning activity of IF1 is not an essential function, since the nucleic acid melting/transcription antitermination activity of IF1 is not essential, at least under the laboratory growth conditions tested here. Overexpression of E. coli IF1 complements the growth defect of a B. subtilis csp double mutant (41). Since nucleic acid melting is an essential function of bacterial Csp's, this result suggests that the nucleic acid melting function of IF1 may indeed be useful to the cell under certain conditions. In addition to establishing the activity of IF1 in modulating transcription, in the present study we were also able to dissect the two functions of IF1 with the help of a specific transcription antitermination-deficient mutant. The results suggest that although the primary role of IF1 is in translation, it can, by virtue of its structural similarity to the CSD, carry out some of the critical functions of CspA homologues. It is interesting that bacteria such as E. coli have multiple CspA homologues with overlapping functions and, in addition, produce proteins that contain domains similar to the CSD and can carry out some of the presumably critical functions of CspA homologues. This suggests that there may exist cross talk and/or functional overlap between the Csp family proteins, known to be involved in transcription regulation upon cold shock, and S1 domain proteins, known to function in translation. It is interesting that IF1 is moderately induced by cold shock (9) and that certain IF1 mutants confer cold sensitivity on cells (5). One can also contemplate that proteins such as IF1 or other S1 domain proteins, while performing essential functions unrelated to their nucleic acid melting activity under normal growth conditions, can carry out other functions such as transcription antitermination under conditions of stress.
Acknowledgments
We thank Nina Bubunenko for help in the preparation of DNA oligonucleotides.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (under contract NO1-CO-12400), and by a Trans-NIH/FDA Intramural Biodefense Program grant from NIAID (to D.L.C.). This project was funded by NIH grant GM59295 to K.S.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bubunenko, M., T. Baker, and D. L. Court. 2007. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 189:2844-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bycroft, M., T. J. Hubbard, M. Proctor, S. M. Freund, and A. G. Murzin. 1997. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell 88:235-242. [DOI] [PubMed] [Google Scholar]
- 4.Court, D. L., J. A. Sawitzke, and L. C. Thomason. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36:361-388. [DOI] [PubMed] [Google Scholar]
- 5.Croitoru, V., M. Bucheli-Witschel, P. Hagg, F. Abdulkarim, and L. A. Isaksson. 2004. Generation and characterization of functional mutants in the translation initiation factor IF1 of Escherichia coli. Eur. J. Biochem. 271:534-544. [DOI] [PubMed] [Google Scholar]
- 6.Croitoru, V., K. Semrad, S. Prenninger, L. Rajkowitsch, M. Vejen, B. S. Laursen, H. U. Sperling-Petersen, and L. A. Isaksson. 2006. RNA chaperone activity of translation initiation factor IF1. Biochimie 88:1875-1882. [DOI] [PubMed] [Google Scholar]
- 7.Cummings, H. S., and J. W. Hershey. 1994. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, W., R. Tejero, D. E. Zimmerman, M. Inouye, and G. T. Montelione. 1998. Solution NMR structure and backbone dynamics of the major cold-shock protein (CspA) from Escherichia coli: evidence for conformational dynamics in the single-stranded RNA-binding site. Biochemistry 37:10881-10896. [DOI] [PubMed] [Google Scholar]
- 9.Giuliodori, A. M., A. Brandi, C. O. Gualerzi, and C. L. Pon. 2004. Preferential translation of cold-shock mRNAs during cold adaptation. RNA 10:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunberg-Manago, M., P. Dessen, D. Pantaloni, T. Godefroy-Colburn, A. D. Wolfe, and J. Dondon. 1975. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol. 94:461-478. [DOI] [PubMed] [Google Scholar]
- 11.Hillier, B. J., H. M. Rodriguez, and L. M. Gregoret. 1998. Coupling protein stability and protein function in Escherichia coli CspA. Fold Des. 3:87-93. [DOI] [PubMed] [Google Scholar]
- 12.Inouye, M. 1983. Multipurpose expression cloning vehicles in Escherichia coli. Academic Press, New York, NY.
- 13.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 14.Knowlton, J. R., M. Bubunenko, M. Andrykovitch, W. Guo, K. M. Routzahn, D. S. Waugh, D. L. Court, and X. Ji. 2003. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry 42:2275-2281. [DOI] [PubMed] [Google Scholar]
- 15.Kolb, A., J. M. Hermoso, J. O. Thomas, and W. Szer. 1977. Nucleic acid helix-unwinding properties of ribosomal protein S1 and the role of S1 in mRNA binding to ribosomes. Proc. Natl. Acad. Sci. USA 74:2379-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korepanov, A. P., G. M. Gongadze, M. B. Garber, D. L. Court, and M. G. Bubunenko. 2007. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J. Mol. Biol. 366:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landick, R., J. Stewart, and D. N. Lee. 1990. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 4:1623-1636. [DOI] [PubMed] [Google Scholar]
- 18.Lerner, C. G., T. Kobayashi, and M. Inouye. 1990. Isolation of subtilisin pro-sequence mutations that affect formation of active protease by localized random polymerase chain reaction mutagenesis. J. Biol. Chem. 265:20085-20086. [PubMed] [Google Scholar]
- 19.Lupski, J. R., J. R. Roth, and G. M. Weinstock. 1996. Chromosomal duplications in bacteria, fruit flies, and humans. Am. J. Hum Genet. 58:21-27. [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 21.Murzin, A. G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 12:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakaminami, K., D. T. Karlson, and R. Imai. 2006. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. USA 103:10122-10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newkirk, K., W. Feng, W. Jiang, R. Tejero, S. D. Emerson, M. Inouye, and G. T. Montelione. 1994. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. USA 91:5114-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phadtare, S., J. Hwang, K. Severinov, and M. Inouye. 2003. CspB and CspL, thermostable cold-shock proteins from Thermotoga maritima. Genes Cells 8:801-810. [DOI] [PubMed] [Google Scholar]
- 25.Phadtare, S., and M. Inouye. 1999. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 33:1004-1014. [DOI] [PubMed] [Google Scholar]
- 26.Phadtare, S., M. Inouye, and K. Severinov. 2004. The mechanism of nucleic acid melting by a CspA family protein. J. Mol. Biol. 337:147-155. [DOI] [PubMed] [Google Scholar]
- 27.Phadtare, S., M. Inouye, and K. Severinov. 2002. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277:7239-7245. [DOI] [PubMed] [Google Scholar]
- 28.Phadtare, S., and K. Severinov. 2005. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 33:5583-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phadtare, S., S. Tyagi, M. Inouye, and K. Severinov. 2002. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277:46706-46711. [DOI] [PubMed] [Google Scholar]
- 30.Pon, C. L., and C. O. Gualerzi. 1984. Mechanism of protein biosynthesis in prokaryotic cells. Effect of initiation factor IF1 on the initial rate of 30 S initiation complex formation. FEBS Lett. 175:203-207. [DOI] [PubMed] [Google Scholar]
- 31.Schindelin, H., W. Jiang, M. Inouye, and U. Heinemann. 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin, H., M. A. Marahiel, and U. Heinemann. 1993. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 364:164-168. [DOI] [PubMed] [Google Scholar]
- 33.Schleich, T., G. L. Verwolf, and K. Twombly. 1980. A circular dichroism study of Escherichia coli initiation factor-1 binding to polynucleotides. Biochim. Biophys. Acta 609:313-320. [DOI] [PubMed] [Google Scholar]
- 34.Schnuchel, A., R. Wiltscheck, M. Czisch, M. Herrler, G. Willimsky, P. Graumann, M. A. Marahiel, and T. A. Holak. 1993. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature 364:169-171. [DOI] [PubMed] [Google Scholar]
- 35.Schroder, K., P. Graumann, A. Schnuchel, T. A. Holak, and M. A. Marahiel. 1995. Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16:699-708. [DOI] [PubMed] [Google Scholar]
- 36.Sette, M., P. van Tilborg, R. Spurio, R. Kaptein, M. Paci, C. O. Gualerzi, and R. Boelens. 1997. The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spee, J. H., W. M. de Vos, and O. P. Kuipers. 1993. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21:777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukhodolets, M. V., S. Garges, and S. Adhya. 2006. Ribosomal protein S1 promotes transcriptional cycling. RNA 12:1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomason, L. C., M. Bubunenko, N. Costantino, H. R. Wilson, A. Oppenheim, S. Datta, and D. L. Court. 2005. Recombineering: genetic engineering in bacteria using homologous recombination. Wiley, New York, NY. [DOI] [PubMed]
- 40.Tzareva, N. V., V. I. Makhno, and I. V. Boni. 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337:189-194. [DOI] [PubMed] [Google Scholar]
- 41.Weber, M. H., C. L. Beckering, and M. A. Marahiel. 2001. Complementation of cold shock proteins by translation initiation factor IF1 in vivo. J. Bacteriol. 183:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wintermeyer, W., and C. Gualerzi. 1983. Effect of Escherichia coli initiation factors on the kinetics of N-AcPhe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry 22:690-694. [DOI] [PubMed] [Google Scholar]
- 43.Xia, B., H. Ke, and M. Inouye. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179-188. [DOI] [PubMed] [Google Scholar]
- 44.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 45.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zakharova, N., I. Bass, E. Arsenieva, V. Nikiforov, and K. Severinov. 1998. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta subunit inhibit transcript cleavage and transcript elongation. J. Biol. Chem. 273:24912-24920. [DOI] [PubMed] [Google Scholar]