Abstract
“Candidatus Chlorothrix halophila” is a recently described halophilic, filamentous, anoxygenic photoautotroph (J. A. Klappenbach and B. K. Pierson, Arch. Microbiol. 181:17-25, 2004) that was enriched from the hypersaline microbial mats at Guerrero Negro, Mexico. Analysis of the photosynthetic apparatus by negative staining, spectroscopy, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated that the photosynthetic apparatus in this organism has similarities to the photosynthetic apparatus in both the Chloroflexi and Chlorobi phyla of green photosynthetic bacteria. The chlorosomes were found to be ellipsoidal and of various sizes, characteristics that are comparable to characteristics of chlorosomes in other species of green photosynthetic bacteria. The absorption spectrum of whole cells was dominated by the chlorosome bacteriochlorophyll c (BChl c) peak at 759 nm, with fluorescence emission at 760 nm. A second fluorescence emission band was observed at 870 nm and was tentatively attributed to a membrane-bound antenna complex. Fluorescence emission spectra obtained at 77 K revealed another complex that fluoresced at 820 nm, which probably resulted from the chlorosome baseplate complex. All of these results suggest that BChl c is present in the chlorosomes of “Ca. Chlorothrix halophila,” that BChl a is present in the baseplate, and that there is a membrane-bound antenna complex. Analysis of the proteins in the chlorosomes revealed an ∼6-kDa band, which was found to be related to the BChl c binding protein CsmA found in other green bacteria. Overall, the absorbance and fluorescence spectra of “Ca. Chlorothrix halophila” revealed an interesting mixture of photosynthetic characteristics that seemed to have properties similar to properties of both phyla of green bacteria when they were compared to the photosynthetic characteristics of Chlorobium tepidum and Chloroflexus aurantiacus.
Photosynthesis is a fundamental biological process that converts light energy into chemical energy. The organic products of photosynthesis are used by nonphotosynthesizing organisms for energy. Photosynthetic products include food products and fossil fuels, the legacy of ancient photosynthesis. There are different types of photosynthesis, including oxygenic photosynthesis, which occurs in cyanobacteria, algae, and plants, and anoxygenic photosynthesis, which occurs in purple bacteria, two groups of green bacteria, and heliobacteria (3).
Green photosynthetic bacteria can perform photosynthesis at the lowest light intensities of all photosynthetic organisms, even at light intensities that occur at the bottom of the sea near hydrothermal vents (1). This feat is accomplished with pigment-packed antenna systems called chlorosomes, which capture photons and transfer their energy to other pigment-protein antenna systems and then to reaction centers where photochemistry takes place (4).
In general, the architecture of chlorosomes is similar in all species of green photosynthetic bacteria. These structures are ellipsoidal, between 70 and 260 nm long, between 40 and 100 nm wide, and between 10 and 60 nm thick (31, 32). The interior of a chlorosome contains approximately 200,000 molecules of bacteriochlorophyll c (BChl c), BChl d, or BChl e packaged as oligomers with some BChl a and carotenoids and a small number of proteins (4). This array of pigments is thought to be surrounded by a lipid monolayer envelope (31). In the cell, the chlorosome attaches to the inner leaflet of the cytoplasmic membrane, where at the site of attachment a proteinaceous baseplate sits on the chlorosome. This baseplate contains the CsmA baseplate protein, which binds BChl a, as well as carotenoids (22). Antenna complexes and reaction centers are stored in the cytoplasmic membrane next to the chlorosome.
There are two phyla of green photosynthetic bacteria: the Chlorobi (14) and the Chloroflexi (13). The Chlorobi are green sulfur bacteria, and the Chloroflexi are filamentous anoxygenic phototrophs, which were previously called the green nonsulfur bacteria (24). While both groups contain chlorosomes, the compositions of their pigment-protein antenna systems and reaction centers differ. The Chlorobi contain an Fe-S type of reaction center similar to photosystem I (12) and a pigment-protein antenna complex called the BChl a protein or the Fenna-Matthews-Olson (FMO) protein (9, 20). The Chloroflexi contain a pheophytin-quinone type of reaction center similar to the reaction centers present in photosystem II and the purple phototrophic bacteria (11). The Chloroflexi also have a pigment-protein antenna complex called the B808-866 antenna protein, which is related to the LH1 complex that occurs in purple bacteria (41, 42). In all species of green photosynthetic bacteria, the energy from absorbed photons is transferred down an energy gradient from the main chlorosome body to the chlorosome baseplate before it moves to the membrane-bound pigment-protein antenna complexes and then the reaction center.
“Candidatus Chlorothrix halophila” is a hypersaline-tolerant photosynthetic bacterium that was obtained from a salt evaporation pond in Guerrero Negro, Mexico, and has since been maintained in laboratory culture (19). This organism was provisionally named pending formal microbiological description. Initial 16S rRNA gene sequence investigations of the phylogeny of “Ca. Chlorothrix halophila” placed this organism in the phylum Chloroflexi (19). Preliminary physiological studies indicated that it is a photoautotroph with an unknown method of carbon fixation and that it requires sulfide for a strongly reducing environment, which is unusual in this phylum. The nature of the photosynthetic apparatus has remained enigmatic as initial investigations suggested that no BChl a was present in the cells and there was no evidence of a reaction center. Recently, the presence of a BChl a variant has been established (23). In this work we characterized the absorbance and fluorescence emission characteristics of whole cells and isolated chlorosomes of “Ca. Chlorothrix halophila” and compared these characteristics to characteristics of representatives of the phyla Chloroflexi and Chlorobi, Chloroflexus aurantiacus and Chlorobium tepidum. The photosynthetic apparatus of “Ca. Chlorothrix halophila” has a combination of features that are present in both phyla of green bacteria; the chlorosome peak is similar to that of C. tepidum, and the likely antenna complex is comparable to the B808-866 antenna in C. aurantiacus.
MATERIALS AND METHODS
Cell culture.
Enriched cultures of “Ca. Chlorothrix halophila,” primarily containing “Ca. Chlorothrix halophila” cells along with cells of a few other unidentified species, were grown in MCLO medium (19) in either 150-ml glass bottles or 75-cm2 culture flasks (BD Biosciences, Bedford, MA). Approximately 5 mM sodium sulfide was added to the medium, and a sand substrate was required. The culture containers were filled to the brim and sealed to maintain anaerobic conditions, and they were also kept still to recreate the motionless environment of the hypersaline pond from which the organism was isolated. The temperature was maintained at 35 to 38°C by using a 60-W soft white incandescent bulb, which was also the light source. The culture flasks were placed at a distance from the light source so that they received a light intensity of 20 μmol photons m−2 s−1. The cells grew within and above the sandy substrate, making the light intensity that the cells received difficult to measure. The other species in the cultures appeared to be prokaryotic organisms that were nonphotosynthetic, which was determined by fluorescent microscopy of autofluorescence in culture samples.
Chloroflexus aurantiacus J-10fl cells were grown in medium D (25) in 1-liter bottles at 55°C under incandescent bulbs that provided a light intensity of 50 μmol photons m−2 s−1. Chlorobium tepidum cells were grown in a medium described by Wahlund et al. (38) at 42°C in 150-ml bottles under incandescent lights that provided a light intensity of 50 μmol photons m−2 s−1.
Isolation of chlorosomes.
Pelleted cells of “Ca. Chlorothrix halophila” were resuspended in chlorosome buffer (15), which contained 50 mM Tris, 10 mM sodium ascorbate, and 2 M sodium thiocyanate (pH 7.5), at a concentration of 1 g cells/ml buffer. The cells in buffer were homogenized briefly and then disrupted three times for 30 s with an ultrasonicator at 30% power. The cells were placed on ice for 30 s between sonication steps. The resulting mixture was centrifuged at 2,000 × g for 3 min to remove any remaining sand and then for 5 min at 3,000 × g to remove whole-cell debris before concentration to a volume of approximately 600 μl using Centricon Plus 100,000-molecular-weight filter units (Millipore, Billerica, MA). To separate the chlorosomes, the concentrated chlorosome-containing mixture was placed on a discontinuous gradient containing 10, 20, and 50% sucrose in chlorosome buffer and centrifuged for 16 h at 140,000 × g (32,000 rpm; Beckman SW 32). The majority of the chlorosomes were located between the 10 and 20% sucrose layers.
C. aurantiacus and C. tepidum chlorosomes were isolated by using sucrose density gradients and gel filtration (10).
Electron microscopy of chlorosomes.
A drop of purified chlorosomes was placed on a copper slot grid coated with Formvar for 30 s. The liquid was wicked away with filter paper, and then a drop of 0.1% uranyl acetate in double-distilled H2O was placed on the chlorosomes for 30 s before it was removed. The chlorosomes were viewed with a CM12S transmission electron microscope (Philips Electronic Instruments Co., Mahwah, NJ) at 80 kV. Images were captured digitally with a 1,024- by 1,024-pixel charged-coupled device camera (Gatan Inc., Pleasanton, CA) using Digital Micrograph software (Gatan).
Absorption and fluorescence spectroscopy.
All absorbance spectra were obtained using a UV-2501 PC UV-VIS recording spectrophotometer (Shimadzu Scientific Instruments, Inc., Columbia, MD), which was operated with the UVPC software (Shimadzu Scientific Instruments, Inc.). For low-temperature (77 K) absorbance, an optical Dewar cryostat (Oxford Instruments, Oxon, United Kingdom) was used.
Fluorescence excitation and fluorescence emission spectra at both room temperature and 77 K were recorded using a fluorometer (Photon Technology International, Birmingham, NJ) equipped with an avalanche photodiode detector for long-wavelength detection (Advanced Photonics Inc., Camarillo, CA). To obtain the excitation fluorescence spectrum at 825 nm, a 520-nm long-pass filter was placed in the excitation beam path and an 825-nm interference filter was placed in the emission path. To obtain the fluorescence emission spectra, a 440-nm interference filter was placed in the excitation beam path, and a 720 nm long-pass filter was placed in the detector beam path to prevent stray light and scattered excitation light from reaching the detector. Data were recorded using the Felix software (Photon Technology). Samples were prepared by diluting a sample to obtain an optical density of 0.1 to 0.2 at the chlorosome peak in 10 mM Tris buffer (pH 8.0). The wavelength of the fluorescence emission of the chlorosome peak varied depending on the species, the temperature, and whether the preparation was prepared with whole cells or isolated chlorosomes. The relative fluorescence of each spectrum was normalized to the fluorescence at the highest point of the chlorosome peak. No reducing agents, such as sodium dithionite, were used for isolated “Ca. Chlorothrix halophila” chlorosomes.
Protein analysis.
The proteins associated with the chlorosomes of “Ca. Chlorothrix halophila,” C. tepidum, and C. aurantiacus were analyzed by Tricine sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (28). Ten microliters of chlorosomes, with an optical density of 0.7 at the maximum wavelength for chlorosome absorbance for the species, was added directly to 10 μl of 2× sample buffer (200 mM Tris-HCl [pH 6.8], 2% SDS, 40% glycerol, 0.04% Coomassie brilliant blue G-250, 20 μl β-mercaptoethanol). The samples were heated for 6 to 8 min in boiled water and separated on a 10 to 20% gradient Tris-Tricine/peptide polyacrylamide gel (Bio-Rad, Hercules, CA). After electrophoresis, the gels were stained with Coomassie blue R-250.
For protein sequencing, chlorosomes were processed for SDS-PAGE as described above and transferred to a polyvinylidene fluoride membrane using 10 mM N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer (pH 11.0) with 20% methanol. The polyvinylidene fluoride membrane was stained with Ponceau S, and the putative CsmA band was cut out. N-terminal sequencing was performed using a Porton 2090E protein sequencer (Beckman Coulter, Fullerton, CA).
RESULTS
Cell culture.
In culture, “Ca. Chlorothrix halophila” grows relatively slowly compared to Chloroflexus aurantiacus and Chlorobium tepidum. The cells could be subcultured only every 1 to 2 weeks, and additional sulfide was required every 1 to 2 weeks to maintain growing cultures. It was not clear whether the sodium sulfide itself was required for growth of “Ca. Chlorothrix halophila” or whether it was the reduced conditions in the culture environment that were needed. In cultures that were reduced and actively growing, the sand in the culture container was black, and the cells were dark green and grew up the sides of the culture flask. If the sand and culture medium became oxidized, the sand became lighter and the “Ca. Chlorothrix halophila” cells became pale green and did not seem to attach as well to the sand. It was not clear whether these cells were dead or in a less active phase of growth. If enough sodium sulfide was added again to the culture medium, the culture began to grow again, suggesting that some cells were still viable. These results suggested that the reduced conditions obtained by adding sodium sulfide to the culture medium were required for growth of “Ca. Chlorothrix halophila” cells.
Electron microscopy of chlorosomes.
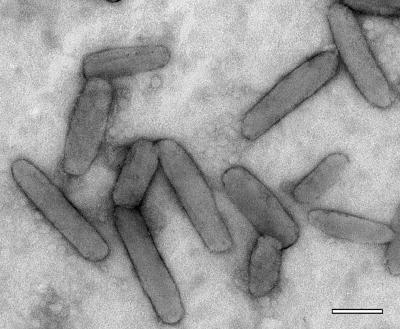
The negatively stained chlorosomes (Fig. 1) were the same shape as chlorosomes observed in other species of green bacteria, such as C. aurantiacus (30) and C. tepidum (31). On average, the chlorosomes were 190 nm long and 52 nm wide, although these dimensions were variable; the length ranged from 60 to 220 nm, and the width ranged from 30 to 70 nm. Fifty chlorosomes were measured.
FIG. 1.
Electron micrograph of negatively stained chlorosomes of “Ca. Chlorothrix halophila.” Scale bar = 100 nm.
Absorption and fluorescence spectroscopy. (i) Whole cells.
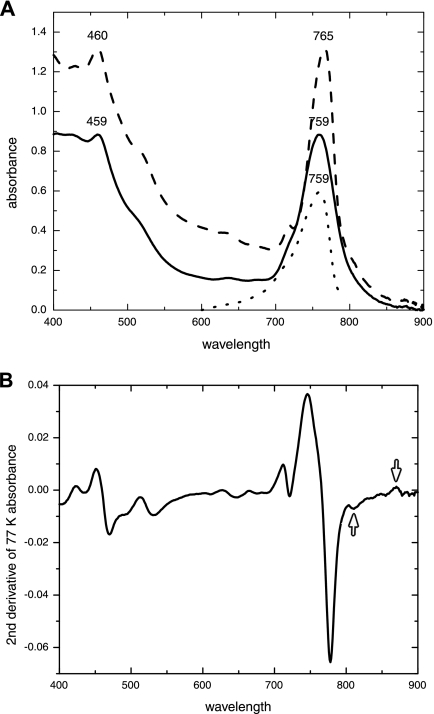
Whole cells of “Ca. Chlorothrix halophila” absorbed at 459, 629, and 759 nm at room temperature and at 460, 630, and 765 nm at 77 K (Fig. 2A). The three peaks in each spectrum reflected the Soret peak, the Qx peak, and the Qy peak, respectively, of the BChl c in the chlorosomes. The BChl c peaks dominated the absorbance spectrum at both room temperature and 77 K, as they do for C. tepidum. C. aurantiacus has a chlorosome peak along with two other peaks at 805 and 873 nm, which are due to BChl a absorbance (2). In “Ca. Chlorothrix halophila,” at 77 K there was evidence of a shoulder around 810 to 825 nm and there was an apparent peak at approximately 870 nm, both of which were probably related to BChl a.
FIG. 2.
(A) Absorbance and fluorescence excitation spectra for whole cells of “Ca. Chlorothrix halophila.” The 459- and 759-nm peaks of the absorbance spectrum at room temperature (solid line) represent the BChl c in the chlorosomes. At 77 K (dashed line), the peaks are at 460 and 765 nm, there is a shoulder at 810 to 825 nm, and there is a small peak at 870 nm. The fluorescence excitation spectrum for whole cells (excitation wavelength, 825 nm) at 77 K (dotted line) has a fluorescence peak at 759 nm. (B) The second-derivative spectrum of the 77 K absorbance spectrum from panel A shows the small peaks at 810 and 870 nm (arrows).
To determine whether these features in the red portion of the spectrum were produced by “Ca. Chlorothrix halophila” or a contaminant organism in the enriched culture, fluorescence excitation at 825 nm was investigated. The results revealed a chlorosome peak at 759 nm (Fig. 2A), clearly indicating that there was energy transfer from BChl c to BChl a. The second derivative of the absorption spectrum at 77 K was calculated, which resulted in detection of small peaks around 810 and 870 nm (Fig. 2B).
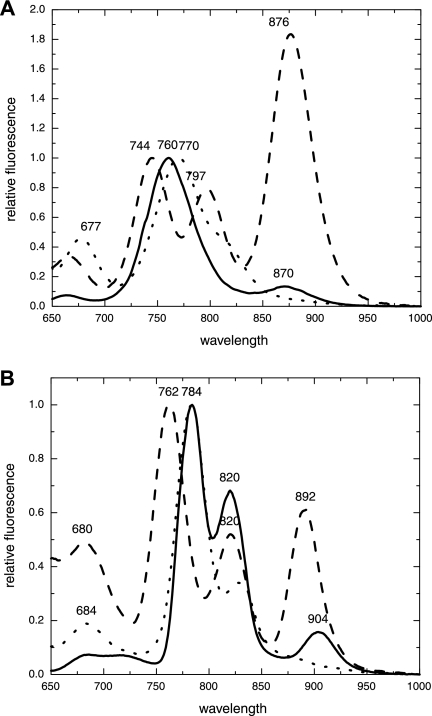
At room temperature, the fluorescence emission of cells of “Ca. Chlorothrix halophila” that were excited at 440 nm produced peaks at 760 and 870 nm (Fig. 3A), indicating that BChl c from chlorosomes and possibly a BChl a-like pigment, respectively, were present. The 870-nm peak was tentatively assigned to an antenna complex that is probably associated with the reaction center. The ratio of the 760-nm peak to the 870-nm peak was 100:13.
FIG. 3.
Fluorescence emission spectra (excitation wavelength, 440 nm) for “Ca. Chlorothrix halophila” (solid line), Chlorobium tepidum (dotted line), and Chloroflexus aurantiacus (dashed line) cells at room temperature (A) and 77 K (B). The chlorosome (BChl c) peaks are normalized. “Ca. Chlorothrix halophila” cells produce a dominant chlorosome peak at 760 nm and a smaller peak at 870 nm at room temperature. At 77 K the chlorosome peak is split in two, with maxima at 784 and 820 nm, and the 870 nm peak is redshifted to 904 nm.
The fluorescence emission spectrum of C. tepidum had a 770-nm chlorosome peak and a shoulder around 820 nm. No similar shoulder was observed for “Ca. Chlorothrix halophila.” The C. aurantiacus spectrum has a chlorosome peak at 744 nm, a 797-nm peak that results from the baseplate of the chlorosomes, and an 876-nm peak that results from the B808-866 nm antenna complex (21). The 870-nm peak of “Ca. Chlorothrix halophila” resembled the C. aurantiacus 876-nm peak. However, the relative fluorescence of the C. aurantiacus 876-nm peak appeared to be far more intense than the relative fluorescence of the 870-nm peak of “Ca. Chlorothrix halophila” after normalization of the spectra at the appropriate chlorosome peaks. No peak equivalent to the 797-nm peak of C. aurantiacus was observed for either “Ca. Chlorothrix halophila” or C. tepidum.
At 77 K for “Ca. Chlorothrix halophila,” the two fluorescence emission peaks were redshifted to 784 and 904 nm, and a third peak was apparent at 820 nm (Fig. 3B). This third peak may have been due to fluorescence from a BChl a-like pigment in the baseplate of the chlorosome. The C. tepidum chlorosome peak was redshifted to fluoresce at 784 nm, the wavelength of the chlorosome peak of “Ca. Chlorothrix halophila.” The C. tepidum shoulder was redshifted to 830 nm at 77 K and became a discrete peak. Whole cells of C. aurantiacus produced fluorescence emission peaks at 762, 820, and 892 nm.
(ii) Isolated chlorosomes.
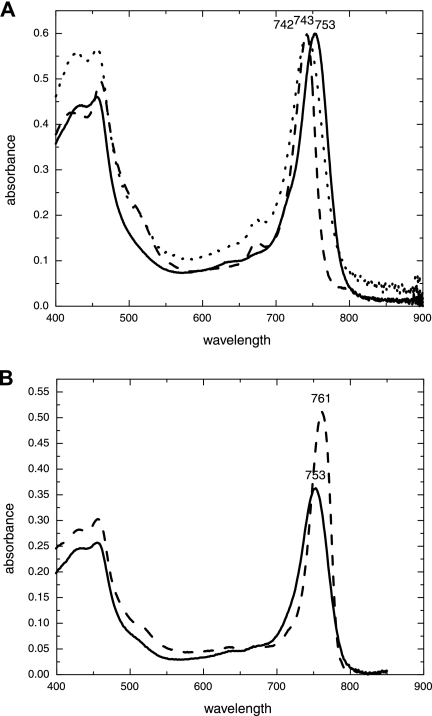
The absorbance spectrum of isolated chlorosomes of “Ca. Chlorothrix halophila” (Fig. 4A) at room temperature had peaks at 460 and 753 nm. The C. tepidum chlorosome peak was at 743 nm, and the C. aurantiacus peak was at 742 nm. No shoulders like those observed for C. aurantiacus were associated with the chlorosome peak of “Ca. Chlorothrix halophila” (2). At 77 K, the 753-nm peak of “Ca. Chlorothrix halophila” was redshifted to 761 nm and was sharper than the room temperature peak (Fig. 4B).
FIG. 4.
(A) Room temperature absorbance of “Ca. Chlorothrix halophila” chlorosomes at 753 nm (solid line), Chlorobium tepidum chlorosomes at 743 nm (dotted line), and Chloroflexus aurantiacus chlorosomes at 742 nm (dashed line). (B) Absorbance of “Ca. Chlorothrix halophila” chlorosomes at room temperature (solid line) and at 77 K (dashed line). The chlorosomes absorb at 753 nm at room temperature, and this absorbance is redshifted by 8 nm at 77 K to 761 nm.
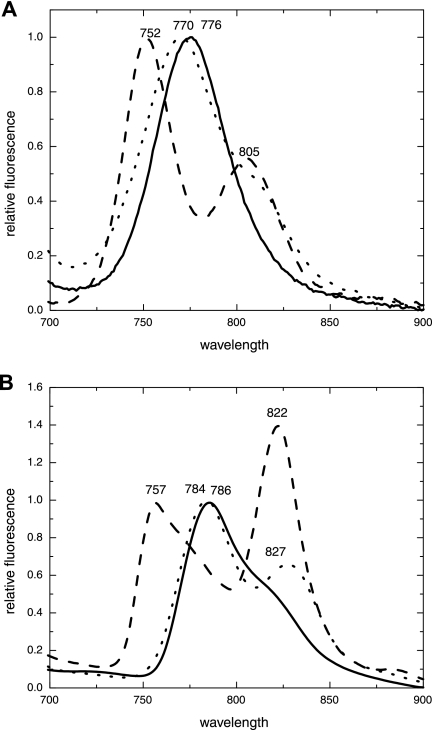
When excited at 440 nm, the “Ca. Chlorothrix halophila” chlorosomes fluoresced at 776 nm at room temperature (Fig. 5A). C. tepidum chlorosomes fluoresced and produced a single peak at 770 nm, while the C. aurantiacus chlorosomes fluoresced and produced a split peak, with the dominant chlorosome peak at 752 nm and a second peak at 805 nm.
FIG. 5.
Fluorescence emission spectra (excitation wavelength, 440 nm) for “Ca. Chlorothrix halophila” (solid line), Chlorobium tepidum (dotted line), and Chloroflexus aurantiacus (dashed line) chlorosomes at room temperature (A) and 77 K (B). The chlorosome (BChl c) peaks are normalized. At room temperature “Ca. Chlorothrix halophila” chlorosomes emit fluorescence at a single wavelength, 776 nm. At 77 K, the chlorosome fluorescence becomes asymmetrical, with emission at 786 nm and a shoulder at around 820 nm.
At 77 K, the “Ca. Chlorothrix halophila” chlorosome fluorescence peak shifted to 786 nm and had a shoulder around 820 nm (Fig. 5B), suggesting that a baseplate is probably associated with the chlorosome. The fluorescence emission spectrum of C. tepidum chlorosomes at 77 K had two peaks with maxima at 784 and 827 nm, while the C. aurantiacus chlorosomes fluoresced at 757 and 822 nm. The intensity of the 822-nm peak was 1.4 times greater than the intensity of the 757-nm peak.
Protein analysis of the chlorosomes.
Isolated chlorosomes of “Ca. Chlorothrix halophila” were electrophoresed on an SDS-PAGE gel (see Fig. S1 in the supplemental material) alongside chlorosomes from C. aurantiacus and C. tepidum. For all three species, there was a band at approximately 6 kDa, which corresponded to the baseplate protein CsmA (7, 27, 37). No other “Ca. Chlorothrix halophila” band appeared to have the same apparent mass as proteins from the other two species.
To determine whether the band at ∼6 kDa was a CsmA-like protein, N-terminal sequencing was performed. This analysis yielded a sequence (Fig. 6) that was clearly related to the C. aurantiacus CsmA sequence (40).
FIG. 6.
Comparison of the first 23 amino acids of the sequence of the putative CsmA protein from “Ca. Chlorothrix halophila” with the known sequences for Chloroflexus aurantiacus and Chlorobium tepidum. N-terminal sequencing did not clearly determine the first amino acid of the sequence. The last four amino acids were not positively identified, nor was residue 17.
DISCUSSION
“Ca. Chlorothrix halophila” is related phylogenetically to the Chloroflexi group of green photosynthetic bacteria based on 16S rRNA gene data (19). However, the photosynthetic apparatus of this organism appears to have characteristics resembling characteristics of the photosynthetic apparatus of both groups of green photosynthetic bacteria. While these characteristics may represent a photosynthetic mechanism that utilizes photosynthetic components from each group, further work needs to be done to determine the type of reaction center that “Ca. Chlorothrix halophila” contains.
The whole-cell absorbance spectrum of “Ca. Chlorothrix halophila” is dominated by the chlorosome peak with a shoulder at approximately 810 to 825 nm and a small peak at 870 nm at 77 K. This is similar to the absorbance spectrum of Chlorobium tepidum, which also has a small shoulder at approximately 810 nm (38). To ensure that the shoulder observed at 77 K for “Ca. Chlorothrix halophila” was not due to a contaminating photosynthetic organism, such as a purple bacterium, a fluorescence excitation spectrum at 825 nm was obtained. The excitation at 825 nm resulted in a clear 759-nm peak, indicating that the BChl c and BChl a pigments were close enough for energy transfer and therefore in the same cell. Thus, the shoulder in the 77 K spectrum was due to “Ca. Chlorothrix halophila” BChl a and not due to a contaminant photosynthetic organism. The presence of the shoulder and the presence of the 870-nm peak were further supported by the second derivative of the 77 K absorbance spectrum, which showed that there were small peaks around 810 and 870 nm.
The chlorosome peak in the whole-cell fluorescence spectrum of “Ca. Chlorothrix halophila” at room temperature was approximately midway between the chlorosome fluorescence of C. tepidum and the chlorosome fluorescence of Chloroflexus aurantiacus. No shoulder like that obtained for C. tepidum chlorosome fluorescence was associated with the “Ca. Chlorothrix halophila” chlorosome fluorescence peak. At 77 K, the “Ca. Chlorothrix halophila” chlorosome peak redshifted to 784 nm, exactly the same maximum wavelength as the C. tepidum chlorosome peak.
While both the absorbance and fluorescence spectra of the “Ca. Chlorothrix halophila” chlorosome peak seem to resemble most closely the absorbance and fluorescence spectra of the chlorosome peak of C. tepidum, the 870-nm fluorescence peak at room temperature resembles more closely the 876-nm peak of C. aurantiacus. This suggests that “Ca. Chlorothrix halophila” may have an antenna complex similar to the B808-866 antenna and a type II reaction center. No evidence of a reaction center in “Ca. Chlorothrix halophila” was obtained in this study. It is likely that the ratio of chlorosomes to reaction centers changes in “Ca. Chlorothrix halophila” with changes in light intensity, which should allow further investigation of the reaction centers. Such a change has been observed in C. aurantiacus, in which a decrease in the light intensity increases the ratio of BChl c to BChl a in the cells (26, 29). Unfortunately, “Ca. Chlorothrix halophila” cells grow both above and below the sandy substrate, making control of the light conditions a challenge. While the greater fluorescence intensity of the C. aurantiacus BChl a 876-nm peak than of the “Ca. Chlorothrix halophila” 870-nm peak may have been due to the BChl c/BChl a ratio caused by light conditions, it may also have been due to differences between the photosynthetic apparatus of the two species. One possibility is that the BChl a content is greater in the antenna complexes of C. aurantiacus than in similar structures in “Ca. Chlorothrix halophila.” Alternatively, it is also possible that the energy transfer from the chlorosomes of C. aurantiacus to the B808-866 antenna complex and the reaction center is less efficient than the energy transfer in “Ca. Chlorothrix halophila.”
At 77 K, the fluorescence spectrum of whole cells of “Ca. Chlorothrix halophila” had three fluorescence emission peaks, at 784, 820, and 904 nm. The 784-nm fluorescence emission peak matched the C. tepidum chlorosome fluorescence emission peak. The 820-nm fluorescence peak was similar to the second C. aurantiacus peak at 820 nm and distinct from the 830-nm fluorescence emission peak of C. tepidum. The redshift of the third peak from 870 to 904 nm was greater than the redshift observed in C. aurantiacus, in which the 876-nm peak shifted to 892 nm. The reason for this difference is not clear, and the difference is not likely to be due to the difference in structure of the BChl a of “Ca. Chlorothrix halophila,” as the tail is modified and the head group is not modified (23).
The chlorosome absorbance spectrum for “Ca. Chlorothrix halophila” has a peak at 753 nm at room temperature, compared to peaks at 742 nm for C. aurantiacus and at 743 nm for C. tepidum. There was no evidence that there was a BChl a-related shoulder in the absorbance spectra of isolated chlorosomes at either room temperature or 77 K, as there was in the absorbance spectra of whole cells. “Ca. Chlorothrix halophila” has two major types of BChl c (23), but this probably does not affect the absorption characteristics of the chlorosomes, and the difference in absorption between the three species is more likely to be due to the wide range of absorbance typical of BChl c.
The room temperature fluorescence emission spectrum of the chlorosomes of “Ca. Chlorothrix halophila” had a single peak at 776 nm. Again, no shoulder like that seen in C. tepidum was present. In C. aurantiacus, the chlorosome fluorescence spectrum had two prominent peaks, at 752 and 805 nm, indicating that BChl c was present in the chlorosome and BChl a was present in the baseplate. At 77 K, the “Ca. Chlorothrix halophila” chlorosome peak had a shoulder at approximately 825 nm, indicating that a baseplate is probably associated with the chlorosomes. The isolated chlorosomes of the other two species have more distinct peaks at 77 K, which suggests that there may be less BChl a in the “Ca. Chlorothrix halophila” baseplate. Alternatively, in “Ca. Chlorothrix halophila” baseplates may not be associated with all chlorosomes.
SDS-PAGE separation of proteins from the chlorosomes of all three species revealed a common, abundant band at ∼6 kDa. Most of the other proteins associated with the chlorosomes from “Ca. Chlorothrix halophila” appeared to have different masses than the chlorosome proteins isolated from C. aurantiacus and C. tepidum. The amino acid sequence of the ∼6-kDa band from “Ca. Chlorothrix halophila” showed that this protein exhibits substantial sequence similarity (65% identity; 15 of 23 residues) to the CsmA protein from C. aurantiacus (40) and a much lower level of similarity to the CsmA sequence from C. tepidum (13% identity; 3 of 23 residues) (8).
The results obtained from absorbance and fluorescence spectra indicate that the chlorosomes of “Ca. Chlorothrix halophila” most closely resemble C. tepidum chlorosomes and that the baseplate and membrane-associated antenna complex are similar to those present in C. aurantiacus. Interestingly, the photosynthetic antenna system of another green photosynthetic bacterium, Oscillochloris trichoides, which also has a combination of different properties of photosynthesis from the Chlorobi and the Chloroflexi, has recently been described (33-36). This species of the Chloroflexi (17) was isolated from a warm hydrogen sulfide spring in southeastern Europe (18); therefore, its habitat and probably its physiology are different than those of the halophilic organism “Ca. Chlorothrix halophila.”
While the specific wavelengths of absorption of O. trichoides (456, 748, and 852 nm) (17, 18) differ from those of “Ca. Chlorothrix halophila,” there are striking similarities between the two organisms. The absorbance spectra of whole cells of both species are dominated by the BChl c chlorosome peak, which resembles the results obtained for Chlorobi species, and a minimal amount of BChl a is present. In O. trichoides, the ratio of BChl c to BChl a has been shown to differ depending on the intensity of the light in which the cells are grown (34). Isolated chlorosomes from O. trichoides also contained only small amounts of BChl a (34), similar to the results obtained for “Ca. Chlorothrix halophila.” A light-harvesting membrane antenna system that absorbed at 805 and 860 nm was also discovered, indicating that this organism has spectral features similar to those of the B808-866 antenna of C. aurantiacus (34). A similar antenna complex seems to be present in “Ca. Chlorothrix halophila.” In both species there is no evidence of the FMO protein.
The fluorescence spectrum of whole cells of O. trichoides is dominated by the BChl c chlorosome peak, and features associated with BChl a are observed at 77 K (34). The chlorosomes of O. trichoides were highly redox sensitive, as observed for species belonging to the Chlorobi (16), and addition of sodium dithionite greatly increased the intensity of fluorescence. “Ca. Chlorothrix halophila” did not show such sensitivity to oxygen, possibly aligning the chlorosomes of this species more closely with the chlorosomes of the Chloroflexi, in which there was only a two- to threefold increase in the intensity of fluorescence when sodium dithionite was added (5, 39).
The similarities of the properties of the photosynthetic apparatus of “Ca. Chlorothrix halophila” and the photosynthetic apparatus of O. trichoides may be due to convergent evolution, where the photosynthetic traits of the two species arose independently due to environmental pressure. It is also possible that chlorosome genes were transferred laterally from the Chlorobi to the two groups in the Chloroflexi in two independent transfer events. Alternatively, “Ca. Chlorothrix halophila” and O. trichoides may represent a single evolutionary group in the Chloroflexi with a photosynthetic antenna system that contains chlorosomes whose spectral properties are similar to the spectral properties of the chlorosomes of Chlorobi and with a membrane-bound antenna system similar to the B808-866 antenna of C. aurantiacus.
Based on the absorbance and fluorescence emission spectra of “Ca. Chlorothrix halophila,” energy transfer in this organism appears to progress from B753 BChl c to ∼B797 BChl a to ∼B870 BChl a, which probably represents the transfer of energy from the chlorosomes to the baseplate to the membrane-bound antenna. The B797 approximation comes from the fluorescence emission peak at 820 nm at 77 K. This peak corresponds to a 820-nm C. aurantiacus peak at 77 K, which is at approximately 797 nm at room temperature. It is also likely that there is an intermediate energy transfer step around 808 nm, like the step observed for both C. tepidum and C. aurantiacus. An isolated sucrose density gradient fraction from “Ca. Chlorothrix halophila” was found to absorb at around 808 nm (results not shown). Consequently, the transfer sequence of “Ca. Chlorothrix halophila” may be B753 BChl c to ∼B797 BChl a to ∼B808 BChl a to ∼B870 BChl a to P8? BChl a reaction center, where P8? is the unknown wavelength, most likely in the >800-nm range, for the reaction center that is probably present in this organism. For comparison, the energy transfer sequence of C. tepidum is B742 BChl c to B794 BChl a to B808 water-soluble BChl a (from FMO) to P840 BChl a reaction center (15), and the energy transfer sequence of C. aurantiacus is B740 BChl c to B792 BChl a to B808 BChl a to B866 BChl a to P870 BChl a reaction center (6).
“Ca. Chlorothrix halophila” is related phylogenetically to the Chloroflexi group of green photosynthetic bacteria according to 16S rRNA gene data (19). However, based on the absorption and fluorescence spectra and the amino acid sequence of the putative CsmA protein, the photosynthetic apparatus of this organism appears to have characteristics resembling characteristics of the photosynthetic apparatus of both groups of green photosynthetic bacteria. Further characterization of this organism is under way.
Supplementary Material
Acknowledgments
This work was supported by DOE grant DE-FG02-04ER15550.
We thank Amanda Cunow for providing the chlorosomes from C. aurantiacus, Heather Matthies for providing the chlorosomes from C. tepidum, Alexander Melkozernov for help with the 77 K absorption spectra, Dan Brune of the Arizona State University Proteomics and Protein Chemistry Lab for N-terminal sequencing, and Yueyong Xin for discussions concerning protein techniques. The electron microscopy was performed at the ASU Life Sciences Electron Microscopy Facility.
Footnotes
Published ahead of print on 16 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Beatty, J. T., J. Overmann, M. T. Lince, A. K. Manske, A. S. Lang, R. E. Blankenship, C. L. Van Dover, T. A. Martinson, and F. G. Plumley. 2005. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc. Natl. Acad. Sci. USA 102:9306-9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betti, J. A., R. E. Blankenship, L. V. Natarajan, L. C. Dickinson, and R. C. Fuller. 1982. Antenna organization and evidence for the function of a new antenna pigment species in the green photosynthetic bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 680:194-201. [Google Scholar]
- 3.Blankenship, R. E. 2002. Molecular mechanisms of photosynthesis. Blackwell, Malden, MA.
- 4.Blankenship, R. E., and K. Matsuura. 2003. Antenna complexes from green photosynthetic bacteria, p. 195-217. In B. R. Green and W. W. Parson (ed.), Light harvesting antennas. Kluwer, Dordrecht, The Netherlands.
- 5.Blankenship, R. E., J. M. Olson, and M. Miller. 1995. Antenna complexes from green photosynthetic bacteria, p. 399-435. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, The Netherlands.
- 6.Brune, D. C., T. Nozawa, and R. E. Blankenship. 1987. Antenna organization in green photosynthetic bacteria. 1. Oligomeric bacteriochlorophyll c as a model for the 740 nm absorbing bacteriochlorophyll c in Chloroflexus aurantiacus chlorosomes. Biochemistry 26:8644-8652. [DOI] [PubMed] [Google Scholar]
- 7.Chung, S., and D. A. Bryant. 1996. Characterization of the csmD and csmE genes from Chlorobium tepidum. The CsmA, CsmC, CsmD, and CsmE proteins are components of the chlorosome envelope. Photosynth. Res. 50:41-59. [DOI] [PubMed] [Google Scholar]
- 8.Chung, S., G. Frank, H. Zuber, and D. A. Bryant. 1994. Genes encoding two chlorosome components from the green sulfur bacteria Chlorobium vibrioforme strain 83271d and Chlorobium tepidum. Photosynth. Res. 41:261-275. [DOI] [PubMed] [Google Scholar]
- 9.Dracheva, S., J. C. Williams, and R. E. Blankenship. 1992. Sequencing of the FMO-protein from Chlorobium tepidum, p. 53-56. In N. Murata (ed.), Research in photosynthesis, vol. 1. Kluwer, Dordrecht, The Netherlands. [Google Scholar]
- 10.Eckhardt, A., R. Brunisholz, G. Frank, and H. Zuber. 1990. Selective solubilization of chlorosome proteins in Chloroflexus aurantiacus. FEBS Lett. 267:199-202. [DOI] [PubMed] [Google Scholar]
- 11.Feick, R. G., J. A. Shiozawa, and A. Ertlmaier. 1995. Biochemical and spectroscopic properties of the reaction center of the green filamentous bacterium Chloroflexus aurantiacus, p. 699-708. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, The Netherlands.
- 12.Feiler, U., and G. Hauska. 1995. The reaction center from green sulfur bacteria, p. 699-708. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, The Netherlands.
- 13.Garrity, G. M., and J. G. Holt. 2001. Phylum BVI. Chloroflexi phy. nov., p. 427-446. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 14.Garrity, G. M., and J. G. Holt. 2001. Phylum BXI. Chlorobi phy. nov., p. 601-623. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 15.Gerola, P. D., and J. M. Olson. 1986. A new bacteriochlorophyll a-protein complex associated with chlorosomes of green sulfur bacteria. Biochim. Biophys. Acta 848:69-76. [DOI] [PubMed] [Google Scholar]
- 16.Karapetyan, N. V., T. Swarthoff, C. P. Rijgersberg, and J. Amesz. 1980. Fluorescence emission spectra of cells and subcellular preparations of a green photosynthetic bacterium. Effects of dithionite on the intensity of the emission bands. Biochim. Biophys. Acta 593:254-260. [DOI] [PubMed] [Google Scholar]
- 17.Keppen, O. I., O. I. Baulina, and E. N. Kondratieva. 1994. Oscillochloris trichoides neotype strain DG-6. Photosynth. Res. 41:29-33. [DOI] [PubMed] [Google Scholar]
- 18.Keppen, O. I., O. I. Baulina, A. M. Lysenko, and E. N. Kondratieva. 1993. New green bacterium belonging to family Chloroflexaceae. Mikrobiologiya 62:267-274. [Google Scholar]
- 19.Klappenbach, J. A., and B. K. Pierson. 2004. Phylogenetic and physiological characterization of a filamentous anoxygenic photoautotrophic bacterium “Candidatus Chlorothrix halophila” gen. nov., sp. nov., recovered from hypersaline microbial mats. Arch. Microbiol. 181:17-25. [DOI] [PubMed] [Google Scholar]
- 20.Matthews, B. W., R. E. Fenna, M. C. Bolognesi, M. F. Schmid, and J. M. Olson. 1979. Structure of a bacteriochlorophyll a-protein from the green photosynthetic bacterium Prosthecochloris aestuarii. J. Mol. Biol. 131:259-285. [DOI] [PubMed] [Google Scholar]
- 21.Mimuro, M., T. Nozawa, N. Tamai, Y. Nishimura, and I. Yamazaki. 1994. Presence and significance of minor antenna components in the energy-transfer sequence of the green photosynthetic bacterium Chloroflexus aurantiacus. FEBS Lett. 340:167-172. [DOI] [PubMed] [Google Scholar]
- 22.Montaño, G. A., H. M. Wu, S. Lin, D. C. Brune, and R. E. Blankenship. 2003. Isolation and characterization of the B798 light-harvesting baseplate from the chlorosomes of Chloroflexus aurantiacus. Biochemistry 42:10246-10251. [DOI] [PubMed] [Google Scholar]
- 23.Olson, T. L., A. M. L. van de Meene, J. N. Francis, B. K. Pierson, and R. E. Blankenship. 2007. Pigment analysis of “Cadidatus Chlorothrix halophila,” a green filamentous anoxygenic phototrophic bacterium. J. Bacteriol. 189:4187-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson, B. K. 2001. Order I. Chloroflexales, p. 427-444. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, NY.
- 25.Pierson, B. K., and R. W. Castenholz. 1974. Phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch. Microbiol. 100:5-24. [DOI] [PubMed] [Google Scholar]
- 26.Pierson, B. K., and R. W. Castenholz. 1974. Studies of pigments and growth in Chloroflexus aurantiacus, a phototropic filamentous bacterium. Arch. Microbiol. 100:283-305. [DOI] [PubMed] [Google Scholar]
- 27.Sakuragi, Y., N. U. Frigaard, K. Shimada, and K. Matsuura. 1999. Association of bacteriochlorophyll a with the CsmA protein in chlorosomes of the photosynthetic green filamentous bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 1413:172-180. [DOI] [PubMed] [Google Scholar]
- 28.Schagger, H., and G. Von Jagow. 1987. Tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt, K., M. Maarzahl, and F. Mayer. 1980. Development and pigmentation of chlorosomes in Chloroflexus aurantiacus Ok-70-fl. Arch. Microbiol. 127:87-97. [Google Scholar]
- 30.Sprague, S. G., and A. R. Varga. 1986. Membrane architecture of anoxygenic photosynthetic bacteria, p. 603-619. In L. A. Staehelin and C. J. Arntzen (ed.), Encyclopedia of plant physiology, photosynthesis. III. Photosynthetic membranes and light harvesting systems, vol. 19. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 31.Staehelin, L. A., J. R. Golecki, and G. Drews. 1980. Supramolecular organization of chlorosomes (Chlorobium vesicles) and of their membrane attachment sites in Chlorobium limicola. Biochim. Biophys. Acta 589:30-45. [DOI] [PubMed] [Google Scholar]
- 32.Stolz, J. F. (ed.). 1991. Structure of phototrophic prokaryotes. CRC Press, Boca Raton, FL.
- 33.Taisova, A. S., O. I. Keppen, and Z. G. Fetisova. 2004. A study of the content of pigments in the light-harvesting antenna of the green bacterium from the new family Oscillochloridaceae. Biofizika 49:1069-1074. [PubMed] [Google Scholar]
- 34.Taisova, A. S., O. I. Keppen, E. P. Lukashev, A. M. Arutyunyan, and Z. G. Fetisova. 2002. Study of the chlorosomal antenna of the green mesophilic filamentous bacterium Oscillochloris trichoides. Photosynth. Res. 74:73-85. [DOI] [PubMed] [Google Scholar]
- 35.Taisova, A. S., O. I. Keppen, A. A. Novikov, M. G. Naumova, and Z. G. Fetisova. 2006. Some factors controlling the biosynthesis of chlorosome antenna bacteriochlorophylls in green filamentous anoxygenic phototrophic bacteria of the family Oscillochloridaceae. Microbiology 75:129-135. [PubMed] [Google Scholar]
- 36.Taisova, A. S., E. P. Lukashev, O. I. Keppen, and Z. G. Fetisova. 2005. A comparative study of the fluorescence properties of the chlorosomal antenna of the green bacterium from the family Oscillochloridaceae and the members from two other families of green bacteria. Biofizika 50:271-276. [PubMed] [Google Scholar]
- 37.Theroux, S. J., T. E. Redlinger, R. C. Fuller, and S. J. Robinson. 1990. Gene encoding the 5.7-kilodalton chlorosome protein of Chloroflexus aurantiacus: regulated message levels and a predicted carboxy-terminal protein extension. J. Bacteriol. 172:4497-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch. Microbiol. 156:81-90. [Google Scholar]
- 39.Wang, J., D. C. Brune, and R. E. Blankenship. 1990. Effects of oxidants and reductants on the efficiency of excitation transfer in green photosynthetic bacteria. Biochim. Biophys. Acta 1015:457-463. [DOI] [PubMed] [Google Scholar]
- 40.Wechsler, T., F. Suter, R. C. Fuller, and H. Zuber. 1985. The complete amino acid sequence of the bacteriochlorophyll-c binding polypeptide from chlorosomes of the green photosynthetic bacterium Chloroflexus aurantiacus. FEBS Lett. 181:173-178. [Google Scholar]
- 41.Wechsler, T. D., R. A. Brunisholz, G. Frank, F. Suter, and H. Zuber. 1987. The complete amino acid sequence of the antenna polypeptide B806-866-beta from the cytoplasmic membrane of the green bacterium Chloroflexus aurantiacus. FEBS Lett. 210:189-194. [Google Scholar]
- 42.Wechsler, T. D., R. A. Brunisholz, G. Frank, and H. Zuber. 1991. Isolation and protein chemical characterization of the B806-866 antenna complex of the green thermophilic bacterium Chloroflexus aurantiacus. J. Photochem. Photobiol. B 8:189-197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.