Abstract
Microcin J25 (MccJ25) uptake by Escherichia coli requires the outer membrane receptor FhuA and the inner membrane proteins TonB, ExbD, ExbB, and SbmA. MccJ25 appears to have two intracellular targets: (i) RNA polymerase (RNAP), which has been described in E. coli and Salmonella enterica serovars, and (ii) the respiratory chain, reported only in S. enterica serovars. In the current study, it is shown that the observed difference between the actions of microcin on the respiratory chain in E. coli and S. enterica is due to the relatively low microcin uptake via the chromosomally encoded FhuA. Higher expression by a plasmid-encoded FhuA allowed greater uptake of MccJ25 by E. coli strains and the consequent inhibition of oxygen consumption. The two mechanisms, inhibition of RNAP and oxygen consumption, are independent of each other. Further analysis revealed for the first time that MccJ25 stimulates the production of reactive oxygen species (O2·−) in bacterial cells, which could be the main reason for the damage produced on the membrane respiratory chain.
Escherichia coli microcin J25 (MccJ25) is a lasso peptide antibiotic of 21 l-amino acid residues (G1-G-A-G-H5-V-P-E-Y-F10-V-G-I-G-T15-P-I-S-F-Y20-G) (3, 42, 56). MccJ25 is active on gram-negative bacteria related to the producer strain, including some pathogenic strains (43, 44, 55). Four plasmid genes, mcjABCD, are involved in MccJ25 production: mcjA, mcjB, and mcjC code for an MccJ25 precursor and two processing enzymes required for the in vivo synthesis of the mature peptide, respectively, and mcjD encodes the McjD immunity protein (53). McjD, a homologous ABC exporter family protein, participates in MccJ25 secretion (52). Thus, immunity is mediated by active efflux of the peptide, keeping its intracellular concentration below a critical level (53). Recently, it has been demonstrated that YojI, a chromosomal protein with ABC-type exporter homology (36), is also able to export MccJ25 from the cells (14). TolC, an E. coli outer membrane protein, is necessary for MccJ25 secretion mediated by either McjD or YojI (11, 14). On the other hand, the uptake of MccJ25 by E. coli is dependent on the outer membrane receptor FhuA (15, 45) and the four inner membrane proteins TonB, ExbD, ExbB, and SbmA, the first three of which constitute the Ton complex (38), while the last one acts as a transporter (46).
Convincing evidence showing that RNA polymerase (RNAP) is the target for MccJ25 action in E. coli was previously provided by our laboratory. The peptide inhibits the enzyme activity by obstructing the secondary channel and consequently preventing access of the substrates to its active sites (1, 12, 34, 57). Later, it was demonstrated that MccJ25 can bind and penetrate into the phospholipid monolayer and disrupt the electric potential of liposomes composed of phospholipids from gram-negative bacteria (5, 40). These results encouraged the study of the effect of MccJ25 on the bacterial membrane. MccJ25 was found to disrupt the membrane potential inhibiting oxygen consumption in Salmonella enterica serovar Newport (41) and S. enterica serovar Typhimurium transformed with a plasmid carrying fhuA from E. coli (55), suggesting the presence of a second target for the peptide. In addition, the chemical amidation of the C-terminal glycine localized in the MccJ25 lariat ring region (MccJ25-GA) specifically blocked the capacity for RNAP inhibition, but not cell respiration inhibition or peptide uptake, in S. enterica serovars (54). These results indicate that MccJ25 has at least two different intracellular targets, depending on the bacterial species. The absence of an MccJ25 effect on the cell respiration of E. coli is a surprising result that raises a question about the general presence of both mechanisms in a single bacterial species (16, 39).
The goal of this study was to elucidate the question concerning the absence or the presence of an MccJ25 effect on cell respiration in E. coli strains. Interestingly, the results reported here indicate that a membrane target is also present in laboratory E. coli strains, in which the effect becomes noticeable when the bacteria overexpress the MccJ25-FhuA receptor. Moreover, it was found that the effect of MccJ25 on the E. coli respiratory-chain enzymes is mediated by increased superoxide production demonstrated by the in vitro reversion effect of superoxide dismutase (SOD). These findings shed light on the dual and independent mechanisms of action of MccJ25.
MATERIALS AND METHODS
Bacterial culture media and growth conditions.
The bacteria and plasmids used are listed in Table 1. Luria broth (LB) and M9 minimal salts were purchased from Sigma Chemical Co. Minimal medium was supplemented with 0.2% glucose, 1 mM MgSO4, and 1 μg ml−1 vitamin B1. Solid media were prepared by adding agar to a final concentration of 1.5%. When required, 50 μg ml−1 ampicillin, 30 μg ml−1 kanamycin (Km), or 30 μg ml−1 chloramphenicol was added. Liquid cultures were grown with agitation in 100-ml flasks. For anaerobic growth, liquid cultures were grown without agitation in filled tubes with the medium overlaid with mineral oil. Growth was monitored by measuring the optical density at 600 nm (OD600) for 24 h. After that, the culture was followed under aerobic conditions in 100-ml flasks with vigorous shaking. All cultures were incubated at 37°C. Aliquots were taken for CFU determination. For growth under low-iron conditions, we used the Tris-buffered medium of Simon and Tessman (50) (T medium) without iron.
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid | Genotype or descriptiona | Reference or sourceb |
|---|---|---|
| E. coli | ||
| MC4100 | F−araD139 Δ(argF-lac)205 λ−rpsL150 flbB5301 relA deoC1 pstF25 | CGSC |
| EZE100 | MC4100 sbmA::Tn5 | R. De Cristóbal |
| AB1133 | F−thr-1 ara-14 leuB6 lacY1 Δ(gpt-proA)62 supE44 galK2 λ−rac hisG4 rfbD1 rpsL31 (Smr) kdgK51 xyl-5 mtl-1 argE3 thi-1 | CGSC |
| SBG231 | AB259 spontaneous MccJ25r mutant (rpoC T931I) | 12 |
| CAG18615 | MG1655 zjb-3179::Tn10kan | 51 |
| PA230 | SBG231 rpoC T931I (P1/CAG18615xSBG231) | This work |
| PA232 | AB1133 rpoC T931I (P1/PA230xAB1133) | This work |
| PA233 | MC4100 rpoC T931I (P1/PA230xMC4100) | This work |
| GS022 | araD139 Δ(argF-lac)169 λ−flhD5301 fruA25 relA1 rpsL150 rbsR22 deoC1 λRS45 φ(katG::lacZ) | 48 |
| S. enterica serovar Newport | Clinical isolate | UNT |
| Plasmids | ||
| pDJJ12 | pBR322 carrying rplJL rpoB rpoC htrC | 20 |
| pGC01 | pBR322 carrying fhuA | 10 |
| pMM73.4 | pBR322 carrying sbmA | 29 |
MccJ25r, resistant to MccJ25.
CGSC, E. coli Genetic Stock Center; UNT, Universidad Nacional de Tucumán.
DNA manipulations and genetic methods.
Plasmid DNA was isolated with the Wizard miniprep DNA purification system (Promega). Transformation of competent cells was carried out by the CaCl2 procedure. Phage P1 vir was used for transduction of the rpoC mutation (rpoC encodes the β′ subunit of RNAP), which confers MccJ25 resistance (12), as follows. First, zjb-3179::Tn10kan from CAG18615 (51) was transduced to SBG231 with selection for Km and MccJ25 resistance. Then, a P1 lysate prepared from one of these transductants, called PA230, was used to cotransduce the zjb-3179::Tn10kan and rpoC mutations to strains AB1133, MC4100, and GS022. The respective mutated strains PA232, PA233, and GS022 rpoC were selected for Km resistance. The presence of the rpoC mutation was confirmed by transforming it with pDJJ12 (pBR322 containing cloned wild-type rpoC) and checking for the restoration of a wild-type level of MccJ25 sensitivity.
β-Galactosidase assays.
Overnight anaerobic cultures of GS022 and GS022 rpoC (strains carrying a transcriptional fusion, katG::lacZ) growing without agitation in filled tubes were passed under aerobic conditions in the absence and presence of 20 μM MccJ25 for β-galactosidase assays (2-ml volume in 100-ml Erlenmeyer flasks) and incubated for 1 h with vigorous shaking. β-Galactosidase activity was measured as described by Miller (33), using cells (GS022 and GS022 rpoC) permeabilized with sodium dodecyl sulfate and chloroform, and it was reported in Miller units. The assays were repeated at least three times for each sample.
Isolation of MccJ25 and MccJ25-GA peptides.
MccJ25 was purified from 2-liter cultures of strain MC4100 harboring pTUC202 as previously described (41). MccJ25 appeared homogeneous in two different systems of analytical reverse-phase high-performance liquid chromatography and showed a single peak of 2,107 Da on matrix-assisted laser desorption ionization-time of flight mass spectrometry (6). The chemical derivation and isolation of MccJ25-GA peptide was performed as described by Bellomio et al. (6). Native MccJ25 and its amidated derivative were dissolved in methanol, and their concentrations were determined by absorbance at 278 nm (ɛ278 = 3,340 M−1 cm−1) (5).
Sensitivity test of microcin activity.
Sensitivity to microcin was tested by a spot-on-lawn assay as follows. To avoid aggregation (7), concentrated methanolic solutions of the peptides (2 mM) were prepared for antibiotic activity assay, and serial doubling dilutions were performed in double-distilled water containing 0.1% Tween 80 in Eppendorf tubes. Samples (10 μl) of each dilution were spotted onto M9 medium plates supplemented with 0.2% tryptone. After the drops had dried, the plates were overlaid with 4 ml of soft agar (0.6%) containing 30 μl of an overnight LB culture of the strain to be assayed. The plates were incubated for 12 h at 37°C and examined for different degrees of inhibition. The concentration (mM) of the last dilution giving a clear or turbid spot was taken as the MIC of MccJ25 and its analogues.
Oxygen consumption.
Cells were grown to exponential phase (OD600 = 0.4 to 0.5) in LB. Samples were diluted in LB to an OD600 of 0.2 and incubated at 37°C for 30 min without MccJ25 (control) and with different concentrations of the antibiotic. The average rate of cell respiration over the subsequent 5 min was polarographically measured with a Clark-type electrode oxygraph and normalized to the OD600.
In vivo transcription assay.
The in vivo transcription assay was carried out as described by Delgado et al. (12). The strains were grown in M9 medium to early exponential phase. Uridine (150 μg) and 1.85 × 105 Bq [3H]uridine were added to the cell suspension (10 ml), and then aliquots were extracted at different incubation times without (control) or with peptides. At 0 and 30 min, 0.5-ml samples were mixed with 1.5 ml of cold 5% trichloroacetic acid, incubated for 1 h on ice, filtered through Millipore HAWP02500 filters, and washed with cold 5% trichloroacetic acid. The radioactivity retained on the filters was estimated in a Beckman LS-1801 liquid scintillation counter.
Enzymatic-activity determination.
The enzyme activities were measured on membrane preparations obtained as previously described by Evans (17). Dehydrogenase activities, in the absence and in the presence of 6 mM potassium cyanide, were determined at 37°C in a total volume of 0.5 ml 50 mM phosphate (pH 7.5) containing 0.5 mM NADH, 10 mM succinate, or 10 mM d-lactate as a substrate. The reaction was followed by reduction of the artificial acceptor thiazolyl blue tetrazolium bromide (MTT) (50 μg ml−1) to its derivative formazan at 570 nm. The MccJ25 effect was studied by preincubation of the membrane sample (10 μg ml−1 protein) for 10 min at 37°C with the antibacterial peptide (10 μM). The oxidase activities were measured by incubating the membrane preparation at 37°C with 10 μM MccJ25 and the corresponding substrates in the absence and in the presence of 40 IU ml−1 superoxide dismutase. The NADH oxidase activity was followed by measuring absorbance at 340 nm in a Beckman DU 7500 spectrophotometer. Succinate and lactate oxidase activities were determined by following the oxygen consumption registered polarographically with a Clark-type electrode oxygraph. The membrane protein concentration was measured by the method of Lowry et al. (30).
Superoxide production rate.
O2·− formation was measured as SOD-sensitive cytochrome c reduction (25). Bacterial membrane from E. coli strain AB1133 (final concentration, 1 mg protein ml−1) was assayed in 0.6-ml reaction volumes of 50 mM phosphate buffer (pH 7.8) containing 40 μM cytochrome c in the absence and in the presence of 20 μM MccJ25. Reduction of cytochrome c was initiated by the addition of 10 mM succinate (final concentration) and monitored in a Beckman DU 7500 spectrophotometer at 550 nm. Duplicate reactions were performed with the addition of 30 units E. coli Mn-SOD (Sigma). The extent of reduced cytochrome c was calculated by using an absorption coefficient (ɛ) of 0.21 mM−1 cm−1.
RESULTS
Relationship between MccJ25 activity and the expression of its outer membrane receptor, FhuA.
The intracellular concentration of MccJ25 in E. coli depends basically on the two active processes that take place simultaneously in the cell, the uptake (15, 45, 46) and the export (13, 14) of the peptide. Table 2 shows the sensitivities to MccJ25 of two E. coli strains overexpressing FhuA. The sensitivities of E. coli MC4100 and AB1133 increased notably when the strains were transformed with pGC01 (carrying the fhuA gene) (compare rows 1 and 2 with 3 and 4). It is interesting that the inhibitory halo of the strains overexpressing FhuA became clear and that the MICs are comparable to that of S. enterica serovar Newport (row 5). The turbid inhibition halo and the apparent major resistance of strain MC4100 in comparison with strain AB1133 is due to major YojI activity and, as a consequence, major efficiency in removing MccJ25 through the putative YojI-TolC pathway (unpublished results). The overexpression of FhuA also increased sensitivity to MccJ25-GA: the resistant MC4100 strain became sensitive (compare row 1 with row 3), and the sensitive AB1133 become eightfold more sensitive (rows 2 and 4). As was previously reported, MccJ25-GA specifically lost the capability to inhibit RNAP in E. coli; afterward, the presence of an additional antibiotic target in E. coli can be assumed, as occurs in S. enterica (41, 54). To further investigate the presence of the additional membrane target in E. coli, two strains harboring an MccJ25-resistant RNAP, PA232 and PA233, which were derived from AB1133 and MC4100, respectively, were constructed. The donor strain in these transduction experiments, SBG231, carries the rpoC T931I mutation (see Materials and Methods) (12). As can be seen in Table 2, PA232 was sensitive while PA233 was resistant to MccJ25 and MccJ25-GA (rows 6 and 8). However, when these strains were transformed with pGC01, PA232 became 100- and 8-fold more sensitive to MccJ25 and MccJ25-GA, respectively, and the resistant PA233 became sensitive to both MccJ25 and MccJ25-GA. Moreover, it was possible to produce more sensitivity in PA233 when FhuA was derepressed under iron-depleted conditions (rows 10 and 11). These results confirmed our previous assumption about the existence of a second MccJ25 target on E. coli.
TABLE 2.
Comparison of the sensitivities of several E. coli strains to MccJ25 and MccJ25-GA relative to the expression of FhuA
| Row | Strain | Sensitivitya
|
|
|---|---|---|---|
| MccJ25 | MccJ25-GA | ||
| 1 | MC4100 | 1.0a | R |
| 2 | AB1133 | 0.24 | 31.2 |
| 3 | MC4100(pGC01) | 0.02 | 3.9 |
| 4 | AB1133(pGC01) | 0.02 | 3.9 |
| 5 | S. enterica serovar Newport | 0.02 | 2.4 |
| 6 | PA232 (AB1133 rpoC T931I) | 2 | 62.5 |
| 7 | PA232(pGC01) | 0.02 | 7.8 |
| 8 | PA233 (MC4100 rpoC T931I) | R | R |
| 9 | PA233(pGC01) | 0.6 | 62.5 |
| 10 | PA233b | R | |
| 11 | PA233c | 0.2 | |
| 12 | EZE100(pGC01) | R | R |
The results are given as the peptide concentration (μM) of the last dilution which produced a clear or turbid inhibitory spot. A fourfold or greater difference was considered significant. R, >2.0 mM.
Growth in T medium with iron.
Growth in T medium without iron.
In contrast to MC4100(pGC01), EZE100(pGC01), which harbors an sbmA mutation, was not sensitive to MccJ25 and MccJ25-GA (compare rows 3 and 10). These results clearly indicate that both peptides must penetrate into the cell in order to exert antibacterial action.
Relationship between the MccJ25 membrane target and oxygen consumption.
As pointed out above, it was previously found that MccJ25 disrupts the membrane potential inhibiting oxygen consumption in S. enterica serovar Newport, but not in E. coli MC4100 (41). Since S. enterica serovar Newport is 50-fold more sensitive than the above-mentioned E. coli strain, it was interesting to verify the presence of the MccJ25 membrane target in the most sensitive E. coli strains, such as AB1133. Thus, the effects of the peptides on the in vivo synthesis of RNA and on oxygen consumption were explored. The results in Table 3 show that 20 μM MccJ25 inhibited nearly 50% of the in vivo RNA synthesis in strains AB1133 and AB1133(pGC01), but not in the PA232 (AB1133 rpoC T931I) and PA232(pGC01) strains. Moreover, a threefold-higher concentration of MccJ25-GA was unable to inhibit the RNA synthesis of any strain. On the other hand, 10 μM MccJ25 or MccJ25-GA inhibited oxygen consumption only in E. coli strains overexpressing FhuA, with consumption remaining unchanged in AB1133 and PA232. Oxygen consumption remained unaffected in E. coli AB1133, even at 20 μM MccJ25, whereas AB1133(pGC01) respiration was clearly inhibited in a concentration-dependent manner. A similar effect was observed with E. coli MC4100(pGC01) (data not shown).
TABLE 3.
Inhibition of RNA synthesis and oxygen consumption by MccJ25 and MccJ25-GAa
| Strain | In vivo RNA synthesis
|
Oxygen consumption
|
||
|---|---|---|---|---|
| MccJ25 (20 μM) | MccJ25-GA (60 μM) | MccJ25 (10 μM) | MccJ25-GA (10 μM) | |
| AB1133 | 50.0 | 0.3 | 2 | 3 |
| AB1133(pGC01) | 55.0 | 0.1 | 29 | 27 |
| PA232 | 0.2 | 0.5 | 3 | 1 |
| PA232(pGC01) | 0.1 | 0.2 | 35 | 25 |
The results are expressed as percentages of the inhibition of a control.
MccJ25 increases superoxide generation in the membrane respiratory chain.
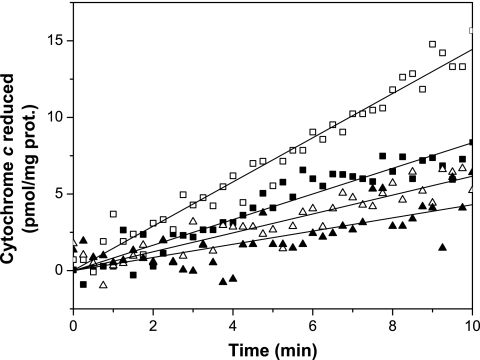
To further assess the MccJ25 target on bacterial membranes, the peptide effects on dehydrogenase and oxidase activities (Table 4) were studied. For this experiment, membrane from E. coli AB1133 was used. As previously observed, MccJ25 (10 μM) did not inhibit NADH, lactate, and succinate dehydrogenase activities when the assays were performed in the presence of 6 mM cyanide. However, when the assays were carried out in the absence of cyanide, clear inhibition was obtained for NADH (21% ± 2.0%) and lactate (44% ± 6.0%) dehydrogenases. In the presence of cyanide, MTT is the obligatory electron acceptor, while in the absence of cyanide, the electrons are captured mainly by oxygen. MccJ25 is suspected to stimulate adventitious electron transfers to oxygen, generating superoxide and hydrogen peroxide; these reactive species can oxidize biomolecules, causing irreversible damage (24). The succinate dehydrogenase activity was not affected by the presence of MccJ25, probably because the iron-sulfur clusters of the succinate dehydrogenase enzyme are not damaged by superoxide (19). To avoid false results, since superoxide overproduction could be obtained when dehydrogenase activities are determined by electron transference to MTT (32), oxidase activities were measured by direct determination of oxygen consumption. The results shown in Table 4 indicated that NADH and lactate activities were inhibited similarly to the corresponding dehydrogenase activities (28.0% ± 0.8% and 31.5% ± 1.5%, respectively). As occurred with the dehydrogenase activity, succinate oxidase was not inhibited by MccJ25. To confirm that reactive oxygen species (ROS) overproduction could be involved in oxidase and dehydrogenase inhibition, the reversion effect by SOD (an O2·− scavenger) was investigated. As shown in Table 4, the inhibitory effect of MccJ25 was reversed by SOD in all cases. Membranes preincubated with MccJ25 plus succinate (as a respiratory substrate) showed remarkable NADH oxidase inhibition. When SOD was added during the preincubation, no inhibition of the NADH oxidase activity was found. These results suggest that the presence of MccJ25 increased superoxide production when the respiratory chain was active, and in turn, the superoxide generated during preincubation affected NADH oxidase activity. To further explore this assumption, the rate of superoxide production in vitro was determined by using succinate as an electron donor (Fig. 1). The rate of SOD-inhibitable cytochrome c reduction (calculated in pmol·min−1, as indicated in Materials and Methods) was incremented by 20 μM MccJ25 from 0.40 ± 0.1 to 0.83 ± 0.08. This increase was abolished when the experiment was carried out in the presence of Mn2+-SOD in the reaction mixture. The addition of 2 mM KCN prevented cytochrome c reduction (0.1 ± 0.06), even in the presence of MccJ25 (0.12 ± 0.05) (data not shown).
TABLE 4.
Effects of MccJ25 on the enzymatic activities of dehydrogenases and oxidases from the respiratory chain
| Enzyme | Enzymatic activitya
|
% Inhibitionb | |
|---|---|---|---|
| Without MccJ25 | With MccJ25 | ||
| Dehydrogenases | |||
| NADH with CN− | 127 | 120 | 0.6 ± 0.1 |
| NADH without CN− | 55 | 44 | 21.1 ± 2.0 |
| Lactate with CN− | 63 | 60 | 5.0 ± 0.3 |
| Lactate without CN− | 53 | 30 | 44.0 ± 6.0 |
| Succinate with CN− | 60 | 60 | 0.4 ± 0.2 |
| Succinate without CN− | 20 | 20 | 0.5 ± 0.2 |
| Oxidases | |||
| NADH | 166 | 120 | 28.0 ± 0.8 |
| NADH + SOD | 113 | 125 | −10.0 ± 6.0 |
| Lactate | 2.95 | 2.1 | 31.5 ± 0.5 |
| Lactate + SOD | 2.16 | 2.2 | 0.1 ± 0.1 |
| Succinate | 17 | 16 | 5.7 ± 3.8 |
| NADHc | 162 | 60 | 63.0 ± 10.0 |
| NADHd | 44 | 50 | −16.2 ± 10.0 |
The absolute values of the enzymatic activities were calculated as OD570 units/min/mg protein for dehydrogenase activities, OD340 units/min/mg protein for NADH oxidase activities, and μM O2/min/mg protein for succinate and lactate oxidase activities. The data are averages of five determinations that did not vary more than 15%.
Negative values indicate activation.
The activity was evaluated after preincubation for 1 h with succinate and 10 μM MccJ25.
The activity was evaluated after preincubation for 1 h with succinate and MccJ25 plus 40 IU/ml SOD.
FIG. 1.
MccJ25 effect on the time course of superoxide production by E. coli membrane. A solution containing membranes (1 mg protein [prot.]·ml−1) and 40 mM cytochrome c in 50 mM phosphate buffer, pH 7.8 (final volume, 0.6 ml), was incubated in the absence (▪) and in the presence (□) of 20 μM MccJ25 at 37°C. The same experiment was carried out with the addition of 30 units Mn2+-SOD in the absence (▴) and in the presence (▵) of 20 μM MccJ25. The time course reaction was initiated with 10 mM succinate (final concentration). The cytochrome c reduction was followed spectrophotometrically at 550 nm, as described in Materials and Methods.
Physiological evidence that MccJ25 increases ROS production in vivo.
To demonstrate the in vivo increase in superoxide production induced by MccJ25, two experimental approaches were developed. First, the effect of MccJ25 on E. coli GS022 was explored, when the strain harbored an RNAP mutation and a fusion with the capacity to sense H2O2 production. Second, the antibiotic activities of MccJ25 on E. coli PA232(pGC01) growing under both anaerobic and aerobic conditions were determined.
The steady-state H2O2 concentration in E. coli is sensed by OxyR. The increase of H2O2 activates the transcription of a defensive regulon that include katG, a gene that codes for hydroperoxidase I catalase (9). Afterward, katG expression served as a reporter for OxyR activity. In fact, katG::lacZ expression was elevated 10- to 15-fold in a strain lacking Ahp (hydroperoxidase), the primary scavenger of endogenous H2O2 in E. coli (49). The strain GS022, harboring a katG::lacZ fusion (48) and transduced with an rpoC T931I mutation (12), was used to investigate MccJ25 stimulation of in vivo H2O2 generation (primarily O2·−). The results shown in Fig. 2 indicate that incubation of the mutated GS022 strain raised by twofold the β-galactosidase activities in three independent experiments. Although modest, this increment is significant, because it was absent in the parental strain (GS022) due to RNAP inhibition by MccJ25 (data not shown).
FIG. 2.
Effect of MccJ25 on the expression of the katG gene from E. coli. Anaerobic cultures of E. coli strain GS022 rpoC T931I growing without agitation in filled tubes were passed to aerobic conditions in the absence (Control) and the presence of 20 μM MccJ25. The bacteria were then incubated for 1 h with vigorous shaking. β-Galactosidase activity was measured on the cells permeabilized with sodium dodecyl sulfate and chloroform as described in Materials and Methods. The results are expressed as the means ± standard deviations of three independent experiments.
In light of these results, it would be relevant to determine whether MccJ25 is capable of inhibiting the growth of E. coli PA232(pGC01) in anaerobiosis. It can be presumed that in the absence of oxygen, the formation of superoxide anions would not occur. The growth curves in the absence and in the presence of 20 μM MccJ25 are shown in Fig. 3. After 24 h, no difference was observed between them. Afterward, the bacterial cultures were changed to aerobic conditions, and cells were unable to grow in the presence of MccJ25. Meanwhile, in the absence of the antibiotic, they continued growing, reaching an OD that the bacteria normally reach under aerobic conditions. It can be concluded that in the absence of oxygen, MccJ25 lost antibiotic activity on RNAP-resistant strains, supporting the hypothesis that the overproduction of superoxide anions is positively involved in the antibiotic action. MccJ25 inhibited the growth of AB1133(pGC01) carrying wild-type rpoC, even under anaerobic conditions (Fig. 3, inset), indicating that MccJ25 acts on RNAP in an O2-independent manner.
FIG. 3.
MccJ25 effects on the growth of E. coli under anaerobic and aerobic conditions. E. coli strain PA232(pGC01) in the absence (▪) and in the presence (□) of 20 μM MccJ25 was grown without agitation in filled tubes with liquid cultures lacking oxygen, and the medium was overlaid with mineral oil. Growth was monitored by measuring the OD600 for 24 h. Next, the culture was changed to aerobic conditions (arrow) in 100-ml flasks with vigorous shaking. All cultures were incubated at 37°C. Aliquots of cultures in the absence (▴) and in the presence (▵) of 20 μM MccJ25 were taken for CFU determination (CFU/ml). (Inset) Growth under anaerobic conditions of E. coli strain AB1133(pGC01) carrying wild-type rpoC in the absence (▪) and in the presence (□) of 20 μM MccJ25.
DISCUSSION
The aim of this study was to further characterize the mechanism of action of MccJ25. Prior analysis of the antimicrobial peptide revealed important inhibition of RNAP (12). However, preliminary studies on model membranes and S. enterica serovar Newport implied an alternative mechanism that has not been detected in E. coli. The present study indicates, as a major finding, that MccJ25 is also responsible for the inhibition of oxygen consumption in E. coli strains throughout the increase in the ROS concentration. E. coli strains harboring an MccJ25-resistant RNAP and MccJ25-GA (a chemically altered MccJ25 that does not inhibit RNAP) were employed to demonstrate that MccJ25 could inhibit the growth of E. coli in an RNAP-independent manner. The in vivo experiment showed that the inhibitory effect of MccJ25 on cell respiration depends on the expression and/or activity of the transporter protein FhuA, which is involved in peptide uptake and consequently is responsible for the cytoplasm peptide concentration. The growth-inhibitory effect of MccJ25-GA on MC4100(pGC01), but not on EZE100(pGC01), led us to conclude that the antibiotic must reach the cytoplasm to act on the cellular respiration. This interesting observation could explain the absence of an additional periplasmic immunity protein in the MccJ25 production system. Such immunity proteins were described by Braun et al. (8) as indispensable for microcins, which act from the periplasm space. We could not determine inhibition of the respiration of AB1133, despite sensitivity to MccJ25-GA. This result could be attributed to a low sensitivity of the oxygen consumption method compared with the spot test method. In fact, an MccJ25 concentration (10 μM) 500-fold higher than the minimal concentration to produce inhibition (0.02 μM) was required to get 30% respiration inhibition in AB1133(pGC01). Furthermore, only respiration inhibition of PA232(pGC01), which was 100-fold more sensitive than PA232, could be detected.
Normal aerobic metabolism gives rise to active oxygen species, such as superoxide radicals and hydrogen peroxide, formed by the partial reduction of molecular oxygen (18, 37). Moreover, redox cycling of various chemical substances, including some antibiotics, affects the reactive species of oxygen produced by cells during oxidative processes. For example, plumbagin and paraquat were used for several years as models to study oxidative stress (21, 22). Recently, a number of antibiotics, including ciprofloxacin, have been demonstrated to stimulate the production of ROS in bacterial cells (2, 4). In this work, evidence that a microcin, MccJ25, perturbs the natural reaction of the respiratory-chain enzymes with oxygen, generating a significant increment in ROS, is reported for the first time. The involvement of superoxide anion in the antibacterial action of MccJ25 was analyzed in vitro with isolated bacterial membrane and also in in vivo experiments (Fig. 2 and 3). The action of MccJ25 on respiratory-chain enzymes was irrefutably demonstrated by a direct measurement of ROS formation (Fig. 1) and SOD damage protection (Table 4). The lack of peptide effect on bacterial growth in the anaerobic state simultaneously with the overexpression of the katG gene supports and decisively confirms the results obtained in vitro.
Recently, Korshunov and Imlay (27) reported that substantial superoxide was released into the periplasmic compartment as an incidental by-product of respiration, apparently due to the adventitious oxidation of menaquinone in E. coli cultures. The rate of periplasmic superoxide formation is quite high, about 3 μM/s, when normalized to the estimated periplasmic volume. That value is comparable to the 5 μM/s that has been estimated for superoxide formation in the cytosol (27). The superoxide produced by cytosolic enzymes cannot cross inner membranes at physiological pH (28, 31). The simple hypothesis that MccJ25 increases the ROS concentration could be postulated, although the site where ROS is overproduced is unknown.
At this point, it is possible to question the physiological relevance of the ROS increase produced by MccJ25, since it is a peptide synthesized by bacterial strains growing in an anaerobic habitat. Although E. coli grows essentially in an oxygen-poor environment (47), it can be abruptly changed to an aerobic one, inducing the sudden formation of ROS (23). In such a new ecosystem, the bacterial strains producing MccJ25 would have a clear advantage in competing with the sensitive bacteria. MccJ25 acts on the most sensitive strains in both directions, increasing ROS and inhibiting the gene transcription needed to synthesize protective enzymes (26). Furthermore, a toxic capability for MccJ25 could not be excluded, since our laboratory described the peptide's deleterious effect on mammalian mitochondria under aerobic conditions (35). In addition, Delgado and Salomon (11) suggested in a previous paper that the plasmid pTUC100, responsible for MccJ25 production and immunity, could have originated in Shigella (a pathogenic bacterium) and then been transmitted to E. coli.
On the basis of these results, it can be concluded that the antibacterial action of MccJ25 involves two independent mechanisms: the RNAP inhibition previously described (12) and the overproductions of ROS, such as superoxide anions. However, the exact mechanism of the latter phenomenon has yet to be worked out. A combination of structural and biochemical approaches are currently being used to identify the active site of MccJ25 on the cell membrane and to discover how the electrons are sequestered by oxygen rather than used to generate the electrochemical gradient.
Acknowledgments
We are indebted to R. Salomon for his help in strain construction and for useful discussions; to Ricardo de Cristóbal, Mónica Delgado, and James A. Imlay for generously providing strains that were used in this paper; and to Luisa Rodriguez Montelongo for assistance with enzyme activity determinations.
This work was supported by FONCYT (grants PICT 17819 and PICTO 843) and CIUNT (grants 26/D235 and 26/D228). A.B. is the recipient of a CONICET postdoctoral fellowship. P.A.V., R.D.M., and R.N.F. are career researchers of CONICET.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Adelman, K., J. Yuzenkova, A. La Porta, N. Zenkin, J. Lee, J. T. Lis, S. Borukhov, M. D. Wang, and K. Severinov. 2004. Molecular mechanism of transcription inhibition by peptide antibiotic microcin J25. Mol. Cell 14:753-762. [DOI] [PubMed] [Google Scholar]
- 2.Albesa, I., M. C. Becerra, P. C. Battan, and P. L. Paez. 2004. Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 317:605-609. [DOI] [PubMed] [Google Scholar]
- 3.Bayro, M. J., J. Mukhopadhyay, G. V. Swapna, J. Y. Huang, L. C. Ma, E. Sineva, P. E. Dawson, G. T. Montelione, and R. H. Ebright. 2003. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J. Am. Chem. Soc. 125:12382-12383. [DOI] [PubMed] [Google Scholar]
- 4.Becerra, M. C., and I. Albesa. 2002. Oxidative stress induced by ciprofloxacin in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 297:1003-1007. [DOI] [PubMed] [Google Scholar]
- 5.Bellomio, A., R. G. Oliveira, B. Maggio, and R. D. Morero. 2005. Penetration and interactions of the antimicrobial peptide, microcin J25, into uncharged phospholipid monolayers. J. Colloid Interface Sci. 285:118-124. [DOI] [PubMed] [Google Scholar]
- 6.Bellomio, A., M. R. Rintoul, and R. D. Morero. 2003. Chemical modification of microcin J25 with diethylpyrocarbonate and carbodiimide: evidence for essential histidyl and carboxyl residues. Biochem. Biophys. Res. Commun. 303:458-462. [DOI] [PubMed] [Google Scholar]
- 7.Blond, A., J. Peduzzi, C. Goulard, M. J. Chiuchiolo, M. Barthelemy, Y. Prigent, R. A. Salomon, R. N. Farias, F. Moreno, and S. Rebuffat. 1999. The cyclic structure of microcin J25, a 21-residue peptide antibiotic from Escherichia coli. Eur. J. Biochem. 259:747-755. [DOI] [PubMed] [Google Scholar]
- 8.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 9.Cabiscol, E., J. Tamarit, and J. Ros. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3:3-8. [PubMed] [Google Scholar]
- 10.Coulton, J. W., P. Mason, and M. S. DuBow. 1983. Molecular cloning of the ferrichrome-iron receptor of Escherichia coli K-12. J. Bacteriol. 156:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado, M. A., and R. A. Salomon. 2005. Molecular characterization of a DNA fragment carrying the basic replicon of pTUC100, the natural plasmid encoding the peptide antibiotic microcin J25 system. Plasmid 53:258-262. [DOI] [PubMed] [Google Scholar]
- 12.Delgado, M. A., M. R. Rintoul, R. N. Farias, and R. A. Salomon. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgado, M. A., J. O. Solbiati, M. J. Chiuchiolo, R. N. Farias, and R. A. Salomon. 1999. Escherichia coli outer membrane protein TolC is involved in production of the peptide antibiotic microcin J25. J. Bacteriol. 181:1968-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado, M. A., P. A. Vincent, R. N. Farias, and R. A. Salomon. 2005. YojI of Escherichia coli functions as a microcin J25 efflux pump. J. Bacteriol. 187:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Destoumieux-Garzon, D., S. Duquesne, J. Peduzzi, C. Goulard, M. Desmadril, L. Letellier, S. Rebuffat, and P. Boulanger. 2005. The iron-siderophore transporter FhuA is the receptor for the antimicrobial peptide microcin J25: role of the microcin Val11-Pro16 beta-hairpin region in the recognition mechanism. Biochem. J. 389:869-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Destoumieux-Garzon, D., J. Peduzzi, and S. Rebuffat. 2002. Focus on modified microcins: structural features and mechanisms of action. Biochimie 84:511-519. [DOI] [PubMed] [Google Scholar]
- 17.Evans, D. J., Jr. 1969. Membrane adenosine triphosphatase of Escherichia coli: activation by calcium ion and inhibition by monovalent cations. J. Bacteriol. 100:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridovich, I. 1983. Superoxide dismutases: regularities and irregularities. Harvey Lect. 79:51-75. [PubMed] [Google Scholar]
- 19.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 20.Gross, C., F. Engbaek, T. Flammang, and R. Burgess. 1976. Rapid micromethod for the purification of Escherichia coli ribonucleic acid polymerase and the preparation of bacterial extracts active in ribonucleic acid synthesis. J. Bacteriol. 128:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan, H. M., and I. Fridovich. 1979. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch. Biochem. Biophys. 196:385-395. [DOI] [PubMed] [Google Scholar]
- 22.Hassan, H. M., and I. Fridovich. 1979. Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254:10846-10852. [PubMed] [Google Scholar]
- 23.Imlay, J. A. 1995. A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J. Biol. Chem. 270:19767-19777. [PubMed] [Google Scholar]
- 24.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 25.Imlay, J. A., and I. Fridovich. 1991. Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem. 266:6957-6965. [PubMed] [Google Scholar]
- 26.Iuchi, S., and L. Weiner. 1996. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J. Biochem. 120:1055-1063. [DOI] [PubMed] [Google Scholar]
- 27.Korshunov, S., and J. A. Imlay. 2006. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J. Bacteriol. 188:6326-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korshunov, S. S., and J. A. Imlay. 2002. A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of Gram-negative bacteria. Mol. Microbiol. 43:95-106. [DOI] [PubMed] [Google Scholar]
- 29.Lavina, M., A. P. Pugsley, and F. Moreno. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J. Gen. Microbiol. 132:1685-1693. [DOI] [PubMed] [Google Scholar]
- 30.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 31.Lynch, R. E., and I. Fridovich. 1978. Permeation of the erythrocyte stroma by superoxide radical. J. Biol. Chem. 253:4697-4699. [PubMed] [Google Scholar]
- 32.Massa, E. M., M. A. Leiva, and R. N. Farias. 1985. Study of a time lag in the assay of Escherichia coli membrane-bound dehydrogenases based on tetrazolium salt reduction. Biochim. Biophys. Acta 827:150-156. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Mukhopadhyay, J., E. Sineva, J. Knight, R. M. Levy, and R. H. Ebright. 2004. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell 14:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niklison Chirou, M. V., C. J. Minahk, and R. D. Morero. 2004. Antimitochondrial activity displayed by the antimicrobial peptide microcin J25. Biochem. Biophys. Res. Commun. 317:882-886. [DOI] [PubMed] [Google Scholar]
- 36.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomposiello, P. J., and B. Demple. 2000. Oxidative stress, p. 526-532. In J. Lederberg et al. (ed.), Encyclopedia of microbiology. Academic Press, San Diego, CA.
- 38.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 39.Rebuffat, S., A. Blond, D. Destoumieux-Garzon, C. Goulard, and J. Peduzzi. 2004. Microcin J25, from the macrocyclic to the lasso structure: implications for biosynthetic, evolutionary and biotechnological perspectives. Curr. Protein Pept. Sci. 5:383-391. [DOI] [PubMed] [Google Scholar]
- 40.Rintoul, M. R., B. F. de Arcuri, and R. D. Morero. 2000. Effects of the antibiotic peptide microcin J25 on liposomes: role of acyl chain length and negatively charged phospholipid. Biochim. Biophys. Acta 1509:65-72. [DOI] [PubMed] [Google Scholar]
- 41.Rintoul, M. R., B. F. de Arcuri, R. A. Salomon, R. N. Farias, and R. D. Morero. 2001. The antibacterial action of microcin J25: evidence for disruption of cytoplasmic membrane energization in Salmonella newport. FEMS Microbiol. Lett. 204:265-270. [DOI] [PubMed] [Google Scholar]
- 42.Rosengren, K. J., R. J. Clark, N. L. Daly, U. Goransson, A. Jones, and D. J. Craik. 2003. Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 125:12464-12474. [DOI] [PubMed] [Google Scholar]
- 43.Sable, S., A. M. Pons, S. Gendron-Gaillard, and G. Cottenceau. 2000. Antibacterial activity evaluation of microcin J25 against diarrheagenic Escherichia coli. Appl. Environ. Microbiol. 66:4595-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salomon, R. A., and R. N. Farias. 1992. Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salomon, R. A., and R. N. Farias. 1993. The FhuA protein is involved in microcin 25 uptake. J. Bacteriol. 175:7741-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salomon, R. A., and R. N. Farias. 1995. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J. Bacteriol. 177:3323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savageau, M. A. 1974. Genetic regulatory mechanisms and the ecological niche of Escherichia coli. Proc. Natl. Acad. Sci. USA 71:2453-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seaver, L. C., and J. A. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seaver, L. C., and J. A. Imlay. 2004. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J. Biol. Chem. 279:48742-48750. [DOI] [PubMed] [Google Scholar]
- 50.Simon, E. H., and I. Tessman. 1963. Thymidine-requiring mutants of phage T4. Proc. Natl. Acad. Sci. USA 50:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solbiati, J. O., M. Ciaccio, R. N. Farias, and R. A. Salomon. 1996. Genetic analysis of plasmid determinants for microcin J25 production and immunity. J. Bacteriol. 178:3661-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Solbiati, J. O., M. Ciaccio, R. N. Farias, J. E. Gonzalez-Pastor, F. Moreno, and R. A. Salomon. 1999. Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J. Bacteriol. 181:2659-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincent, P. A., A. Bellomio, B. F. de Arcuri, R. N. Farias, and R. D. Morero. 2005. MccJ25 C-terminal is involved in RNA-polymerase inhibition but not in respiration inhibition. Biochem. Biophys. Res. Commun. 331:549-551. [DOI] [PubMed] [Google Scholar]
- 55.Vincent, P. A., M. A. Delgado, R. N. Farias, and R. A. Salomon. 2004. Inhibition of Salmonella enterica serovars by microcin J25. FEMS Microbiol. Lett. 236:103-107. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, K. A., M. Kalkum, J. Ottesen, J. Yuzenkova, B. T. Chait, R. Landick, T. Muir, K. Severinov, and S. A. Darst. 2003. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J. Am. Chem. Soc. 125:12475-12483. [DOI] [PubMed] [Google Scholar]
- 57.Yuzenkova, J., M. Delgado, S. Nechaev, D. Savalia, V. Epshtein, I. Artsimovitch, R. A. Mooney, R. Landick, R. N. Farias, R. Salomon, and K. Severinov. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]