Abstract
Over 90% of Methanothermobacter thermautotrophicus mutants isolated as spontaneously resistant to 5-methyl tryptophan had mutations in trpY. Most were single-base-pair substitutions that identified separate DNA- and tryptophan-binding regions in TrpY. In vivo and in vitro studies revealed that DNA binding was sufficient for TrpY repression of trpY transcription but that TrpY must bind DNA and tryptophan to assemble a complex that represses trpEGCFBAD.
Tryptophan biosynthesis is metabolically very expensive, and expression of trp genes that encode enzymes that catalyze tryptophan biosynthesis is always very tightly regulated (25). The trp genes in Bacteria, Archaea, and eukaryotes have a common ancestry, but very different mechanisms regulate their expression. Investigations of trp gene regulation have, in fact, led to our current understanding of many of the regulatory mechanisms that control gene expression in Bacteria and eukaryotes (2, 5, 13, 23, 25, 26). With this in mind, we have chosen to determine how trp gene expression is regulated in an archaeon. We have established that in Methanothermobacter thermautotrophicus, the trpEGCFBAD biosynthetic operon is transcribed divergently from trpY (Fig. 1A), a gene that encodes a tryptophan-sensing regulator that binds to TRP-box sequences (consensus, 5′-TGTACA [8]) located between trpY and trpEGCFBAD (24). In the absence of tryptophan, TrpY autorepresses trpY transcription, but in the presence of tryptophan, TrpY also represses trpEGCFBAD transcription. We predicted, therefore, that TrpY has separate DNA- and tryptophan-binding domains and that tryptophan binding modulates DNA binding, directly and/or via TrpY-TrpY interactions. Consistent with this, the structure predicted for TrpY has both an α helix-turn-α helix (HTH) DNA-binding domain (3) and a small molecule-binding ACT domain (1). While there is no precedent for an ACT domain that binds tryptophan, most ACT domains do bind amino acids and binding allosterically regulates metabolic enzymes and transcription factors (4, 6, 10, 16). To investigate TrpY further, we isolated 5-methyl tryptophan (5MT)-resistant mutants of M. thermautotrophicus based on the observation that two 5MT-resistant mutants of a close relative, Methanothermobacter marburgensis, appeared to be defective in TrpY repression of trpEGCFBAD expression (7). Sequencing of the trpY gene amplified from 100 spontaneously 5MT-resistant mutants has confirmed the specificity of this selection for mutations in trpY, and as described below, TrpY variants that are defective in DNA and/or tryptophan binding have been obtained. In vivo and in vitro assays of these variants have confirmed that TrpY must bind DNA and tryptophan to repress trpEGCFBAD transcription but have also revealed that this is not per se sufficient for repression.
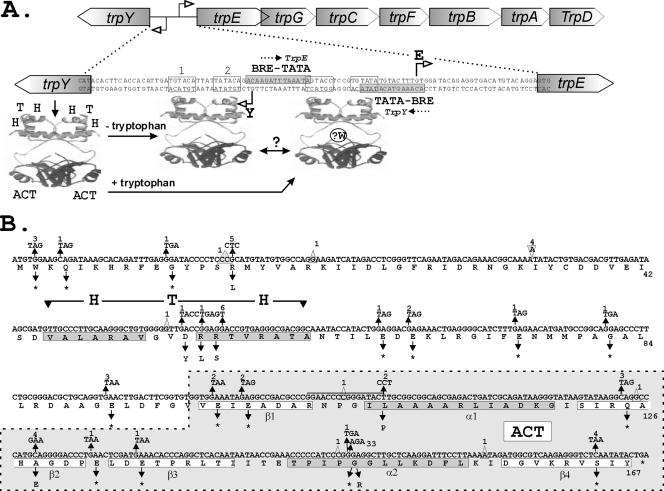
FIG. 1.
Organization of the trpY-trpEGCFBAD region and TrpY variants. (A) Sequence of the intergenic region separating trpY and trpE, with the TFB-responsive element (BRE) and TATA box sequences (shaded) that direct trpY and trpEGCFBAD transcription initiation at the sites identified by open arrows. In the absence of tryptophan, TrpY binds to the TRP boxes, designated 1 and 2, and inhibits trpY transcription (8). When complexed with tryptophan, TrpY binds to additional sites and also inhibits trpEGCFBAD transcription (24). The protein structure shown (PDB1I1G) is that of LrpA, a transcription regulator from Pyrococcus furiosus (6) that is provided to illustrate the proposed separation of DNA- and tryptophan-binding domains in TrpY. LrpA does have an ACT-related RAM domain (6) but otherwise has no recognizable relationship to TrpY. Repression of trpEGCFBAD requires a step in addition to DNA and tryptophan binding, most likely a TrpY-TrpY interaction, as indicated (?). The site(s) of tryptophan binding (?W) in TrpY also remains to be determined. (B) Mutations in trpY and residue substitutions in TrpY (*, translation stop) resulting from selection for 5MT resistance. Residues predicted to form the α helices of the DNA-binding HTH domain (3) and the α-helical and β-sheet regions (β1, α1, β2, β3, α2, β4) of the ACT domain (1, 4, 10) are boxed. The number of times each mutation occurred in the 100 trpY genes amplified from 5MT-resistant mutants is shown above the mutation. Deletion (Δ) and insertion (∇) mutations are identified with the deleted/inserted residue(s) inside the symbol. In two mutants, a 16-bp sequence (indicated by a line above the sequence) was tandem duplicated.
Selection and sequences of mutations in trpY.
Aliquots (100 μl) of four cultures of M. thermautotrophicus cultures grown independently from single-colony inocula to late exponential phase, in a minimal salts medium at 65°C (15), were diluted into 3 ml of the same medium held at 70°C that contained 10 μM 5MT and 1.5% agar. The cell suspensions were poured onto the surface of plates that contained the same agar-solidified minimal medium plus 10 μM 5MT. After the overlays had solidified, the plates were incubated at 65°C for 5 days in sealed containers pressurized to 2 × 105 Pa with 89% H2 and 11% CO2. M. thermautotrophicus mutants resistant to 5MT formed colonies at a frequency of approximately one per 107 plated cells, based on direct cell counts. Individual colonies were restreaked on 5MT-containing medium, and genomic DNA was isolated from 100 reconfirmed 5MT-resistant mutants. Oligonucleotides with the M. thermautotrophicus genome sequences from 1518157 to 1518180 and 1519126 to 1519150 (21) were used as primers to PCR amplify the trpY gene from each genomic DNA and to sequence both strands of the amplified DNA. A mutation(s) was present in 91 of the 100 amplified trpY genes, amounting to 31 different mutations (Fig. 1B). There were 22 single-base-pair substitutions (BPS), 6 single-base-pair deletions, one 2-bp deletion, one 1-bp insertion, and one 16-bp duplication. This is a very different spectrum of mutations from those reported for spontaneous mutations in pyrE and pyrF that confer 5-fluoroorotic acid resistance on Sulfolobus acidocaldarius (12) and Sulfolobus solfataricus (18). In S. acidocaldarius and S. solfataricus, only 12 of 101 and 9 of 111 mutations were BPS, respectively. The majority of the spontaneous mutations in S. acidocaldarius were short deletions or insertions, whereas IS element insertions predominated in S. solfataricus (11, 12, 18).
The same mutation, a G-C to A-T transition, was present in 33 of the amplified trpY genes. This changed codon 149 from GGA to AGA and so resulted in a TrpY variant with a glycine-to-arginine substitution, designated TrpYG149R. Some of the 5MT-resistant colonies could have originated from 5MT-resistant sibling cells, but in each separate repetition of the selection procedure, ∼35% of the trpY genes amplified from separate colonies had this mutation. Such G-C to A-T transitions likely result from cytosine deamination followed by replication of the resulting G-U base pairs, although M. thermautotrophicus does have an endonuclease that removes deoxyuridines from DNA (9). Sixteen of the BPS were G-C to T-A transversions, and 14 of these were T-for-G substitutions in the DNA strand shown in Fig. 1B, despite Gs on this strand outnumbering Gs on the opposite strand by only 155 to 114. This strand bias could reflect the trpY sequence (MTH1654) shown in Fig. 1B being on the lagging strand for DNA replication (22) or being transcribed in the opposite direction from chromosome replication (21).
Most of the BPS and all of the deletions and insertions resulted in premature nonsense codons in trpY and encoded truncated TrpY variants that were therefore nonfunctional and/or too unstable in vivo to maintain trp operon repression. At seven locations, missense mutations in trpY conferred 5MT resistance (Fig. 1B), specifically in three adjacent codons that changed residues within the predicted HTH DNA-binding motif (D54Y, R55L, and R56S), in three codons that changed residues in different regions of the ACT domain (L109P in α1, A128E in β2, and G149R in α2), and in codon 15, which removed a positive charge (R15L) near the N terminus.
Synthesis and purification of recombinant TrpY and TrpY variants.
trpY (MT1654) and the mutated trpY genes encoding TrpYR15L, TrpYD54Y, TrpYR55L, TrpYR56S, TrpYL109P, TrpYA128E, and TrpYG149R were PCR amplified from genomic DNA using primers (sequences available from K.S.) that added flanking NdeI and BamHI sites that facilitated cloning of the amplified DNA into NdeI- and BamHI-digested pKS773, a derivative of pT7-7 (U.S. Biochemicals). Eight in-frame codons were added to the cloned trpY genes that encoded a C-terminal GSHHHHHH (His tag) sequence. These plasmids were transformed into Escherichia coli Rosetta (DE3) (Novagen), and synthesis of the encoded recombinant wild-type TrpY (TrpYWT) and TrpY variants was induced in cultures growing in Luria-Bertani medium by the addition of isopropyl-β-d-thiogalactopyranoside (1 mM final concentration). After continued incubation at 37°C for 3 h, cells were harvested by centrifugation, resuspended in TSG buffer (0.1 M Tris-HCl [pH 8], 0.15 M NaCl, 15% [vol/vol] glycerol) at ∼0.2 g wet weight/ml, and lysed by passage through a French pressure cell at 20,000 lb/in2. The resulting lysates, clarified by centrifugation at 4°C for 30 min at 30,000 × g and then for 90 min at 114,000 × g, were loaded on to a Co2+-conjugated chelating column (Pharmacia). Bound proteins were washed with TSG containing 75 mM imidazole and then eluted with TSG containing 50 mM EDTA (pH 8). The presence of TrpYWT or a TrpY variant in eluted fractions was confirmed by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. These fractions were pooled and loaded on to a heparin column (Pharmacia) equilibrated in TSG. Proteins that bound were washed with TSG and then eluted with a 0.15 to 0.5 M NaCl gradient dissolved in 0.1 M Tris-HCl (pH 8)-15% (vol/vol) glycerol. Fractions that contained only TrpYWT or a TrpY variant, based on the absence of additional Coomassie blue-staining bands after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were pooled, concentrated, and quantified by Bradford assays.
The effector ligands in phosphoglycerate dehydrogenase and aspartate kinase, serine and lysine, respectively, are bound between the β1 and α2 elements of ACT domains (10, 14, 20). Site-specific mutagenesis (Quik Change; Stratagene) was therefore used to change the trpY sequence in pKS772 to obtain TrpY variants (TrpYD102A, TrpYN105A, and TrpYI108A) with residue changes in the β1-α2 region of TrpY (Fig. 1B). The only tryptophan in TrpY is at position 2, and translation of this codon may play a sensing role in determining trp operon expression (24). We therefore used site-directed mutagenesis to delete and change codon 2 to obtain TrpYW2Δ, TrpYW2A, and TrpYW2F. TrpYS14A was generated to obtain a variant with a residue substitution directly adjacent that in TrpYR15L selected by 5MT resistance, and TrpYI123A changed a residue highly conserved in the β2 region of ACT domains. All of these TrpY variants were generated in E. coli and purified as described above.
DNA binding by TrpY variants.
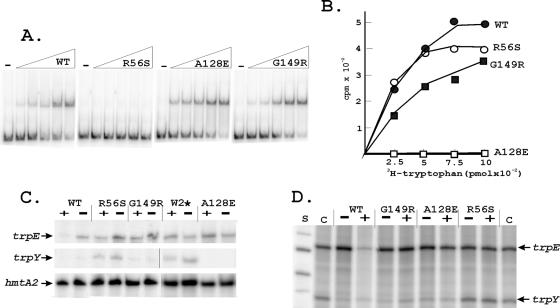
We established previously that TrpY binds to TRP-box sequences located between trpY and trpE (Fig. 1A) (24). This intergenic region (102 bp) plus 41 bp from the 5′ region of trpY was therefore amplified, with flanking NcoI sites that facilitated cloning into NcoI-digested pLITMUS28 (New England Biolabs), for use as the substrate in DNA-binding assays. One plasmid (pKS795) had two tandem copies of the desired DNA. This was amplified, and preparative amounts of the DNA were obtained by NcoI digestion. The single-stranded NcoI ends were partially filled and 32P labeled by incubation with [32P]dATP, dCTP, and Klenow DNA polymerase. DNA-binding reaction mixtures (15 μl) contained the substrate DNA (0.5 ng), poly(dI·dC) (50 ng), and increasing amounts of TrpY or a TrpY variant in transcription buffer (20 mM Tris [pH 8], 7.5 mM MgCl2, 5 mM dithiothreitol, 120 mM KCl [19]). The complexes formed were separated by electrophoresis through nondenaturing 8% (wt/vol) polyacrylamide gels and visualized by phosphorimaging (Storm 840; Pharmacia). TrpYWT bound and retarded the mobility of the DNA, giving a gel shift, as previously documented (24), and essentially identical gel shifts were obtained with TrpYWΔ2, TrpYS14A, TrpYD102A, TrpYN105A, TrpYI108A, TrpYI123A, TrpYA128E, and TrpYG149R (illustrated in Fig. 2A by TrpYA128E and TrpYG149R). Gel shifts were also obtained with TrpYW2A, TrpYW2F, TrpYD54Y, and TrpYL109P, although only at higher protein-to-DNA ratios than with TrpYWT, and the complexes formed by TrpYD54Y had a different mobility from those formed by TrpYWT. In contrast, gel shifts were never observed with TrpYR56S (Fig. 2A), TrpYR15L, or TrpYR55L regardless of the protein-to-DNA ratio. For TrpYR55L and TrpYR56S, the loss of DNA-binding ability was consistent with the prediction that R55 and R56 are components of an HTH DNA-binding motif (Fig. 1B). The DNA-binding proficiency of TrpYA128E and TrpYG149R demonstrated that a loss of DNA binding was not the only TrpY defect that resulted in loss of trp operon repression.
FIG. 2.
DNA and tryptophan binding and transcription regulation by TrpY and TrpY variants. (A) Electrophoretic mobility shift assays of TrpYWT and TrpY variants binding to the trpY-trpE intergenic region. Reaction mixtures contained 32P-labeled DNA (0.5 ng), 50 ng poly(dI·dC) and 0 (−), 1.25, 2.5, 5, 10, or 20 ng of TrpYWT or a TrpY variant. (B) [3H]tryptophan binding by TrpYWT and TrpY variants. (C) Primer extension products generated by reverse transcription of the trpY, trpEGCFBAD (trpE), and hmtA2 transcripts in RNA preparations (17) from wild-type (WT) and TrpY variant-containing M. thermautotrophicus cells grown with (+) or without (−) tryptophan. (D) Runoff trpY and trpE transcripts synthesized in vitro from a template DNA (previously designated T1 [24]) with the entire trpY-trpE intergenic region. Reaction mixtures contained 200 μM (each) ATP, CTP, and GTP; 20 μM UTP plus 10 μCi of [α-32P]UTP; 40 nM M. thermautotrophicus RNA polymerase; 80 nM TFB; 80 nM TBP; and 100 nM TrpYWT or the TrpY variant listed, with (+) or without (−) 24 μM tryptophan (18). After incubation at 60°C for 30 min, the accumulated transcripts were separated by gel electrophoresis and visualized by phosphorimaging, as described in detail previously (24). Control (C) lanes contained the transcripts synthesized in reaction mixtures with no TrpY or TrpY variant added. Single-stranded DNA molecules provided size standards (S).
Tryptophan binding by TrpY variants.
To assay tryptophan binding, TrpYWT or a TrpY variant (50 pmol) was mixed with [3H]-l-tryptophan (0.25 to 1 nmol, 0.128 μCi/nmol; Amersham), and the reaction mixture (100 μl) was then slowly filtered through a 0.45-μm HAWP filter (Millipore) presaturated with 100 μM unlabeled l-tryptophan. The filters were washed twice with transcription buffer and dried, and the [3H]tryptophan retained on the filters was determined by scintillation counting. TrpYWT, TrpW2Δ, TrpYS14A, TrpYR15L, TrpYD54Y, TrpYR55L, TrpYR56S, and TrpYG149R bound tryptophan similarly (Fig. 2B). TrpYD102A and TrpYI123A also bound tryptophan, although with lower affinity than TrpYWT. In contrast, TrpYW2A, TrpYW2F, TrpYI108A, TrpYL109P, and TrpYA128E did not bind tryptophan (Fig. 2B), and TrpYN105A bound tryptophan with higher affinity than TrpYWT, consistent with the β1-α1 region of the ACT domain (residues 105 to 107 in TrpY [Fig. 1B] [10]) being critical for ligand binding. Other than long-range folding differences, it is unclear why residue substitutions for W2 (TrpYW2A, TrpYW2F) would reduce tryptophan binding when deletion of W2 (TrpYW2Δ) had no detrimental effect on tryptophan binding.
The properties of TrpYA128E, the ability to bind DNA (Fig. 2A) but inability to bind tryptophan (Fig. 2B), are consistent with a TrpY-tryptophan complex being required for trpEGCFBAD repression (Fig. 1A). However, as TrpYG149R binds both DNA and tryptophan (Fig. 2A and B), these binding abilities alone are not apparently sufficient for trpEGCFBAD repression.
Regulation of trpY and trpEGCFBAD transcription in vivo.
To confirm that the trp operon was, in fact, derepressed in M. thermautotrophicus 5MT-resistant mutants, RNA preparations were isolated (17) from cultures grown to late exponential phase in minimal medium with or without 5 mM tryptophan. 32P-labeled oligonucleotide primers (sequences available from L.C.) complementary to sequences near the 5′ termini of the trpY, trpEGCFBAD, and hmtA2 transcripts (a constitutively expressed archaeal histone-encoding gene [12]) were incubated with the RNAs for 10 min at 65°C in 0.2 M Tris-HCl-2 M NaCl-5 mM EDTA (pH 7.5). Hybridized primers were extended by SuperscriptII reverse transcriptase (200 U; Invitrogen) in reaction mixtures that contained 1 mM deoxynucleoside triphosphates, 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 10 mM dithiothreitol and that were incubated for 60 min at 42°C. The 32P-labeled extension products were separated by electrophoresis and visualized by phosphorimaging (24). In all RNA preparations, the hmtA2 transcript was readily detectable, whereas the trpY and trpE transcripts were below detectable levels in RNA preparations from wild-type M. thermautotrophicus cells grown with tryptophan. The trpY transcript was also barely detectable in RNA preparations from wild-type cells grown without tryptophan, but these RNAs did contain much higher levels of trpE, consistent with trpEGCFBAD derepression (Fig. 2C). Both the trpY and trpE transcripts were abundant in all RNA preparations isolated from 5MT-resistant mutants that lacked TrpY (TrpYW2*) or contained a variant (TrpYR56S) that lacked DNA-binding ability. TrpY binding in wild-type cells apparently therefore maintains trpY transcription below the fully derepressed level that occurs in M. thermoautotrophicum mutants that entirely lack TrpY, regardless of tryptophan availability. RNA preparations from mutants containing TrpYA128E or TrpYG149R, grown with or without tryptophan, contained the trpE transcript but barely detectable levels of the trpY transcript. These TrpY variants retain the ability to bind DNA (Fig. 2A), and this ability was apparently sufficient to repress trpY transcription in vivo but insufficient for trp operon repression. TrpYA128E has lost the ability to bind tryptophan (Fig. 2B), and the presence of trpE transcripts in cells containing TrpYA128E was consistent with trp operon repression requiring a TrpY-tryptophan complex. However, as TrpYG149R retains both DNA- and tryptophan-binding ability, the presence of trpE transcripts in cells containing TrpYG149R argues that a reaction, in addition to DNA and tryptophan binding, is required for TrpY repression of trpEGCFBAD transcription.
TrpY regulation of trpY and trpE transcription in vitro.
TrpY was shown previously to regulate trpY and trpE transcription in vitro, as observed in vivo, in reaction mixtures that contain the intergenic region as template DNA plus M. thermautotrophicus RNA polymerase and the basal transcription factors TATA box-binding protein (TBP) and transcription factor B (TFB) (24). As shown in Fig. 2D, TrpYR56S, TrpYA128E, and TrpYG149R also regulated trpY and trpE transcription in vitro, as documented in vivo (Fig. 2C). TrpYWT, TrpYA128E, and TrpYG149R inhibited trpY transcription in the absence or presence of tryptophan, but only TrpYWT inhibited trpE transcription in the presence of tryptophan. Consistent with the loss of DNA-binding ability, the addition of TrpYR56S had no effect on trpY or trpE transcription in vitro.
Conclusions.
The results reported add to our knowledge of both the nature of spontaneous mutations and the mechanics of trp gene regulation in a thermophilic euryarchaeon. As almost all (>90%) of the 5MT-resistant mutants of M. thermautotrophicus had mutations in trpY, this selection is remarkably gene specific and so should be very useful for archaeal mutagenesis studies. In contrast to the dominance of insertions and deletions that inactivated pyrE and pyrF in spontaneous mutants of the crenarchaeal Sulfolobus species (11, 12, 18), most of the spontaneous mutations in trpY in M. thermautotrophicus were single BPS.
The properties of the TrpY variants generated have confirmed the presence of DNA- and tryptophan-binding domains in this archaeal regulator and are consistent with TrpY and TrpY-tryptophan complexes repressing trpY and trpEGCFBAD transcription by binding to the intergenic region, as shown in Fig. 1A (24). TrpY binding to this region is apparently sufficient for trpY repression, whereas DNA and tryptophan binding and an additional reaction are required for repression of trpEGCFBAD transcription. Establishing the nature of the additional event, most likely a higher-order interaction of TrpY-tryptophan complexes, should be facilitated by investigations of TrpYG149R, as this variant retains the ability to bind DNA and tryptophan but lacks the ability to repress trpEGCFBAD transcription.
Acknowledgments
This research was supported by grant DE-FG02-87ER13731 from the Department of Energy (to J.N.R.) and by an NSF Postdoctoral Fellowship (0400072) awarded to E.A.K. We thank T. J. Santangelo, R. Samson, and M. Nelson for advice and for help with in vitro transcription, RNA purification, and the isolation of 5MT-resistant mutants, respectively.
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1999. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287:1023-1040. [DOI] [PubMed] [Google Scholar]
- 2.Babitzke, P. 2004. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr. Opin. Microbiol. 7:132-139. [DOI] [PubMed] [Google Scholar]
- 3.Brandon, C., and J. Tooze. 1991. Introduction to protein structure. Garland Publishing, Inc., London, United Kingdom.
- 4.Chipman, D. M., and B. Shaanan. 2001. The ACT domain family. Curr. Opin. Struct. Biol. 11:694-700. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, I. P. 1989. Evolution of a biosynthetic pathway: the tryptophan paradigm. Annu. Rev. Microbiol. 43:567-600. [DOI] [PubMed] [Google Scholar]
- 6.Ettema, T. J. G., A. B. Brinkman, T. H. Tani, J. B. Rafferty, and J. van der Oost. 2002. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 277:37464-37468. [DOI] [PubMed] [Google Scholar]
- 7.Gast, D. L., A. Wasserfallen, P. Pfister, S. Ragettli, and T. Leisinger. 1997. Characterization of Methanobacterium thermoautotrophicum Marburg mutants defective in regulation of l-tryptophan biosynthesis. J. Bacteriol. 179:3664-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelfand, M. S., E. V. Koonin, and A. A. Mironov. 2000. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 28:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georg, J., L. Schomacher, J. P. J. Chong, A. I. Majernik, M. Raabe, H. Urlaub, S. Müller, E. Ciirdaeava, W. Kramer, and H.-J. Fritz. 2006. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res. 34:5325-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant, A. G. 2006. The ACT domain: a small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 281:33825-33829. [DOI] [PubMed] [Google Scholar]
- 11.Grogan, D. W. 2004. Stability and repair of DNA in hyperthermophilic Archaea. Curr. Issues Mol. Biol. 6:137-144. [PubMed] [Google Scholar]
- 12.Grogan, D. W., G. T. Carver, and J. W. Drake. 2001. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 98:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutter, R., P. Niederberger, and J. A. DeMoss. 1986. Tryptophan biosynthetic genes in eukaryotic microorganisms. Annu. Rev. Microbiol. 40:55-77. [DOI] [PubMed] [Google Scholar]
- 14.Mas-Droux, C., G. Curien, M. Robert-Genthon, M. Laurencin, J.-L. Ferrer, and R. Dumas. 2006. A novel organization of ACT domains in allosteric enzymes revealed by the crystal structure of Arabidopsis aspartate kinase. Plant Cell 18:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nölling, J., T. D. Pihl, A. Vriesema, and J. N. Reeve. 1995. Organization and growth phase-dependent transcription of methane genes in two regions of the Methanobacterium thermoautotrophicum genome. J. Bacteriol. 177:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittard, J., H. Camakaris, and J. Yang. 2005. The TyrR regulon. Mol. Microbiol. 55:16-26. [DOI] [PubMed] [Google Scholar]
- 17.Prætorius-Ibba, M., T. E. Rogers, R. Samson, Z. Kelman, and M. Ibba. 2005. Association between archaeal prolyl- and leucyl-tRNA synthetases enhances tRNAPro aminoacylation. J. Biol. Chem. 280:26099-26104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redder, P., and R. A. Garrett. 2006. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 188:4198-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santangelo, T. J., and J. N. Reeve. 2006. Archaeal RNA polymerase is sensitive to intrinsic termination directed by transcribed and remote sequences. J. Mol. Biol. 355:196-210. [DOI] [PubMed] [Google Scholar]
- 20.Schuller, D., G. A. Grant, and L. Banaszak. 1995. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat. Struct. Biol. 2:69-76. [DOI] [PubMed] [Google Scholar]
- 21.Smith, D. R., L. A. Doucette-Stamm, C. DeLoughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrokovski, G. Church, C. J. Daniels, J. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. The complete genome sequence of Methanobacterium thermoautotrophicum strain ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worning, P., L. J. Jensen, P. F. Hallin, H.-H. Stærfeldt, and D. W. Ussery. 2006. Origin of replication in circular prokaryotic chromosomes. Environ. Microbiol. 8:353-361. [DOI] [PubMed] [Google Scholar]
- 23.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67:303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie, Y., and J. N. Reeve. 2005. Regulation of tryptophan operon expression in the archaeon Methanothermobacter thermautotrophicus. J. Bacteriol. 187:6419-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanofsky, C. 2003. Using studies of tryptophan metabolism to answer basic biology questions. J. Biol. Chem. 278:10859-10878. [DOI] [PubMed] [Google Scholar]
- 26.Yanofsky, C. 2004. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 20:367-374. [DOI] [PubMed] [Google Scholar]