Abstract
Individual gene-targeted hpn and hpn-like mutants and a mutant with mutations in both hpn genes were more sensitive to nickel, cobalt, and cadmium toxicity than was the parent strain, with the hpn-like strain showing the most metal sensitivity of the two individual His-rich protein mutants. The mutant strains contained up to eightfold more urease activity than the parent under nickel-deficient conditions, and the parent strain was able to achieve mutant strain activity levels by nickel supplementation. The mutants contained 3- to 4-fold more and the double mutant about 10-fold more Ni associated with their total urease pools, even though all of the strains expressed similar levels of total urease protein. Hydrogenase activities in the mutants were like those in the parent strain; thus, hydrogenase is fully activated under nickel-deficient conditions. The histidine-rich proteins appear to compete with the Ni-dependent urease maturation machinery under low-nickel conditions. Upon lowering the pH of the growth medium from 7.3 to 5, the wild-type urease activity increased threefold, but the activity in the three mutant strains was relatively unaffected. This pH effect was attributed to a nickel storage role for the His-rich proteins. Under low-nickel conditions, the addition of a nickel chelator did not significantly affect the urease activity of the wild type but decreased the activity of all of the mutants, supporting a role for the His-rich proteins as Ni reservoirs. These nickel reservoirs significantly impact the active urease activities achieved. The His-rich proteins play dual roles, as Ni storage and as metal detoxification proteins, depending on the exogenous nickel levels.
Helicobacter pylori is a well-studied, gram-negative, motile, microaerophilic pathogen that colonizes the human gastric mucosa and causes chronic gastritis, duodenal ulcers, and certain kinds of gastric cancers (4, 18, 22, 23). H. pylori produces the Ni enzymes urease and hydrogenase (10, 17). Urease accounts for up to 10% of the total cellular protein made and is essential for colonization and virulence (1, 13, 32, 33). The hydrogen-utilizing hydrogenase provides the bacterium an alternative respiratory-based energy generation mechanism independent of carbon substrates (17). These two enzymes represent the major final sinks for nickel within the cell. Other than the nickel enzymes, H. pylori also possesses a NixA nickel-specific permease (2); accessory proteins, UreEFGH and HypABCDEF (some of which bind nickel), required for proper maturation of the two nickel enzymes (19, 30); a nickel-dependent regulator, NikR (6, 8, 35, 36); a recently identified nickel efflux system (CznABC) (28); and a histidine-rich heat shock protein, HspA, which is a GroES homologue (14).
A protein rich in histidine residues (47% histidine residues; TIGR annotation HP1427) was named Hpn because it was first identified in H. pylori and had affinity for nickel ions (12). The presence of Hpn was also noted in the ferret and cat gastric pathogens Helicobacter mustelae and Helicobacter felis, respectively (12). H. pylori mutants lacking hpn were less tolerant to nickel and bismuth than was the wild type (20). Recent biophysical characterizations showed that Hpn exists mainly as a multimer in solution, with each monomer of 7 kDa reversibly binding five nickel ions at pH 7.4. Nickel is released by a decrease in pH or in the presence of nickel-chelating agents such as EDTA (11). The level of nickel in the H. pylori strain 26695 cytoplasm was also found to be slightly higher than in an hpn deletion mutant (11), and nickel levels in Escherichia coli cells expressing H. pylori Hpn from an inducible plasmid were higher than in cells lacking the plasmid (11). No difference in urease activity in the hpn mutant compared to the wild type was noted (11, 12).
The H. pylori sequence reveals the presence of another His-rich protein, termed the “Hpn-like protein,” which is annotated as a histidine- and glutamine-rich protein (HP1432) (30). Sequence analysis indicates that 25% of the amino acids are histidine residues, including a stretch of six consecutive histidine residues, while 30 of the 72 amino acid residues are glutamine residues (42%). The N-terminal sequence (46 residues) of HP1432 shows 56% identity to Hpn; the protein was thus termed Hpn-like. The H. pylori 43504 Hpn-like protein is slightly different from HP1432 and JHP1321. It was sequenced, and the GenBank accession number is EF203427.
Both the Hpn and Hpn-like proteins are transcriptionally activated in the presence of nickel by the nickel sensor NikR (6). Additionally, the Hpn-like protein was shown to be upregulated at pH 5.0, in comparison to pH 7.0, by the two-component system ArsRS (HP165-HP166) (5, 24, 25). Studies thus far suggest that Hpn may be involved in nickel detoxification and/or nickel storage for use at nickel-deficient times. Our results extend a role for detoxification to the Hpn-like protein and indicate that both proteins play a role in storing nickel that significantly affects urease expression when nickel supplies are highly limited. Under low-nickel conditions, chelation of extracellular nickel significantly decreased the urease activities in the mutant strains but not in the wild type, supporting a nickel storage role for these proteins. Also, the results herein imply that H. pylori synthesizes much more (apo)urease than its nickel reserves can satisfy.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are described in Table 1. H. pylori ATCC strain 43504 was used as the wild-type strain and for mutant strain construction. This strain was used as the parent strain, as many nickel metabolism mutant strains are in this genetic background (3, 21). H. pylori cells were grown on brucella agar plates supplemented with 10% defibrinated sheep blood (BA) at 37°C. The plates were supplemented with either 30 μg ml−1 of chloramphenicol or 30 μg ml−1 of kanamycin when needed. Cultures were grown in a 5% CO2 incubator with 4% O2 or were grown in anaerobic jars with anaerobic mix (5% CO2, 10% H2, balance N2).
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Genotype, description, or sequence | Source or referencea |
|---|---|---|
| Helicobacter pylori strains | ||
| 43504 (wild type) | H. pylori ATCC 43504 | ATCC |
| hpn mutant | Kanr; 43504 with hpn kanamycin resistance cassette | This study |
| hpn-like mutant | Cmr; 43504 with hpn-like chloramphenicol resistance cassette | This study |
| Double mutant | Kanr Cmr; 43504 with hpn kanamycin resistance cassette and hpn-like chloramphenicol resistance cassette | This study |
| Escherichia coli DH5α | Cloning strain | Invitrogen |
| Plasmids | ||
| pHP1 | Plasmid containing the kanamycin resistance gene aphA3 | 15 |
| pUC20cat | Plasmid containing the chloramphenicol acetyltransferase gene cat | 37 |
| pGEM-T | Ampr; cloning vector | Promega |
| pGEM-hpn | pGEM-T vector with the hpn gene from H. pylori 43504 | This study |
| pGEM-hpn-like | pGEM-T vector with the hpn-like gene from H. pylori 43504 | This study |
| pGEM-hpn: kanR | pGEM-hpn with the Kanr cassette inserted into the XcmI site of hpn | This study |
| pGEM-hpn-like: cmR | pGEM-hpn-like with the Cmr cassette inserted into the BlpI site of the hpn-like gene | This study |
| Primers | ||
| HPN-F | 5′-TGTTGCGGTTGGATTGG-3′ | IDT |
| HPN-R | 5′-CAAGTGGGTTGCTCGTTTTGTTTC-3′ | IDT |
| 1432-F | 5′-AGCCAACGCCTTTTCTTTCAG-3′ | IDT |
| 1432-R | 5′-TTTTACACCCCATTACGACCACTC-3′ | IDT |
IDT, Integrated DNA Technologies, Inc., Coralville, IA.
For liquid cultures, cells were inoculated at an optical density at 600 nm of 0.07 into serum vials containing 20 ml of brain heart infusion (BHI) broth supplemented with 5% fetal bovine serum. The bottles were sparged with anaerobic mix for 15 min, and 5% oxygen was added to the sealed bottles via a sterile needle and syringe. The cultures were grown at 37°C with shaking at 200 rpm. The pH of the culture medium was set to either 7.3 or 5.0 as desired; after growth, the pH was about 6.8 and 5.3, respectively. A nickel-free medium was obtained as described previously by the addition of dimethyl glyoxime (22).
Escherichia coli strain DH5α was grown at 37°C on Luria-Bertani plates supplemented with no antibiotic, 100 μg ml−1 ampicillin, 30 μg ml−1 of chloramphenicol, 30 μg ml−1 of kanamycin, or a combination of these antibiotics as required.
Mutant construction.
The sequence of hpn from H. pylori 43504 is identical to that of the hpn gene from the genome-sequenced strain H. pylori 26695. For the hpn-like gene, the sequence differs slightly. An approximately 1.2-kb region containing the hpn gene (183 bp) or the hpn-like gene (214 bp) and a 500-bp flanking region on either side was amplified by PCR using the primer pairs described in Table 1 and cloned into the pGEM-T vector to give plasmid pGEM-hpn or pGEM-hpn-like, respectively. A kanamycin resistance cassette, aphA3, was introduced into the XcmI site of the hpn gene, and a chloramphenicol acetyltransferase cassette (cat) was inserted into the BlpI site within the hpn-like gene. The plasmids containing the disrupted genes were introduced into H. pylori by natural transformation to promote allelic exchange via homologous recombination, and antibiotic resistance was used to select colonies that contained the disrupted genes. The insertion of the cassette in each gene was confirmed by PCR using DNA from each mutant as a template (data not shown). The double mutant was obtained by introducing the pGEM-hpn-like: cmR plasmid into the H. pylori hpn mutant background; colonies resistant to both antibiotic markers were isolated, and the presence of the antibiotic cassette in both loci was confirmed by PCR (data not shown).
Metal toxicity.
A starting cell suspension of ∼5 × 108 cells/ml of each strain was suspended in sterile phosphate-buffered saline (PBS) and was incubated with either 1 mM nickel chloride (NiCl2), 20 μM cobalt chloride (CoCl2), 10 μM cadmium chloride (CdCl2) or no metal. The tubes were sealed with stoppers and sparged with anaerobic mix (5% CO2, 10% H2, balance N2) by use of inflow and outflow needles; oxygen was then added from a 100% O2 tank via syringe to a final 4% partial pressure. The sealed test tubes were incubated at 37°C, and dilutions were plated from the suspensions at regular intervals over a 12-h period onto BA plates.
Urease activities.
Cell extracts from 2-day-old cultures grown on BA plates or on BA plates with the indicated nickel supplementation were used. When the urease activities of liquid cultures were assessed, 24-h-old cultures were centrifuged at 7,000 × g and urease assays were carried out. Briefly, cells were washed and resuspended in 50 mM HEPES buffer, pH 7.5. Supernatants from cells broken by two passages through a French pressure cell (12,000 lb/in2) were incubated for 20 min with the same buffer containing 25 mM urea, and the ammonia evolved was measured via a phenol hypochlorite assay as described previously (38). A standard ammonium chloride concentration curve was used to convert the absorbance at 625 nm to nanomoles of ammonia.
Urease immunoblots.
Crude cell extracts were treated with sodium dodecyl sulfate (SDS) buffer, boiled for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) according to the method of Laemmli (16). Separated proteins were transferred electrophoretically onto a nitrocellulose membrane, as described previously (31). Immunoblotting was carried out as described previously, with the only difference being that Anti-UreA antibody was used as the primary antibody at a dilution of 1:1,000 (19).
Nickel content in urease fractions.
Urease was purified as described previously (21), with the exception that YM10 centricons (Millipore, Bedford, MA) were used to concentrate the urease fractions and to exchange the high salt content in the elution buffer for a lower salt content. Purification was carried out with identical protocols for each strain, and the elution profiles obtained for each strain were identical (data not shown). The peak fractions were subjected to SDS-PAGE; the UreA and UreB bands were the most prominent bands from all strains and were judged to represent at least 95% of the total protein (data not shown). Equal protein amounts of each fraction were subjected to graphite furnace atomic absorption spectrophotometry, and the nickel content of the various samples was calculated as described previously (21). Briefly, this involved creating a standard curve with various known nickel concentrations (0 to 0.5 μM nickel) and measuring the nickel content of the various urease-containing samples within this standard curve. Where nickel was added to the growth medium, relatively high but not toxic levels (500 μM) were used, to attempt to obtain the greatest amount of Ni-activated enzyme. Levels as high as 2 mM have been previously used to study H. pylori Ni physiology (20).
RESULTS
Mutant construction.
To explore the roles of the hpn and hpn-like genes, we created individual gene disruption mutants in H. pylori ATCC strain 43504. Since the hpn and hpn-like genes may have overlapping roles in the cell, and the presence of one of the genes might mask the effects of disrupting the other, we also created a double mutant deficient in both hpn genes. The growth rates of the three mutants were determined in BHI medium supplemented with 5% fetal bovine serum by monitoring the absorbance at 600 nm over a 24-h period. No differences in growth rates were observed between the mutants and the parent, indicating that the disruption of the hpn genes did not affect products that are essential for growth (data not shown).
Reduced metal tolerance in the mutant strains.
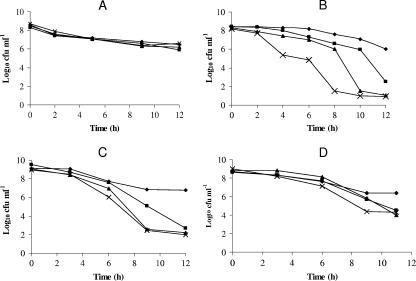
In order to examine the possible roles of the hpn and hpn-like genes in nickel detoxification, we determined the survival ability of the hpn, hpn-like, and hpn/hpn-like double mutant strains in the presence of added nickel in comparison to wild-type H. pylori. This was accomplished by suspending equal numbers of cells in nongrowing conditions with excess nickel (PBS with 1 mM NiCl2). Samples were taken at regular intervals, serially diluted, and plated onto BA plates. The colony counts were then determined, and the results are expressed as log10 CFU ml−1 (Fig. 1B). The results show that the nickel tolerance of the hpn mutant is less than that of the wild type. Decreased tolerance of the hpn mutant to nickel was also reported by Mobley et al. (20). We observed a greater Ni-dependent viability loss for the hpn-like mutant and the double mutant. The effects could be connected to nickel, as all four strains showed similar survival rates (in PBS) in the absence of nickel stress (Fig. 1A). Also, a chloride ion effect was ruled out, as CaCl2 (1 mM) did not cause a viability loss in the strains (data not shown). The nickel stress experiment suggests that along with Hpn, the Hpn-like protein also has a role in nickel detoxification in H. pylori.
FIG. 1.
Viability loss due to metal toxicity. Strains: H. pylori 43504 (♦), hpn (▪), hpn-like (▴), and the double-mutant strain (×). (A) Survival in PBS with no added metals. The experiment was performed in triplicate, and the data shown are means from all three data sets for each strain. No significant differences among strains were observed. (B) Survival in the presence of 1 mM NiCl2. The data are from four independent experiments, each one sampled three times; thus, each data point is the mean of 12 readings. The standard deviation ranged between 3 to 20% of the mean values; all hpn mutant strain results are significantly different (P < 0.05) from the wild type at 6 h and all later time points, while the other two mutant strains are significantly different from the wild type (P < 0.05) at 4 h and all later plating points. (C) Survival in the presence of 10 μM CdCl2. The data are from two independent experiments, each one sampled twice; thus, each point is the mean of four values. The values for the hpn-like mutant and the double mutant are significantly different (P < 0.05) from the wild type at 6 h and later, whereas the hpn mutant is different at the 9- and 12-h time points (P < 0.05). (D) Survival in the presence of 20 μM CoCl2. The data are from two independent experiments, each one sampled twice; thus, each point is the mean of four values. The His-rich protein mutants are significantly less tolerant than the wild type at the 9- and 11-h points (P < 0.05), and the double mutant is significantly less tolerant than the wild type at 6 h and later sampling points.
To determine whether the Hpn and Hpn-like proteins play a role in tolerance of other divalent metal ions, similar experiments were carried out with zinc (data not shown) and with cadmium and cobalt (Fig. 1C and D, respectively). As with nickel, loss of viability was observed for the hpn, hpn-like protein, and double mutants in the presence of cobalt and cadmium but not when exposed to excess zinc. The Hpn-like protein appears to play a larger role in metal resistance, at least for cadmium and nickel. Nevertheless, both Hpn and the Hpn-like protein play a role in nickel, cobalt, and cadmium detoxification.
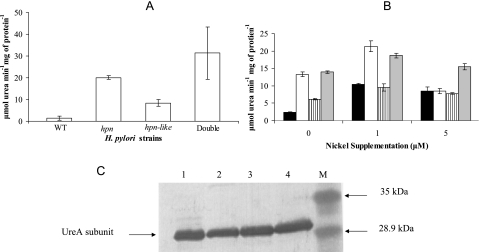
Urease activities are higher in the mutant strains.
To assess whether mutations in the putative nickel detoxification/nickel storage proteins have an effect on the major nickel sink in the cell (each UreB subunit can bind two Ni+ ions), the urease activity of the mutant strains was compared to that of the wild type (Fig. 2A). After growth in non-Ni-supplemented medium, urease activity was increased over wild-type levels by at least eightfold in the hpn mutant, threefold in the hpn-like mutant, and more than eightfold in the hpn/hpn-like double-mutant strain. Upon 1 μM nickel supplementation, urease activities of the mutants increased slightly, whereas the urease activities of the wild-type strain increased about fourfold with 1 μM or 5 μM nickel supplementation (compared to unsupplemented conditions [Fig. 2B]). Supplemental nickel commonly induces H. pylori urease expression (35, 36). The decreased activity in the hpn mutant at 5 μM compared to 1 μM is likely due to the increased susceptibility of this strain to nickel toxicity.
FIG. 2.
Urease activities and protein as a function of the nickel content of the medium. (A) Urease activities of cells grown without nickel supplementation on BA plates. The results shown are means and standard deviations of six replicate samples (these six represent three measurements from each of two independent cultures); the experiment was performed four additional times, with similar results. Among the five experiments, the double-mutant strain mean exceeded the Hpn strain in all cases, by a total range of 5 to 65%. All three mutant strains have significantly greater urease activities than do the wild type (P < 0.01), based on Student t test analysis. (B) Urease activities of cells grown with (1 or 5 μM NiCl2) or without nickel supplementation. Cells were grown on BA plates. The strains are as follows: wild-type H. pylori (black), the hpn strain (white), the hpn-like strain (striped), and the double-mutant strain (gray). The measurements were performed in triplicate (one culture assayed three times); the wild-type urease activities are greater under Ni supplementation conditions than without Ni supplementation (P < 0.01), based on Student t test analysis, and the mutant strain activities exceed the wild-type activity under zero nickel conditions (P < 0.05). The entire experiment was done two other times, with results similar to those shown. (C) Anti-UreA immunoblots performed on gel-resolved peptides of crude cell extracts from cells grown in non-Ni-supplemented medium. Crude extracts were loaded (5 μg protein) with the double-mutant (lane 1), hpn-like (lane 2), hpn (lane 3), or wild type (lane 4) strain. Lane M is the low-range prestained molecular mass marker, with sizes indicated on the right. Densitometric scanning of each lane revealed no significant differences in (UreA) protein amounts between strains.
As a nickel storage role is ascribed to these His-rich proteins, we presumed that gene disruption may display a urease-deficient phenotype. Thus, the observed urease activity results were not expected. The higher urease activities in the mutants could be due to the availability of a larger cytoplasmic nickel pool for the urease maturation enzymes; this would create a higher amount of active urease species in comparison to the (nickel-deficient) wild-type strain. Alternatively, the increase in urease activities could be due to Hpn and the Hpn-like protein affecting (increasing) urease transcription. These possibilities were explored by comparing urease protein levels and nickel levels in the urease fractions from various strains.
Hydrogenase activity is not affected in the mutant strains.
Hydrogenase is the other nickel-containing enzyme in H. pylori (17). To assess whether mutations in the hpn and hpn-like genes were affecting the hydrogenase activities in the cell, hydrogenase assays were performed amperometrically by detecting the hydrogen uptake activities in whole cells, as previously described (17). Significant differences in hydrogenase activities were not observed in the various mutant strains compared to the wild-type strain (data not shown). Hydrogenase is a minor Ni sink compared to urease, as it is made in much lesser amounts and contains much less nickel per enzyme molecule than does urease. Also, the two Ni enzyme sinks (hydrogenase and urease) contain different accessory maturation proteins; these would be expected to affect nickel allocation between the two Ni enzymes based on their individual affinities for the metal. The absence of a hydrogenase phenotype effect by loss of the nickel-sequestering proteins (i.e., in the mutants) likely means that hydrogenase is fully nickel activated even under low-nickel conditions in all of the strains.
Urease protein levels are similar in all strains.
To explore whether the increase in urease activities in the mutant strains was due to a corresponding increase in the amount of urease produced, immunoblotting was done, as previously described (19). Anti-UreA antiserum was used to immunoblot crude extracts from the wild-type, hpn, hpn-like, and double-mutant strains (Fig. 2C). From the results, it was determined that the amount of urease protein made by the mutants is not greater than that made by the parent strain. This suggested that the differences in urease activities were not due to differences in urease protein synthesis.
Nickel in the urease fractions is higher in the mutant strains.
Urease activities in the mutant strains were higher than for the wild type, and yet there was not an increase in the amount of urease protein made in the cell. It was possible that the urease activity differences could be accounted for by the amount of nickel associated with the urease pool in the strains. If this was true, the amount of nickel bound per unit of urease protein would be higher in the mutants than in the wild type. Indeed, the urease fractions from the two single-mutant strains showed threefold- to almost fourfold-greater nickel content per milligram of urease protein. The amount of nickel measured in our studies for wild-type H. pylori is 32 ng/mg protein (Table 2), which is very close to the nickel content reported for wild-type H. pylori in a previous study from our laboratory (21). In a separate experiment (Table 2), the double mutant was found to contain almost 10-fold more Ni per milligram of urease protein than did the parent. A similar result (higher nickel content in the urease of mutant strains) was observed in experiments performed by adding radioactive 63Ni to the liquid growth medium (Mueller-Hinton Broth with 5% fetal bovine serum in sealed serum vials); the vials were incubated in an atmosphere composed of 4% O2, 5% CO2, and 10% H2 (balance N2), and the counts per minute among the strains were compared by scintillation spectrometry of the partially purified urease fractions (data not shown).
TABLE 2.
Nickel content in the urease fraction
| Straina | Ni (ng/mg of protein)b |
|---|---|
| Wild type* | 32 ± 4 |
| Wild type‡ | 11 ± 2 |
| Wild type (with nickel | |
| supplementation)† | 69 ± 6 |
| hpn mutant* | 139 ± 11 |
| hpn-like mutant* | 104 ± 11 |
| Double mutant‡ | 109 ± 12 |
Symbols: *, cells grown under non-nickel-supplemented conditions; †, H. pylori wild-type cells grown with 500 μM NiCl2 in the growth medium; ‡, nickel urease content of the double mutant determined in comparison to the wild type in an independent experiment. The difference between the values for the two wild-type strains is likely due to variations in the nickel levels of the different sources of (blood) media. The value for the double mutant is significantly greater (P < 0.05) than that for the wild type.
Results are means ± standard deviations from at least three measurements. Both results for the mutant strains (hpn and hpn-like) are significantly greater than that for the wild type (P < 0.05 [Student's t test]).
The nickel content in the urease fractions from wild-type H. pylori cells grown with 500 μM nickel supplementation (see Materials and Methods) was found to be about threefold greater than the nickel content of urease from cells grown on the regular (non-Ni-supplemented) growth medium. This indicates that without supplementation the wild type synthesizes more apo-urease than its nickel pools can satisfy. It has been proposed previously that, at Ni-deficient times, only a minor percentage of H. pylori urease is Ni activated (29) and nickel can be a factor sometimes limiting urease activity rather than activity being limited by the amount of urease protein (35). It was previously observed that the addition of 1 μM nickel to the growth medium increased the urease activities but did not cause an increase in urease expression levels (36), and disruption of H. pylori genes encoding a metal efflux pump (cznABC) led to an increase in urease activity (28), presumably by making cytosolic nickel available.
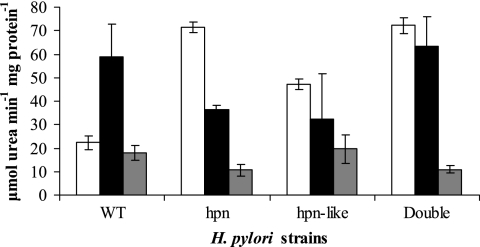
Effects of His-rich proteins on urease activities as a function of pH.
It is known that Hpn releases nickel upon lowering the pH (11). Therefore, upon lowering of the pH of the medium, one may expect to see an increase in urease activities in the wild-type strain (due to the availability of nickel) but no effect or a lesser effect on urease in the hpn or hpn-like strain. Figure 3 shows that the wild-type urease was threefold greater at pH 5 than at pH 7.3, whereas the strains lacking hpn, the hpn-like gene, or both did not have increased activity at the preferred pH for Ni accessibility. The greater absolute activities here (compared to the data in Fig. 2) are attributed to the use of broth cultures. The pH experiment shown (Fig. 3) was also conducted in BHI medium with 50 mM 3-(N-morpholino)propanesulfonic acid and 50 mM potassium acetate to closely maintain the desired pH. The pH of this well-buffered medium after growth had changed only slightly (pH 5 to 5.08 ± 0.08 and pH 7.3 to 7.18 ± 0.12, based on eight measurements at each pH). The results were similar to those in Fig. 3, but all activities were lower than the activities from the regular BHI medium.
FIG. 3.
Effects of pH and dimethyl glyoxime on urease activities. Urease activities of strains grown in BHI medium with 5% fetal bovine serum at pH 7.3 (white) or pH 5 (black) or with the nickel chelator dimethyl glyoxime at pH 7.3 (gray) are shown. The results shown are means and standard deviations of six replicate samples; these six replicates are from two independent cultures, each sampled three times. The entire experiment was performed two additional times (again with two cultures for each experiment), with results similar to those shown. At pH 5, the urease activities of the wild type (WT) are significantly higher (P < 0.05) than at pH 7.3, and the chelator-added condition was not different from that without chelator (pH 7.3). The activities of the chelator-added condition for the mutants are significantly lower than without chelator (P < 0.01).
Chelator effects.
Dimethyl glyoxime is a chelator with high affinity for nickel. The chelator had little effect on the urease activity achieved in the wild type (Fig. 3). This was also true for the wild type when the chelator was added at pH 5.0 (data not shown). However, the mutant strain activities were significantly affected by the chelator (Fig. 3). These results support a role for the Hpn and Hpn-like proteins in nickel storage, as the mutant strains' Ni urease activities were much more dependent on exogenous (available) nickel than was that of the wild type.
DISCUSSION
Previous studies of H. pylori hpn deletion mutants reported no change in urease activities compared to the parent strains (11, 12). However, even slightly higher amounts of available nickel in the complex growth medium (which also contains Ni-sequestering serum components) could abolish phenotypic differences observed in the urease activities. The parent strains from which the hpn mutants were obtained previously (various clinical strains [12] and H. pylori ATCC 26695 [11]) are also different from ours, and variation in results with regard to urease activities with different strains is well known in the H. pylori field. Due to an efficient high-affinity Ni permease, Ni levels were able to reach toxic levels, and the His-rich proteins are proposed to then render nickel tolerance to H. pylori. Even under low-Ni conditions, stringent sequestering of (limiting) nickel rather than allowing activation of all available apo-ureases may have nickel storage advantages.
Under some conditions, H. pylori apparently synthesizes more urease apo-enzyme than it can (Ni) activate (24). For example, a three- to fourfold increase in urease activity upon decreasing the pH for 2 to 3 h was demonstrated previously (27). Also, the Hpn protein releases nickel under low-pH conditions, and E. coli cells expressing Hpn contain fourfold-higher nickel levels than E. coli containing vector alone (11). These results suggest that Hpn proteins may play an important role in sequestering nickel for the cell. In addition, the studies herein demonstrate that both Hpn and the Hpn-like protein play a role in cobalt and cadmium tolerance. The greatest metal tolerance was attributed to the Hpn-like protein. Also, we observed a striking increase in urease activities in the hpn, hpn-like, and hpn/hpn-like double-mutant strains than in the wild type, but there was no corresponding increase in the amount of urease protein in the different mutant strains. The urease activity differences could be attributed to the nickel level associated with purified urease, so that these Hpn proteins appear to compete with Ni-dependent urease maturation under nickel-deficient conditions.
We found that at a lower pH, the urease activity of the wild-type strain increased, probably due to release of nickel from the intact His-rich proteins. Urease activity did not increase in the single mutants or the double mutant. This means that the His-rich nickel proteins serve as stores for the metal, probably in a pH-dependent manner. A pH dependence is expected if the His-rich proteins bind Ni at neutral pH and release Ni at pH 5.0. For Hpn, the nickel was reported to be less than half saturated at pH below 6.3 (11). Although the pH experiments can be explained from the perspective of the nickel binding properties of pure Hpn (11), these physiology experiments must be subject to other interpretations. For example, our experiments involve whole-cell physiology in which many other Ni metabolism parameters (NixA, intracellular pH regulation affecting Ni availability, urease transcription rates) could change during growth. Such physiological changes could be affected by the lack of Hpn or Hpn-like protein. Also, each Hpn or Hpn-like protein is subject to independent regulation, and one of these two is of course still present in each single mutant. Certainly, other Ni level-influencing models and mechanisms need to be considered to obtain an integrated picture of nickel metabolism and urease activity in the gastric pathogen.
In the presence of a nickel chelator, the wild-type urease activity (in contrast to that of the mutants) was not significantly reduced compared to the no-chelator condition, indicating that internal Ni reservoirs supply nickel to urease in the parent strain. Chelation had a significant inhibitory effect on the urease activities of the mutants, indicating that they are dependent on exogenous nickel for urease activation. Therefore, the Hpn and Hpn-like proteins are important for supplying nickel to urease. The results provide physiological evidence that the His-rich proteins act as nickel stores for Helicobacter pylori.
The work presented here answers some questions about Ni homeostasis and detoxification, while raising others. For instance, while we have shown that the Hpn and Hpn-like proteins play a role in nickel (and other metal) detoxification, the role of these proteins in concert with other metal toxicity prevention mechanisms is not understood. High nickel levels repress expression of the NixA transporter and the other Ni-responsive outer membrane protein studied (7, 9, 39), presumably to aid in preventing nickel toxicity; perhaps the His-rich proteins provide a more rapid (detoxification) response to transient nickel fluctuations before the (transporter) expression changes can have an affect. Also, Ni efflux pumps (CznABC) that are proposed to pump out excess nickel have been identified (28); this adds another feature to the overall Ni homeostasis/detoxification picture. Why multiple mechanisms are needed is not clear. Also, whether intracellular Ni (or other metal) concentrations become high enough in vivo that the Hpn and Hpn-like proteins are crucial for survival is not yet known.
Strains that are deficient in functional Hpn, the Hpn-like protein, or both have higher urease activity. This can be explained if Ni binds with a higher affinity to the His-rich proteins than to Ni binding urease maturation enzymes at neutral pH. This could represent a mechanism to limit, via nickel levels, the amount of active urease inside the cell at physiological pH. Upon encountering acidic pH, as in the gastric mucosa during the initial stages of colonization, an increase in the amount of apo-urease is observed (26, 27). Yet NixA and the nickel-responsive outer membrane protein HP1512 are both downregulated at pH 5 in comparison to pH 7 (5, 34). This is where the His-rich proteins would be expected to play a key role as nickel storage reservoirs. Our data that support such roles for Hpn and the Hpn-like protein are shown in Fig. 3, where urease activity is significantly greater in the wild type at low pH than at neutrality, while the activity of mutants is decreased or remains the same at low pH incubation compared to neutrality. Still, the mechanism of Ni release from the Hpn and Hpn-like proteins and whether Ni affinity is regulated by factors other than pH need to be addressed. It would be interesting to determine whether Ni is directly released from the His-rich proteins freely into the cytoplasm or if other proteins are involved in (directed) nickel transfer/sequestering processes.
The His-rich proteins play dual roles depending on the nickel level: nickel storage and nickel detoxification. Nevertheless, these proteins are just two of the many Ni binding proteins H. pylori uses to maintain Ni homeostasis and Ni enzyme function.
Acknowledgments
This work was supported by NIH grant DK06285201.
Antiserum was kindly provided by H. L. Mobley, University of Michigan Medical School, Ann Arbor, MI. We thank the reviewers for valuable suggestions and criticisms.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Bauerfeind, P., R. Garner, B. E. Dunn, and H. L. Mobley. 1997. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauerfeind, P., R. M. Garner, and H. L. Mobley. 1996. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 64:2877-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit, S., and R. J. Maier. 2003. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J. Bacteriol. 185:4787-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser, M. J. 1990. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect. Dis. 161:626-633. [DOI] [PubMed] [Google Scholar]
- 5.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 6.Contreras, M., J. M. Thiberge, M. A. Mandrand-Berthelot, and A. Labigne. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49:947-963. [DOI] [PubMed] [Google Scholar]
- 7.Davis, G. S., E. L. Flannery, and H. L. Mobley. 2006. Helicobacter pylori HP1512 is a nickel-responsive NikR-regulated outer membrane protein. Infect. Immun. 74:6811-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dosanjh, N. S., and S. L. Michel. 2006. Microbial nickel metalloregulation: NikRs for nickel ions. Curr. Opin. Chem. Biol. 10:123-130. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, F. D., E. J. Kuipers, A. Heijens, R. Sarwari, J. Stoof, C. W. Penn, J. G. Kusters, and A. H. van Vliet. 2005. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73:7252-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D. J., Jr., D. G. Evans, S. S. Kirkpatrick, and D. Y. Graham. 1991. Characterization of the Helicobacter pylori urease and purification of its subunits. Microb. Pathog. 10:15-26. [DOI] [PubMed] [Google Scholar]
- 11.Ge, R., R. M. Watt, X. Sun, J. A. Tanner, Q. Y. He, J. D. Huang, and H. Sun. 2006. Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochem. J. 393:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert, J. V., J. Ramakrishna, F. W. Sunderman, Jr., A. Wright, and A. G. Plaut. 1995. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect. Immun. 63:2682-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, L. T., and H. L. Mobley. 1990. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 58:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kansau, I., F. Guillain, J. M. Thiberge, and A. Labigne. 1996. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA). Mol. Microbiol. 22:1013-1023. [DOI] [PubMed] [Google Scholar]
- 15.Kleanthous, H., C. L. Clayton, and S. Tabaqchali. 1991. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol. Microbiol. 5:2377-2389. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Maier, R. J., C. Fu, J. Gilbert, F. Moshiri, J. Olson, and A. G. Plaut. 1996. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 141:71-76. [DOI] [PubMed] [Google Scholar]
- 18.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed] [Google Scholar]
- 19.Mehta, N., J. W. Olson, and R. J. Maier. 2003. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185:726-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mobley, H. L., R. M. Garner, G. R. Chippendale, J. V. Gilbert, A. V. Kane, and A. G. Plaut. 1999. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter 4:162-169. [DOI] [PubMed] [Google Scholar]
- 21.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 22.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 23.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 24.Pflock, M., P. Dietz, J. Schar, and D. Beier. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol. Lett. 234:51-61. [DOI] [PubMed] [Google Scholar]
- 25.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pflock, M., S. Kennard, I. Delany, V. Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 28.Stahler, F. N., S. Odenbreit, R. Haas, J. Wilrich, A. H. Vliet, J. G. Kusters, M. Kist, and S. Bereswill. 2006. The novel Helicobacter pylori CznABC metal efflux pump is required for cadmium, zinc, and nickel resistance, urease modulation, and gastric colonization. Infect. Immun. 74:3845-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stingl, K., and H. De Reuse. 2005. Staying alive overdosed: how does Helicobacter pylori control urease activity? Int. J. Med. Microbiol. 295:307-315. [DOI] [PubMed] [Google Scholar]
- 30.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 31.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuda, M., M. Karita, T. Mizote, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. Essential role of Helicobacter pylori urease in gastric colonization: definite proof using a urease-negative mutant constructed by gene replacement. Eur. J. Gastroenterol. Hepatol. 6(Suppl. 1):S49-S52. [PubMed] [Google Scholar]
- 33.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vliet, A. H., F. D. Ernst, and J. G. Kusters. 2004. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 12:489-494. [DOI] [PubMed] [Google Scholar]
- 35.van Vliet, A. H., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Vliet, A. H., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 38.Weatherburn, M. W. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971-974. [Google Scholar]
- 39.Wolfram, L., E. Haas, and P. Bauerfeind. 2006. Nickel represses the synthesis of the nickel permease NixA of Helicobacter pylori. J. Bacteriol. 188:1245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]