Abstract
Assembly of B-band lipopolysaccharide (LPS) in Pseudomonas aeruginosa follows a Wzy-dependent pathway, requiring the O-antigen polymerase Wzy and other proteins. The peptide sequences of the wzyα product from strains of serotypes O2, O5, and O16 are identical, but the O units in O5 are α-glycosidically linked, while those in O2 and O16 are β-linked. We hypothesized that a derivative of the D3 bacteriophage wzyβ is present in the chromosomes of O2 and O16 and that this gene is responsible for the β-linkage. By a combination of PCR and primer walking, wzyβ genes of both serotypes have been amplified and cloned. They are identical but share only 87.42% sequence identity with their xenolog in D3. A chromosomal knockout mutant of O16 wzyβ was made, and it produces semirough LPS devoid of B-band O antigen. The cloned wzyβ is capable of complementing the O16 wzyβ mutant, as well as cross-complementing a wzyα knockout mutant. However, in the latter case, the restored O antigen was β-linked. Using reverse transcription-PCR, we showed that wzyα was transcribed in O2 and O16 strains and was functional, since both of these genes could complement the wzyα mutant of O5. With the coexistence of wzyα and wzyβ in O2 and O16 and the B-band O polysaccharides in these being β-linked, we hypothesized that iap, an inhibitor of the alpha-polymerase gene, must be present in these serotypes. Indeed, through PCR, TOPO-cloning, and nucleotide-sequencing results, we verified the presence of iap in both O2 and O16 serotypes.
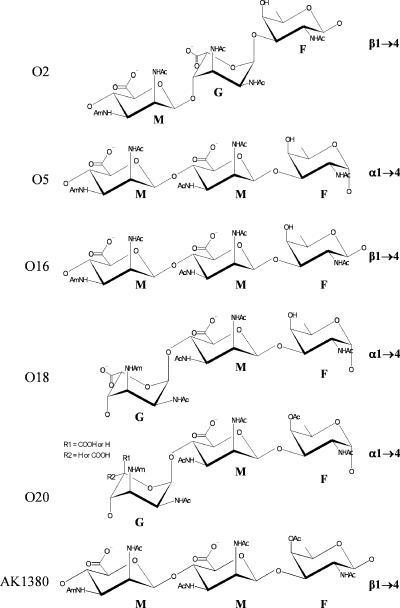
Lipopolysaccharide (LPS) is an important virulence factor of the opportunistic pathogen Pseudomonas aeruginosa. This glycolipid is found in the outer leaflet of the outer membrane and is composed of three main parts: lipid A, core, and O antigen. P. aeruginosa produces two distinct forms of LPS, called A band and B band. The O antigen of A band is composed of a homopolymeric repeating chain of d-rhamnose. B-band LPS, on the other hand, consists of heteropolymeric O-antigen units (28). Differences in the B-band O antigen divide P. aeruginosa into 20 distinct serotypes, as defined by the International Antigenic Typing Scheme (33, 34). The gene cluster for the biosynthesis of B-band LPS has been designated wbp (32). Based on results from Southern hybridization, Burrows et al. (9) found that the wbp cluster is highly conserved among serotypes O2, O5, O16, O18, and O20, which form the O2 serogroup (46). The cloning and sequencing of the LPS biosynthetic clusters from each of these serotypes by Raymond et al. (40) further substantiated the conserved nature of the LPS genes from members of this serogroup. The O antigens of these serotypes show immunochemical cross-reactivity when typed by slide agglutination using polyclonal rabbit antisera (29), but they are distinct from each other based on the type of glycosidic linkage or isomers/epimers of monosaccharides present and the presence or absence of acetyl substituent in their O-antigen sugars (23, 24) (Fig. 1).
FIG. 1.
Chemical structures of the P. aeruginosa serogroup O2 O-antigen repeat units. Serotypes O2, O5, O16, O18, and O20, which make up the O2 serogroup, as well as the D3 lysogen (AK1380), have highly similar O-antigenic sugar structures. Differences among them were due to the type of glycosidic linkage (α-d- versus β-d-N-acetylfucosamine) or isomer (β-d-mannuronic versus α-l-guluronic) or the presence/absence of O-acetyl groups at the C-4 of the fucosamine moiety (23, 26, 39). G, guluronic acid; M, mannuronic acid; F, fucosamine; OAc, O-acetyl group; NHAc, acetamido group; NHAm, acetamidino group.
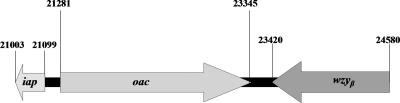
wzyα (formerly called rfc) has been shown to encode the B-band O-antigen polymerase and was responsible for the formation of α-linked O-antigen repeat units in serotype O5, strain PAO1 (14). Its nucleotide sequence differed from that of wzyα of serotype O2 by one nucleotide, but the amino acid encoded within the same codon remained unchanged. Since the O antigens of serotype O5 are α-linked while those of serotype O2 are β-linked, it was puzzling how a single protein could catalyze glycosidic linkages of opposite stereochemistry (41). By examining the D3 phage genome sequence, which was annotated in 2000 (25), our group had identified a 3.6-kb fragment in the D3 genome capable of mediating serotypic conversion identical to that observed in a D3 lysogen strain (Fig. 2) (39). Three open reading frames (ORFs) were identified in this DNA fragment, and they encode a putative α-polymerase inhibitor (Iap), an O acetylase (Oac), and a β-polymerase (Wzyβ). The Wzyβ protein has a size of 42.6 kDa and contains nine predicted transmembrane segments, with a large periplasmic loop near the C terminus (39). It showed no primary amino acid sequence homology to other proteins, a characteristic that is consistent with Wzy/Rfc proteins from many gram-negative bacteria. wzyβ from D3 was used to complement a PAO1-derived (serotype O5) wzyα null mutant that produced semirough LPS. Interestingly, the complemented strain exhibited an LPS banding pattern resembling that of serotype O16. In Western immunoblotting, the LPS bands reacted with O16-specific monoclonal antibody (MAb) (MF47-4), but not with O5-specific MAb (MF15-4), indicating that the restored O-antigen repeat units were β-linked. These data suggested that the D3 wzyβ encodes a functional O-antigen polymerase capable of catalyzing β-glycosidic linkages.
FIG. 2.
The 3.6-kb seroconverting cassette of the D3 bacteriophage genome. The three genes involved in serotype conversion encode the Wzyα inhibitor (iap; 96 bp), O-acetylase (oac; 2,064 bp), and O-antigen β-polymerase (wzyβ; 1,160 bp). The iap, oac, and wzyβ genes correspond to nucleotides 21099 to 21003, 21281 to 23345, and 24580 to 23420 of the D3 genome, respectively (25, 39).
Two other ORFs in the 3.6-kb D3 DNA fragment, iap and oac, were found to encode proteins that also contribute to the mechanism of serotype conversion in P. aeruginosa strain PAO1. A plasmid containing the 96-bp iap ORF was capable of inhibiting long-chain B-band LPS production when transformed into serotypes O5 and O18 (39). The tranformants were still capable of producing A-band LPS. Transformation of the iap plasmid construct into serotype O16, which has β-linked units, showed no effect. These data indicate that Iap is an inhibitor of O-antigen polymerase with specificity for serotypes possessing α-linked O-antigen repeat units (39).
The oac ORF was the only one of the three genes in the 3.6-kb D3 DNA fragment that showed sequence similarity to other proteins. This 2,064-bp ORF was homologous to known O-acetyltransferase proteins (11). When PAO1 was transformed with a plasmid containing all three seroconverting genes, the resulting LPS produced by the transformant showed strong reaction with polyclonal anti-O20 antiserum that had been adsorbed with bacterial cells of O5 and O18 to make it specific for O-acetylated LPS, indicating that like serotype O20, the O antigen of the transformant contained O-acetylated sugars. These observations were substantiated by high-performance liquid chromatography analysis, which showed that LPS isolated from the transformant contained 45-fold more mild base-labile acetate than the wild-type control, PAO1 LPS (39). This increase corresponds to levels found in LPS from the D3 lysogen, P. aeruginosa strain 1380. When the oac gene was disrupted, LPS of the transformants did not show any detectable levels of O acetylation. The oac gene could not be used individually in the complementation tests with the PAO1 background strain, since it was discovered that Oac can acetylate only β-linked O-antigen repeat units (39).
In light of these findings, Newton et al. (39) proposed that P. aeruginosa serotypes O2 and O16 may have acquired a β-polymerase gene from bacteriophage D3 or another closely related phage and that this gene lies outside of the wbp cluster. This P. aeruginosa Wzyβ protein would then be responsible for the β-linkages observed in these serotypes. Southern hybridization experiments showed that a probe generated from D3 wzyβ bound to P. aeruginosa genomic DNA from serotypes O2 and O16, but not O5, O18, or O20 (39). Interestingly, an iap probe did not hybridize with any of the wild-type P. aeruginosa O2 serogroup serotypes. The only strain that bound the iap probe was the transformant containing the entire seroconverting cassette. Therefore, it was postulated that the native α-polymerase (Wzyα) in the wbp cluster of serotypes O2 and O16 is either not functional or not being expressed (39).
Until this study, there was no direct experimental evidence that the O2 and O16 serotypes possess a functional β-polymerase in their chromosomes. We were able to identify, clone, and characterize wzyβ from the chromosomal DNA of O2 and O16. We also obtained evidence that wzyα is transcribed in both serotypes O2 and O16 and that this gene is functional, i.e., capable of complementing a wzyα mutant derived from serotype O5 (strain PAO1). Although previous studies suggested that iap was absent from the P. aeruginosa genome, we were successful in amplifying it by PCR using O2 and O16 chromosomal DNAs as templates. These observations further the understanding of the mechanisms responsible for the diversity in P. aeruginosa serotypes, in particular, the cause of α- versus β-glycosidic linkages in the O antigen.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Serotypes were confirmed by slide agglutination tests as described previously (29), using polyclonal antisera from the serotyping kit for P. aeruginosa (manufactured by Chengdu Institute of Biological Products, Ministry of Public Health, Chengdu, Sichuan, People's Republic of China) (34). Bacterial strains were cultured in Luria-Bertani broth (Invitrogen Canada Inc., Burlington, Ontario, Canada) at 37°C. Pseudomonas isolation agar (Difco) and resolving media were used for mating experiments. The resolving media consisted of 1.0% Bacto tryptone (Difco), 0.5% Bacto yeast extract (Difco), 1.5% Bacto agar (Difco), 5% sucrose, and gentamicin. The following antibiotics were used: kanamycin, 50 μg/ml for Escherichia coli; tetracycline, 10 μg/ml for E. coli and 100 μg/ml for P. aeruginosa; gentamicin, 150 and 300 μg/ml for P. aeruginosa; and carbenicillin, 600 μg/ml for P. aeruginosa (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype, phenotype or properties | Reference or source |
|---|---|---|
| E. coli | ||
| JM109 | endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | 51 |
| ONE Shot Mach 1 T1 | F− φ80(lacZ)ΔM15 ΔlacX74 hsdR(rK− mK−) ΔrecA1398 endA1 tonA (confers resistance to phage T1); chemically competent | Invitrogen |
| SubCloning efficiency DH5α competent cells | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| SM10 | thi-1 thr leu tonA lacy supE recA RP4-2-Tc::Mu Kmr | 44 |
| P. aeruginosa | ||
| O5 | Wild-type strain PAO1; IATS serotype O5; A+ B+ | 17 |
| O2 | Wild-type strain 5934; IATS serotype O2 (ATCC 33349); A+ B+ | 33 |
| O16 | Wild-type strain 170003; IATS serotype O16 (ATCC 33363); A− B+ | 33 |
| O5-wzyα-GmR | O5 wzyα::Gmr; O5 mutant with a gentamicin cassette inserted into the wzyα gene | 14 |
| O16-wzyβ-GmR | O16 wzyβ::Gmr; O16 mutant with a gentamicin cassette inserted into the wzyβ gene | This study |
| O5-wzyα-GmR-27 | O5-wzyα mutant transformed with the pUCP27 plasmid | This study |
| O5-wzyα-GmR-comp-O5wzyα | O5-wzyα mutant complemented with pUCP27 containing wzyα from O5 | This study |
| O5-wzyα-GmR-comp-O2wzyα | O5-wzyα mutant complemented with pUCP27 containing wzyα from O2 | This study |
| O5-wzyα-GmR-comp-O16wzyα | O5-wzyα mutant complemented with pUCP27 containing wzyα from O16 | This study |
| O5-wzyα-GmR-comp-O2wzyβ | O5-wzyα mutant complemented with pUCP27 containing wzyβ from O2 | This study |
| O16-wzyβ-GmR-27 | O16 wzyβ::Gmr mutant transformed with the pUCP27 plasmid | This study |
| O16-wzyβ-GmR-comp-O2wzyα | O16 wzyβ::Gmr mutant complemented with pUCP27 containing wzyα from O2 | This study |
| O16-wzyβ-GmR-comp-O2wzyβ | O16 wzyβ::Gmr mutant complemented with pUCP27 containing wzyβ from O2 | This study |
| Plasmid | ||
| pEX18Ap | Apr/CbroriT+sacB; gene replacement vector with multiple cloning site from pUC18 | 20 |
| pEX18Ap-O2wzyβ | pEX18Ap with wzyβ inserted into SacI and PstI sites | This study |
| pEX18Ap-O16 wzyβ-GmR | pEX18Ap with wzyβ with gentamicin cassette inserted in the EcoRI site of wzyβ | This study |
| pPS856 | Apr Gmr; 0.83-kb blunt-ended SacI fragment from pUCGM ligated into the EcoRV site of pPS854 | 20 |
| pQE80 | 4.8-kb histidine-tagged expression vector with RBS; Apr | QIAGEN |
| pUCP27 | 4.9-kb pUC18-based broad-host-range vectors; Tcr | 50 |
| pUCP27-RBS-O5wzyα | wzyα from O5 with RBS from pQE80 cloned into EcoRI and PstI sites of pUCP27 | This study |
| pUCP27-RBS-O2wzyα | wzyα from O2 with RBS from pQE80 cloned into EcoRI and PstI sites of pUCP27 | This study |
| pUCP27-RBS-O16wzyα | wzyα from O16 with RBS from pQE80 cloned into EcoRI and PstI sites of pUCP27 | This study |
| pUCP27-O2wzyβ | wzyβ from O2 with RBS from pQE80 cloned into EcoRI and PstI sites of pUCP27 | This study |
| TOPO-O2wzyβ-topfrag | 5′ PCR fragment of O2 wzyβ cloned into pCR-Blunt II-TOPO vector | This study |
| TOPO-O16wzyβ-topfrag | 5′ PCR fragment of O16 wzyβ cloned into pCR-Blunt II-TOPO vector | This study |
| TOPO-O2wzyβ-bottomfrag | 3′ PCR fragment of O2 wzyβ cloned into pCR-Blunt II-TOPO vector | This study |
| TOPO-O16wzyβ-bottomfrag | 3′ PCR fragment of O16 wzyβ cloned into pCR-Blunt II-TOPO vector | This study |
| TOPO-O2iap | iap from P. aeruginosa O2 cloned into pCR-Blunt II-TOPO vector | This study |
| TOPO-O16iap | iap from P. aeruginosa O16 cloned into pCR-Blunt II-TOPO vector | This study |
DNA procedures.
Plasmid DNA was isolated using the GenElute plasmid miniprep kit (Sigma-Aldrich) following procedures recommended by the manufacturer. Calf intestinal alkaline phosphatase was purchased from New England Biolabs Ltd. (Mississauga, Ontario, Canada). Ligation of digested DNA was performed using T4 DNA ligase (New England Biolabs) at 16°C overnight (16 h). Alternatively, ligations were performed using the Rapid DNA ligation kit (Roche Applied Science, Laval, Quebec, Canada). The Zero-Blunt TOPO PCR cloning kit (Invitrogen) was used for cloning blunt-ended PCR-amplified DNA into the pCR-Blunt II-TOPO vector. PCRs were performed in GeneAmpPCR System thermocyclers (models 2400 and 9700; Applied Biosystems) using PwoI polymerase (Roche Applied Science, Quebec, Canada). PCR amplification of the iap (for inhibitor of α-polymerase) genes from P. aeruginosa serotypes O2 and O16 was performed using primers specific for the D3 iap 5′ and 3′ sequences: iapFwd (5′-GTGCATTTAAAATTTTCAATCATAG-3′) and iapRev (5′-CATATGTCTTGGTAGTAAGTTGC-3′), respectively. Oligonucleotide primers were purchased from the University of Guelph Molecular Supercenter. DNA extraction from agarose gels was performed using the UltraClean 15 DNA purification kit (Mo Bio Laboratories, Inc., Carlsbad, CA). DNA in solution was purified using the High Pure PCR product purification kit (Roche) and eluted in 50 to 100 μl H2O. Plasmid DNA was introduced into P. aeruginosa serotype O5 by electroporation using a Gene Pulser instrument (Bio-Rad).
Sequencing and in silico analysis.
Sequencing of plasmid constructs, PCR products, and genomic DNA was performed at the Guelph Molecular Supercenter (University of Guelph, Guelph, Ontario, Canada). Sequences were analyzed using GENERUNNER for Windows (Hastings Software, NY), as well as the online servers Basic Local Alignment Search Tool (3, 4), CLUSTALW (48), EMBL-EBI (European Bioinformatics Institute), TMHMM Server v. 2.0 (45), SOSUI (18), and the MEMSAT3 prediction method of the PSIPRED Protein Structure Prediction Server (7, 21, 37).
Construction of plasmids for complementation.
Complementation vectors were made by PCR amplifying the P. aeruginosa wzyα gene using wzyfwd (5′-CGTTGACGAATTCTAGAATGTATATAC-3′) and Rev/wzy/stopO5,O2,O16 (5′-GGTTGATAAAAGCTGCAGTCATAG-3′) (restriction sites are shown in boldface). The fragment was then cloned into the pUCP27 vector (50). Once cloning was verified by sequencing, the fragment was excised with SstI and PstI and cloned into pQE80. To obtain the ribosome binding site (RBS) from pQE80, the fragment was excised with EcoRI and PstI and cloned into pUCP27. The same method was used for wzyα from O5, O2, and O16. Since wzyβ contains an internal EcoRI site, this strategy could not be used for that gene. Instead, plasmid pUCP27-RBS-O5wzyα was digested with XbaI and PstI to remove the O5 wzyα gene, and a wzyβ gene amplified with flanking XbaI and PstI sites was cloned in.
Generation of a wzyβ chromosomal knockout mutant.
The P. aeruginosa chromosomal mutant was generated using the method of Hoang et al. (20). The P. aeruginosa O2 wzyβ gene was amplified with flanking SacI and PstI restriction sites using wzybetafwdSacI (5′-AATAGAGCTCATGAATAGGACCAAGCTTCCG-3′) and wzybetarevPstI (5′-GACTGCAGTTAATTATCCTCGATTTTAAGATTGG-3′). The gene was then cloned into the suicide vector pEX18Ap, which contains the sacB gene of Bacillus subtilis. A gentamicin resistance (Gmr) cassette from pPS856 was excised by SmaI digestion and then subcloned into the EcoRV site of the wzyβ insert DNA. The orientation of the Gmr cassette was confirmed by restriction digests and sequencing. The resulting plasmid was transformed into the mobilizer strain E. coli SM10 and then conjugally transferred into P. aeruginosa serotype O16 according to the method of Simon et al. (44). After they had mated, the cells were spread plated on Pseudomonas Isolation Agar containing 150 μg/ml gentamicin. Colonies that grew on the gentamicin-containing medium were picked and streaked on resolving media.
LPS preparation for SDS-PAGE, silver staining, and Western blotting.
LPS was prepared using the method of Hitchcock and Brown (19) and analyzed by separation on standard discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12.5% running gels (27). LPS was visualized by silver staining using the method of Fomsgaard et al. (15) and by Western blotting using the standard procedure described by Towbin et al. (49) and Burnette (8), with minor modifications. The primary antibodies used in immunoblotting were MAb MF15-4 (specific for O5), MAb MF71-2 (specific for O2), and MAb MF47-4 (specific for O16). All the MAbs used have been described previously (30, 31). The secondary antibody was a goat anti-mouse F(ab′)2-alkaline phosphatase conjugate (Jackson ImmunoResearch). The blots were developed using a substrate containing 0.033% nitroblue tetrazolium and 0.016% BCIP (5-bromo-4-chloro-3-indolyl phosphate) in 0.1 M sodium bicarbonate buffer, pH 9.8.
Immunofluorescence microscopy.
Immunofluorescence microscopy was performed using the method described by Clarke et al. (12). Bacteria were viewed on a Zeiss Axiovert 200 microscope using a 100× objective lens and oil immersion, and the images were processed using Openlab software (Improvision). MAb MF15-4 was used for visualization of LPS antigen of O5, MAb MF71-2 for O2, and MAb MF47-4 for O16. The secondary antibody used was fluorescein isothiocyanate (FITC)-conjugated AffiniPure F(ab′)2, at a 1:50 dilution in 0.85% NaCl containing 1% bovine serum albumin (Bio/Can Scientific).
Reverse transcription (RT) PCR.
RNA was harvested from P. aeruginosa serotypes O5, O2, and O16 using the PURESCRIPT RNA isolation kit (Gentra Systems, Minneapolis, MN) as recommended by the manufacturer. To eliminate contaminating genomic DNA, the RNA solution was incubated at 37°C overnight with RQ1 RNase-Free DNase (Promega). RNA was converted to cDNA using SuperScript II reverse transcriptase (Invitrogen) and random primers (Invitrogen) as recommended by the manufacturer. PCR analysis was performed using primers specific for a 264-bp internal fragment of wzyα. The primers used were alphaRTPCRfwd (5′-GAGGTGTTTGGCTATTCATTCTTG-3′) and alphaRTPCRrev (5′-CCCCCACATAACCCAACTGC-3′). A BLAST search was performed to confirm that the primers would bind specifically to wzyα. The PCR products were resolved by agarose gel electrophoresis and stained with ethidium bromide for visualization under UV light. One microliter of extracted RNA was used as a negative control, while positive controls for the PCR consisted of P. aeruginosa genomic DNA as the template. Further, positive controls used primers specific for a 177-bp fragment of the P. aeruginosa housekeeping gene, rpoD. The primers used were rpoDf (5′-GGGCGAAGAAGGAAATGGTC-3′) and rpoDr (5′-CAGGTGGCGTAGGTGGAGAA-3′) (42). It has been shown that the rpoD gene is constitutively transcribed in all P. aeruginosa serotypes (42).
Nucleotide sequence accession numbers.
The nucleotide sequences of wzyβ from P. aeruginosa O2 and O16 strains have been submitted to the NCBI-GenBank databases under the accession numbers EF153452 and EF153453, respectively.
RESULTS
wzyα genes from P. aeruginosa serotypes O2 and O16 are functional in vivo and encode O-antigen polymerases.
The wzyα gene was amplified from serotypes O2, O5, and O16 and sequenced. The nucleotide sequence of wzyα from O16 had a different sequence than that reported in the NCBI databases. The sequence was identical to that of the O2 wzyα (accession number AF498412). Results from antibody agglutination tests, as well as analysis of LPS by SDS-PAGE and Western blotting, confirmed that our strain was indeed serotype O16. Genomic DNA was reisolated from this strain, and wzyα was amplified, cloned, and resequenced. Once again, the sequence corresponded to the O2 wzyα sequence, indicating that our strain (ATCC 33363) was different than the O16 strain sequenced by Raymond et al. (40), accession number AF498408.
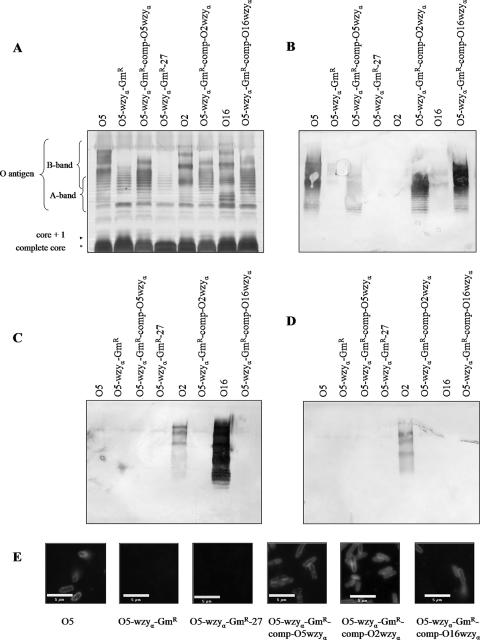
The O5 wzyα mutant was then complemented using plasmids containing wzyα genes from P. aeruginosa serotypes O2 and O16, as well as from serotype O5 as a positive control. wzyα genes from all three strains were able to restore the smooth (S) LPS phenotype, as shown by the banding patterns of LPS of the complemented strains, which resembled that of the wild-type O5 strain (Fig. 3A). Results from Western immunoblotting revealed that LPS from wild-type serotype O5 and the complemented strain showed reactivity only with an O5-specific MAb (MF15-4). An O2-specific MAb (MF71-2) reacted strongly with LPS from wild-type serotype O2 but no other LPS. Similarly, an O16-specific MAb (MF47-4) reacted strongly with LPS from wild-type serotype O16 and weakly with serotype O2 LPS and showed no reactivity with the complemented strains. These results showed that regardless of the origin of the wzyα gene from any of the three serotypes used in the complementation experiments, α-linked O antigen was restored in the O5 wzyα mutant (Fig. 3B to D).
FIG. 3.
wzyα genes from P. aeruginosa O5, O2, and O16 restored alpha-linked O antigen in a wzyα knockout mutant. (A) Silver-stained SDS-PAGE gel of LPS from wild-type serotype O5 (strain PAO1), an O5 wzyα knockout mutant, a wzyα mutant complemented with O5 wzyα, a wzyα mutant transformed with pUCP27 (control), serotype O2 (wild type), a wzyα mutant complemented with O2 wzyα, serotype O16 (wild type), and a wzyα mutant complemented with O16 wzyα. Note that regardless of the origin of the wzyα gene used in the complementation experiment, an O5 LPS banding modality was restored. (B) Western blots of the genes from panel A reacted with O5-specific MAb. (C) Western blots of the genes from panel A reacted with O16-specific MAb. (D) Western blots of the genes from panel A reacted with O2-specific MAb. Only O5-specific MAbs reacted with LPS from the complemented strains, indicating that they have α-linked O antigen. (E) Immunofluorescence micrographs showing that wzyα genes from P. aeruginosa O5, O2, and O16 could restore α-linked O antigen in a wzyα knockout mutant. All cells were reacted with O5-specific primary MAb (MF15-4) and FITC-linked secondary antibody. Only those cells with a functional wzyα gene fluoresced. Each of the scale bars represents 5 μm.
Immunofluorescence microscopy was used to further substantiate the results obtained by Western blotting. The complemented strains showed fluorescence when reacted with O5-specific MAb (MF15-4), followed by incubation with the FITC-labeled goat-anti-mouse Fab2 secondary antibody. Immunofluorescence was observed on the outer surfaces of cells, which is the location of the receptor for the serotype-specific antibodies. Cells of the knockout mutant and the negative control did not react with any antibodies and consequently did not show any fluorescence (Fig. 3E).
Analysis of the transcription of wzyα of serotypes O2 and O16.
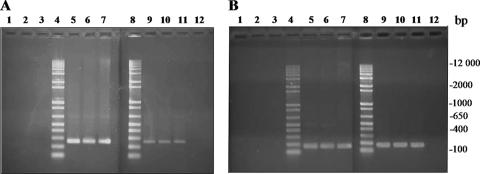
To determine whether wzyα was being transcribed in P. aeruginosa serotypes O2 and O16, RT-PCR was performed. RNA prepared from serotype O5 was used as a positive control. A band at approximately 264 bp, which is the predicted size of the wzyα fragment, was amplified from all three serotypes. This indicated that wzyα was being transcribed in all three serotypes (Fig. 4A).
FIG. 4.
RT-PCR detection of wzyα and rpoD mRNA transcripts. (A) RT-PCR mRNA detection of wzyα (264-bp) transcripts. (B) RT-PCR mRNA detection of rpoD (177-bp) transcripts. Lanes 1, PCR of O5 RNA (negative control); lanes 2, PCR of O2 RNA (negative control); lanes 3, PCR of O16 RNA (negative control); lanes 4, 1-kb Plus DNA ladder; lanes 5, PCR of O5 genomic DNA (positive control); lanes 6, PCR of O2 genomic DNA (positive control); lanes 7, PCR of O16 genomic DNA (positive control); lanes 8, 1-kb Plus DNA ladder; lanes 9, PCR of O5 cDNA; lanes 10, PCR of O2 cDNA; lanes 11, PCR of O16 cDNA; lanes 12, PCR without template (negative control).
Amplification and cloning of P. aeruginosa chromosomal wzyβ.
Using primers specific for the D3 wzyβ gene, amplification of the 5′ end of the gene from serotypes O2 and O16 was easily achieved. The 3′ end, however, could not be amplified, likely due to the fact that D3-specific primers did not bind in that region, suggesting that the 3′ O2/O16 wzyβ sequence was significantly different than the 3′ D3 wzyβ gene sequence. A number of strategies were attempted to determine the sequence of the 3′ region of wzyβ, including designing new primers specific for various regions of the 3′ end, as well as downstream regions (primer sequences available upon request). Ultimately, data obtained from sequencing analysis performed by the Guelph Molecular Supercenter (University of Guelph, Guelph, Ontario, Canada) generated a partial sequence of the 3′ end of the P. aeruginosa wzyβ gene, as well as a short sequence downstream. Despite gaps in the sequencing results, sufficient information was obtained to allow the design of a downstream primer, wzybetaRev8 (5′-GCCCACCAAGAGCGGAATC-3′), that was used along with wzy-betaFwd3 (5′-CGCCCTCTGTTACTTGC-3′) to amplify the remainder of wzyβ from both O2 and O16 P. aeruginosa. PCR products from these reactions were obtained and cloned into the pCR-Blunt II-TOPO vector. Sequencing of both the O2 and O16 wzyβ genes revealed that they share 100% sequence identity. As expected, attempts to amplify wzyβ from P. aeruginosa serotype O5 did not yield a product DNA band. These results substantiate the absence of a wzyβ gene within the whole genome sequence database of PAO1 (http://v2.pseudomonas.com; 47).
A region of 60 bp upstream of wzyβ in O2 and O16 was sequenced and found to have high similarity to the same locus in the D3 genome. This region is not part of the seroconverting operon but appears to have been horizontally transferred along with wzyβ. The sequence of 170 bp downstream of wzyβ in O2 and O16 was also determined but did not show similarity to D3 or any other known sequence. No gene homologous to D3 oac could be identified in this region, even though in the D3 seroconverting operon, oac is located 75 bp downstream of wzyβ. This indicated that modifications had occurred after the genes involved in seroconversion were integrated into the chromosomes of O2 and O16 strains.
In silico analysis of P. aeruginosa wzyβ.
The P. aeruginosa O2/O16 wzyβ gene is predicted to encode a protein of 41.8 kDa. When subjected to a BLAST database search, the only DNA sequence that showed similarity was the D3 phage wzyβ. The P. aeruginosa wzyβ nucleotide sequence was 87.42% identical to that of the D3 wzyβ. The primary amino acid sequence of P. aeruginosa Wzyβ was 90.41% identical and 97.15% similar to that of Wzyβ from the D3 phage. The mol% G+C for wzyβ from P. aeruginosa is 43.99%, which is strikingly similar to the 43.32% G+C from D3 wzyβ. A PSI-BLAST search was performed with the O2/O16 Wzyβ sequence, and iteration 1 showed similarities to the hypothetical protein H16 B0028 of Ralstonia eutropha H16 (accession number YP_728199), with an E value of 1e−15, as well as similarity to the hypothetical protein ELI 13300 from Erythrobacter litoralis HTCC2592 (accession number YP_459549), with an E value of 2e−13. Further iterations using the default parameters did not produce any new hits.
Analyses of the protein topography of Wzyβ were performed by the online programs TMHMM, SOSUI, and MEMSAT3. All three programs predicted high degrees of similarity between the P. aeruginosa and D3 Wzyβ topographies. Comparisons of the P. aeruginosa Wzyα protein to the P. aeruginosa and D3 Wzyβ proteins are summarized in Table 2.
TABLE 2.
Comparison of the P. aeruginosa Wzyα, P. aeruginosa Wzyβ, and D3 bacteriophage Wzyβ O-antigen polymerases
| Property | Value
|
||
|---|---|---|---|
| P. aeruginosa Wzyα | P. aeruginosa Wzyβ | D3 phage Wzyβ | |
| Total no. of bp | 1,317 | 1,155 | 1,161 |
| Total no. of amino acids | 438 | 384 | 386 |
| Molecular mass (kDa)a | 48.9 | 41.8 | 42.6 |
| % G+Ca | 44.8 | 44.0 | 43.3 |
| % Leucine, isoleucine, and phenylalaninea | 30.82 | 31.00 | 29.28 |
| Predicted no. of transmembrane segments | |||
| TMHMM | 12 | 9 | 9 |
| SOSUI | 10 | 10 | 9 |
| MEMSAT3 | 10 | 11 | 11 |
Based on nucleotide sequence.
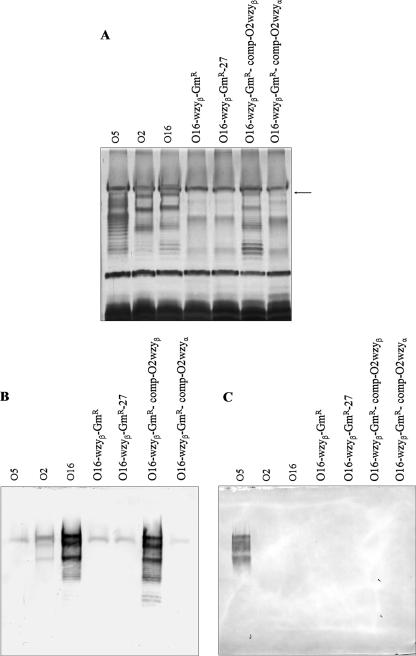
A wzyβ knockout mutant does not produce O antigen.
A chromosomal mutant of wzyβ in serotype O16 was produced. The insertion of the gentamicin cassette into the O16 wzyβ gene was verified by PCR-based screening (data not shown). LPS was isolated from this mutant strain (O16-wzyβ-Gmr) and analyzed using SDS-PAGE and silver staining (Fig. 5A) and Western blotting (Fig. 5B and C). The mutant was found to lack high-molecular-weight (HMW) B-band LPS, which is consistent with the semirough phenotype. The LPS from wild-type O16 showed an LPS banding pattern with molecules of heterogeneous sizes. This is consistent with the S-LPS phenotype. When the wzyβ mutant was complemented in trans with a wzyβ gene, S-LPS was restored. When anti-O16 MAb (MF47-4) was used in Western blotting, there was a strong reaction with both the wild-type O16 LPS and the LPS from the O16 wzyβ mutant complemented with wzyβ (O16-wzyβ-GmR-comp-O2wzyβ). The anti-O16 MAb (MF47-4) showed weak cross-reactivity with wild-type serotype O2 LPS and no reactivity with the O16 wzyβ mutant LPS (see Fig. 5B). Anti-O5 MAb (MF15-4) reacted only with wild-type O5 LPS bands (Fig. 5C). Likewise, anti-O2 MAb (MF71-2) reacted only with wild-type serotype O2 LPS bands (data not shown).
FIG. 5.
B-band LPS production can be restored by a wzyβ gene, but not by a wzyα gene, in a wzyβ knockout mutant. (A) Silver-stained SDS-PAGE gel of LPS from serotype O5 (wild type), serotype O2 (wild type), serotype O16 (wild type), an O16 wzyβ knockout mutant, an O16 wzyβ mutant transformed with pUCP27 (control), an O16 wzyβ mutant complemented with O2 wzyβ, and an O16 wzyβ mutant complemented with O2 wzyα. Note that the banding modality of the O16 wzyβ mutant complemented with O2 wzyβ is very similar to that of wild-type O16, indicating that S-LPS was being restored. O2 wzyα could not restore S-LPS in the O16 wzyβ mutant strain, except for one HMW band, which is indicated by an arrow. (B) Western blots of the samples in panel A reacted with O16-specific MAb. O16-specific MAb reacts with LPS from the O16 wzyβ mutant complemented with O2 wzyβ, but not with LPS from the O16 wzyβ mutant complemented with O2 wzyα, indicating that wzyα cannot restore B-band production in the O16 wzyβ knockout mutant. (C) Western blots of the samples in panel A reacted with O5-specific MAb. O5-specific MAb can react only with α-linked O antigen, which in this blot was found exclusively in the LPS from O5.
Transformation of the O16 wzyβ mutant with a plasmid containing wzyα did not restore S-LPS.
Attempts to complement the O16 wzyβ knockout mutant with wzyα from P. aeruginosa serotype O2 did not restore B-band O-antigen production. Upon careful examination of the LPS bands by SDS-PAGE and silver staining, one HMW LPS band was observed, but the overall LPS banding modality did not resemble that of wild-type O16. Instead, the LPS banding pattern was most similar to those of the O16-wzyβ mutant and the negative control (O16-wzyβ-GmR-27) (Fig. 5A). Results from Western blotting showed that none of the LPS bands observed in the SDS-PAGE gels were B-band LPS, as they were not reactive with either the O5-specific MAb (MF15-4) or the O16-specific MAb (MF47-4) (Fig. 5B and C).
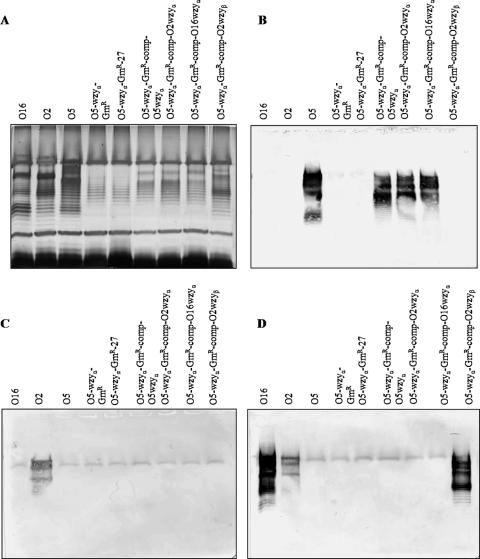
Transformation of the O5 wzyα mutant with wzyβ produced β-linked O antigen.
The O5-wzyα-GmR strain was transformed with a pUCP27-wzyβ gene, and LPS from the cross-complemented strain was analyzed by silver staining (Fig. 6A) and Western blotting (Fig. 6B to D). LPS from the complemented strain exhibited a banding modality more similar to that of wild-type serotype O16 than to wild-type O5, and the LPS bands reacted with an O16-specific MAb (MF47-4). This indicates that a β-linked O antigen was restored. Furthermore, an O5-specific MAb (MF15-4) reacted only with wild-type O5 and the wzyα mutant strains that had been complemented with wzyα.
FIG. 6.
Complementation of a serotype O5-derived wzyα knockout mutant by wzyβ restored a LPS banding modality similar to that of wild-type serotype O16. (A) Silver-stained SDS-PAGE gel of LPS from serotype O16 (wild type), serotype O2 (wild type), serotype O5 (wild type), a wzyα knockout mutant, a wzyα mutant transformed with pUCP27 (control), a wzyα mutant complemented with O5 wzyα, a wzyα mutant complemented with O2 wzyα, a wzyα mutant complemented with O16 wzyα, and a wzyα mutant complemented with O2 wzyβ. (B) Western blots of LPS reacted with O5-specific MAb. (C) Western blots of LPS reacted with O2-specific MAb. (D) Western blots of LPS reacted with O16-specific MAb.
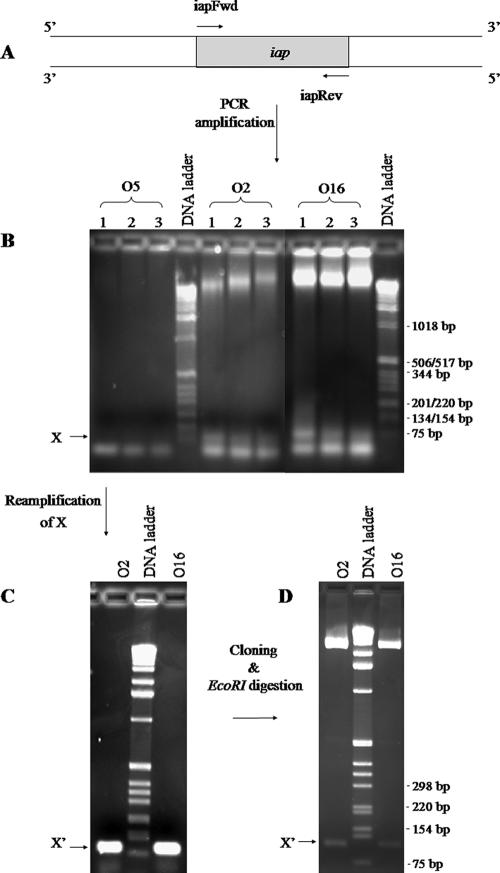
Amplification and cloning of P. aeruginosa chromosomal iap.
Using primers designed based on the D3 bacteriophage iap sequence (Fig. 7A), a faint band at approximately 95 bp was amplified from chromosomal DNA of O2 and O16, but not from O5 (Fig. 7B). When the faint band was excised and the DNA was extracted and reamplified using the same primers, a bright band at approximately 95 bp, consistent with the predicted size of iap, was clearly visible (Fig. 7C). After gel purification, TOPO cloning (Fig. 7D), and sequencing, our results showed that the entire region between primer-binding locations (47 bp) was identical in O2 and O16 and that it was also identical to that of the D3 iap gene. In addition, the D3-specific iapFwd and iapRev primers were able to anneal to O2 and O16 chromosomal DNA in the PCR; it is likely that the 5′ and 3′ ends of the O2 and O16 iap genes are similar, if not identical, to those of the D3 iap.
FIG. 7.
PCR amplification of iap. (A) Primers iapFwd and iapRev were designed to bind to the 5′ and 3′ ends of D3 iap, respectively. (B) Genomic DNAs from serotypes O5, O2, and O16 were used as templates for PCRs. Three different MgSO4 concentrations (1 mM, 1.5 mM, and 2 mM) were used and are indicated by the numbers 1, 2, and 3, respectively. The arrow points to bands corresponding to sizes of ∼95 bp that were present in the O2 and O16 samples but missing in O5. (C) These bands from the O2 and O16 lanes were gel excised and reamplified using primers iapFwd and iapRev. (D) The product, labeled X′, was cloned into a pCR-Blunt II-TOPO vector. Plasmids were extracted from two clones, purified, and digested with EcoRI.
DISCUSSION
The results of the complementation studies with wzyα from serotypes O2 and O16 supported the hypothesis that another gene or process must be involved in the formation of β-linked O antigen in the O2 and O16 serotypes, since the wzyα genes in both serotypes are clearly responsible for the α-linkages in O polysaccharides in P. aeruginosa. The wzyα genes from O2 and O16 are identical, while the O5 wzyα differs from these two by only 1 nucleotide, with the amino acid encoded by that codon being conserved. Thus, they are the same protein with the same substrate specificity.
Results from RT-PCR experiments of wzyα from P. aeruginosa serotypes O5, O2, and O16 clearly showed that wzyα is transcribed in all three serotypes. Transcription of wzyα in O5 was expected, since the O antigen of this serotype contains α-linkages, but it was surprising that transcription was also occurring in serotypes O2 and O16, since these serotypes produce LPS with β-linked O antigens. One possible explanation is that the O-antigen polymerase activity of Wzyα is inhibited in serotypes O2 and O16. We hypothesized that the D3 bacteriophage iap gene, which has been shown to inhibit Wzyα in P. aeruginosa (39), was also horizontally transferred into these two serotypes. Although the presence of iap could not be detected in P. aeruginosa by Southern blotting (39), it is important to consider that iap is only 96 bp in length, which may have been too short for adequate probe binding. Indeed, by means of PCR, we were able to successfully amplify iap from P. aeruginosa serotype O2 and O16 chromosomal DNAs. The presence of iap in these two serotypes supported our hypothesis that the product Iap is responsible for the inhibition of Wzyα. The inability to amplify iap from serotype O5 DNA was expected, since serotype O5 produces α-linked O antigen. It is predicted that inhibition occurs at the translation or protein function level, since the wzyα genes continue to be successfully transcribed in all serotypes tested. The rfb locus of Salmonella enterica serovar Anatum group E1 has been shown to contain two putative wzy genes, namely, orf9.6 and orf17.4, by McConnell et al. (36). orf9.6 apparently encodes Wzyα, responsible for the α1-6 glycosidic linkage in the O10 O antigen of group E1 bacteria. The latter gene, orf17.4, when transformed into a spontaneous Salmonella mutant defective in O polymerase, caused the transformant to produce O15 LPS with β-linked O-antigen units. The authors suggested that orf17.4 might be derived from a phage origin, but the mechanism for only orf9.6 to be functional in vivo was not investigated. In contrast, wzyβ in serotypes O2 and O16 is clearly localized elsewhere in the P. aeruginosa chromosome and not within the wbp-rfb locus. Evidence that wzyβ might have a D3 bacteriophage origin has been presented.
A chromosomal knockout mutant of wzyβ in P. aeruginosa O16 was produced in this study, and this mutant is important for characterizing the function of the gene. Despite the experience of our group in generating knockout mutants of various genes in this species, making the O16 wzyβ mutant was not straightforward. It appeared that the P. aeruginosa O16 strain was spontaneously developing resistance to gentamicin, compromising the screening technique of the protocol. This spontaneous resistance was also accompanied by a loss of fluorescence of the bacterial colonies. It has since been discovered that lower concentrations of gentamicin, such as 30 μg/ml and 40 μg/ml, are more effective at selecting true mutants because they discourage the occurrence of spontaneous gentamicin-resistant mutations in P. aeruginosa (Craig Daniels, Toronto Sick Children Hospital, personal communication). LPS prepared from the O16 wzyβ mutant complemented with wzyβ and reacted exclusively with O16-specific MAb (MF47-4) in Western immunoblotting. These results showed that Wzyβ is responsible for the formation of β-linked O antigen. In addition, an O2 wzyβ gene could be used for the complementation, since it is 100% identical to the O16 wzyβ gene. The LPS banding modality of the O5 wzyα knockout mutant cross-complemented with O2 wzyβ was similar to that of wild-type O16. The minor differences can be explained by the fact that wzyβ was introduced in a high-copy number plasmid, which could affect the stoichiometry of the ratio between Wzyβ and Wzz in the membrane complex for LPS assembly. An altered ratio of the two proteins is expected to affect the length of the O antigen, thereby causing a change in the modality of the LPS bands visualized in the SDS-PAGE gels (13). If Wzyβ was restoring β-linked O antigen in the mutant, it is not surprising that the LPS banding modality of the complemented strain resembled that of wild-type O16, since the serotype O5 and O16 O antigens have the same sugar composition and differ only by linkage type. Essentially, a switch to β-linkage caused the conversion of serotype O5 to O16. This was substantiated by results from Western immunoblotting experiments, since the O5 wzyα mutant cross-complemented with wzyβ was reactive with the anti-O16 MAb (MF47-4).
Transformation of the O16 wzyβ mutant with wzyα did not restore the production of HMW B-band LPS. Only one of the HMW bands appeared to have been restored upon analysis by SDS-PAGE and silver staining, but the overall modality did not resemble that of wild-type O16 (Fig. 5A). The partial restoration of O antigen is consistent with the theory that the iap gene we were able to amplify by PCR encodes a functional O antigen inhibitor in this serotype. The O16 wzyβ knockout mutant already has a functional wzyα gene that is being inhibited. Since the exogenous wzyα gene was being introduced in a high-copy-number plasmid (pUCP27), the amount of Wzyα being expressed in the cell would have been much higher than normal. This supersaturation of Wzyα likely exhausted all Wzyα inhibitor (Iap) binding sites, allowing a small degree of α-linked O antigen to be produced. Analysis of the Western blots, however, revealed no reaction between this complemented strain and the O5-specific MAb (MF15-4) (Fig. 5C). The reason for this is not known.
The P. aeruginosa wzyβ gene is most likely a xenolog of the D3 phage wzyβ gene, since a BLAST search revealed no homology to other known genes or proteins. Likewise, the P. aeruginosa iap gene is also predicted to be a xenolog of the D3 iap gene, since our sequencing results show that they are identical. It is important to consider that the G+C content of the D3 genome is 59%, which is considerably higher than that of the D3 3.6-kb serotype-converting cassette (43%) containing wzyβ and iap (39). The possibility that D3 acquired wzyβ and iap from P. aeruginosa is unlikely, since the G+C content of the P. aeruginosa genome is 67% (39). Newton et al. (39) have also speculated that the D3 serotype conversion genes may be “morons,” a term used to describe portable expression units in the genomes of lambda-like phages with reduced G+C percentages (22). These genes, therefore, may have been acquired from yet another source at a relatively recent point in evolution, with D3 acting as a shuttle vector between them (39). Based on PSI-BLAST results, it is plausible that Wzyβ shares an evolutionary relationship with the hypothetical proteins of R. eutropha H16, E. litoralis HTCC2594, or the phage that infect these gram-negative bacterial species.
The differences in sequence between P. aeruginosa wzyβ and D3 wzyβ indicate that evolutionary modification occurred after wzyβ was integrated into the P. aeruginosa chromosome. Based on the sequences of the O2 and O16 wbp gene clusters reported by Raymond et al. (40), we are certain that wzyβ and iap are elsewhere. Until the whole genome sequences of these strains are available, their exact locus in the chromosome is presently unknown. At this time, it is not clear whether the wzyβ and iap genes integrated on their own or as part of the 3.6-kb seroconverting operon. Although the upstream sequence of wzyβ in O2 and O16 is similar to that of the wzyβ upstream sequence in D3, the downstream sequence showed no similarity. If the entire seroconverting operon had been integrated into O2 and O16, the oac gene would have been detectable in this downstream sequence. However, it is also plausible that the oac gene has undergone drastic changes in sequence and may no longer be easily identifiable by PCR or Southern hybridization. Further sequencing of the regions flanking wzyβ in P. aeruginosa, as well as sequencing of the P. aeruginosa iap upstream and downstream regions, would be of great interest, as it may uncover attP sites, int genes, or insertion sequences. These would suggest that a select few genes from D3 have been permanently acquired, or “fixed,” by P. aeruginosa. Alternatively, it is possible that the entire D3 genome is present in O2 and O16 as a viable prophage. This could be investigated by induction and characterization of phage from serotypes O2 and O16. Our initial attempts to induce D3 from these serotypes with mitomycin C were unsuccessful.
The fixation of the seroconverting genes into a bacterial host chromosome has been observed before. Shigella flexneri serotype 1a strain Y53 has been found to contain a fixed chromosomal version of an SfV phage-derived serotype conversion cassette. It was found that this cassette encoded factors that mediate the conversion of a serotype Y strain to serotype 1a (2, 5). The organization of the 5.8-kb fragment, as well as its upstream and downstream regions (10.6 kb in total), suggested that this operon was originally an Sf1 lysogen, although it appears that a large portion of the phage genome was deleted (2). Southern hybridization analyses revealed that this fragment was also present in three other natural isolates of S. flexneri (2). Another study by Adams et al. (1) revealed that a 3.8-kb fragment from S. flexneri serotype 4a strain NCTC 8296 contains three putative O-antigen modification genes. Phage-derived seroconversion genes have also been found in the chromosomes of E. coli strains (6, 16, 35).
It has become increasingly apparent that phage and their bacterial hosts influence each others' evolution (10, 22, 38, 43). Many important bacterial virulence factors are encoded by prophage or have a phage origin. This makes P. aeruginosa Wzyβ particularly interesting to study, as it likely contributes greatly to P. aeruginosa virulence. The question of whether Wzyβ plays a role in virulence or other physiological functions provides an important avenue for future research. It is imperative to locate the oac gene in the P. aeruginosa chromosome and to characterize its function. To provide further clues to Iap's function, wild-type O5 could be transformed with the wzyβ gene to see if both α- and β-linked O antigens are concomitantly produced. In the long run, these studies can be expanded to include serotypes outside of the O2 serogroup. Ultimately, the serotype-converting genes from the D3 phage can be used together with other genes to create a strain expressing multiple (possibly all 20) types of O antigen. This strain could potentially be useful for the development of a multivalent vaccine.
Acknowledgments
This work was supported by an operating grant to J.S.L from the Canadian Cystic Fibrosis Foundation (CCFF). P.D.A. is the recipient of a CCFF postdoctoral fellowship, and J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Adams, M. M., G. E. Allison, and N. K. Verma. 2001. Type IV O antigen modification genes in the genome of Shigella flexneri NCTC 8296. Microbiology 147:851-860. [DOI] [PubMed] [Google Scholar]
- 2.Adhikari, P., G. Allison, B. Whittle, and N. K. Verma. 1999. Serotype 1a O-antigen modification: molecular characterization of the genes involved and their novel organization in the Shigella flexneri chromosome. J. Bacteriol. 181:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastin, D. A., A. Lord, and N. K. Verma. 1997. Cloning and analysis of the glucosyl transferase gene encoding type I antigen in Shigella flexneri. FEMS Microbiol. Lett. 156:133-139. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36-W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 9.Burrows, L. L., D. F. Charter, and J. S. Lam. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481-495. [DOI] [PubMed] [Google Scholar]
- 10.Canchaya, C., G. Fournous, and H. Brussow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 11.Clarke, A. J., N. T. Blackburn, and H. Strating. 2000. Pathways for the O-acetylation of bacterial cell wall polysaccharides. Kluwer Academic Publishers, New York, NY.
- 12.Clarke, B. R., L. Cuthbertson, and C. Whitfield. 2004. Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J. Biol. Chem. 279:35709-35718. [DOI] [PubMed] [Google Scholar]
- 13.Daniels, C., C. Griffiths, B. Cowles, and J. S. Lam. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4:883-897. [DOI] [PubMed] [Google Scholar]
- 14.de Kievit, T. R., T. Dasgupta, H. Schweizer, and J. S. Lam. 1995. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5). Mol. Microbiol. 16:565-574. [DOI] [PubMed] [Google Scholar]
- 15.Fomsgaard, A., M. A. Freudenberg, and C. Galanos. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J. Clin. Microbiol. 28:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, S., and N. K. Verma. 1998. Serotype conversion of a Shigella flexneri candidate vaccine strain via a novel site-specific chromosome-integration system. FEMS Microbiol. Lett. 166:79-87. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, R. E., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat-2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 19.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 22.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 23.Knirel Yu, A., and N. K. Kochetkov. 1994. Structure of lipopolysaccharides from gram-negative bacteria. III. Structure of O-specific polysaccharides. Biokhimiia 59:1784-1851. [PubMed] [Google Scholar]
- 24.Knirel Yu, A., E. V. Vinogradov, N. A. Kocharova, N. A. Paramonov, N. K. Kochetkov, B. A. Dmitriev, E. S. Stanislavsky, and B. Lanyi. 1988. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa. Acta Microbiol. Hung. 35:3-24. [PubMed] [Google Scholar]
- 25.Kropinski, A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuzio, J., and A. M. Kropinski. 1983. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J. Bacteriol. 155:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lam, J. S., L. L. Graham, J. Lightfoot, T. Dasgupta, and T. J. Beveridge. 1992. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J. Bacteriol. 174:7159-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam, J. S., M. Y. Handelsman, T. R. Chivers, and L. A. MacDonald. 1992. Monoclonal antibodies as probes to examine serotype-specific and cross-reactive epitopes of lipopolysaccharides from serotypes O2, O5, and O16 of Pseudomonas aeruginosa. J. Bacteriol. 174:2178-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam, J. S., L. A. MacDonald, and M. Y. Lam. 1987. Production of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect. Immun. 55:2854-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, J. S., L. A. MacDonald, M. Y. Lam, L. G. Duchesne, and G. G. Southam. 1987. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect. Immun. 55:1051-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lightfoot, J., and J. S. Lam. 1993. Chromosomal mapping, expression and synthesis of lipopolysaccharide in Pseudomonas aeruginosa: a role for guanosine diphospho (GDP)-d-mannose. Mol. Microbiol. 8:771-782. [DOI] [PubMed] [Google Scholar]
- 33.Liu, P. V., H. Matsumoto, H. Kusama, and T. Bergan. 1983. Survey of heat-stable, major somatic antigens of Pseudomonas aeruginosa. Int. J. Syst. Bacteriol. 33:256-264. [Google Scholar]
- 34.Liu, P. V., and S. Wang. 1990. Three new major somatic antigens of Pseudomonas aeruginosa. J. Clin. Microbiol. 28:922-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavris, M., P. A. Manning, and R. Morona. 1997. Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol. 26:939-950. [DOI] [PubMed] [Google Scholar]
- 36.McConnell, M. R., K. R. Oakes, A. N. Patrick, and D. M. Mills. 2001. Two functional O-polysaccharide polymerase wzy (rfc) genes are present in the rfb gene cluster of Group E1 Salmonella enterica serovar Anatum. FEMS Microbiol. Lett. 199:235-240. [DOI] [PubMed] [Google Scholar]
- 37.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 38.Miao, E. A., and S. I. Miller. 1999. Bacteriophages in the evolution of pathogen-host interactions. Proc. Natl. Acad. Sci. USA 96:9452-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton, G. J., C. Daniels, L. L. Burrows, A. M. Kropinski, A. J. Clarke, and J. S. Lam. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 39:1237-1247. [DOI] [PubMed] [Google Scholar]
- 40.Raymond, C. K., E. H. Sims, A. Kas, D. H. Spencer, T. V. Kutyavin, R. G. Ivey, Y. Zhou, R. Kaul, J. B. Clendenning, and M. V. Olson. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savli, H., A. Karadenizli, F. Kolayli, S. Gundes, U. Ozbek, and H. Vahaboglu. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403-408. [DOI] [PubMed] [Google Scholar]
- 43.Shi, S. Y., X. H. Cai, and D. F. Ding. 2005. Identification and categorization of horizontally transferred genes in prokaryotic genomes. Acta Biochim. Biophys. Sin. (Shanghai) 37:561-566. [DOI] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Puhler. 1983. A broad-host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotecchnology 1:784-791. [Google Scholar]
- 45.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 46.Stanislavsky, E. S., and J. S. Lam. 1997. Pseudomonas aeruginosa antigens as potential vaccines. FEMS Microbiol. Rev. 21:243-277. [DOI] [PubMed] [Google Scholar]
- 47.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 51.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]