Abstract
RybB is a small, Hfq-binding noncoding RNA originally identified in a screen of conserved intergenic regions in Escherichia coli. Fusions of the rybB promoter to lacZ were used to screen plasmid genomic libraries and genomic transposon mutants for regulators of rybB expression. A number of plasmids, including some carrying rybB, negatively regulated the fusion. An insertion in the rep helicase and one upstream of dnaK decreased expression of the fusion. Multicopy suppressors of these insertions led to identification of two plasmids that stimulated the fusion. One contained the gene for the response regulator OmpR; the second contained mipA, encoding a murein hydrolase. The involvement of MipA and OmpR in cell surface synthesis suggested that the rybB promoter might be dependent on σE. The sequence upstream of the +1 of rybB contains a consensus σE promoter. The activity of rybB-lacZ was increased in cells lacking the RseA anti-sigma factor and when σE was overproduced from a heterologous promoter. The activity of rybB-lacZ and the detection of RybB were totally abolished in an rpoE-null strain. In vitro, σE efficiently transcribes from this promoter. Both a rybB mutation and an hfq mutation significantly increased expression of both rybB-lacZ and rpoE-lacZ fusions, consistent with negative regulation of the σE response by RybB and other small RNAs. Based on the plasmid screens, NsrR, a repressor sensitive to nitric oxide, was also found to negatively regulate σE-dependent promoters in an RseA-independent fashion.
Prokaryotic small regulatory RNA molecules have long been known to regulate plasmid replication and phage development (60). Recent studies have found that, in addition, regulatory RNAs have important roles for bacterial cell growth and physiology. A number of genome-wide searches performed by various groups using computational and biochemical methods have uncovered close to 100 small RNAs (sRNAs) in Escherichia coli (6, 8, 27, 47, 57, 62). More than a dozen of these RNAs have been characterized thus far and have been found to affect various physiological pathways. Characterization of additional sRNAs continues to yield surprising insights into cell physiology.
In Escherichia coli, the largest class of small regulatory RNAs binds to the RNA chaperone Hfq and regulates the stability and translation of specific mRNAs. The Hfq-binding sRNAs act by pairing with regions of complementarity in their target mRNAs. The vast majority of the Hfq-binding sRNAs act negatively, to either inhibit translation or stimulate the degradation of the target mRNA, also resulting in degradation of the sRNA itself (26, 33, 56). In some cases, sRNAs of this class can act positively. For instance, DsrA and RprA stimulate translation of the stationary-phase/general stress sigma factor RpoS (sigma S or σS) (32, 51).
RybB, an 81-nucleotide sRNA, was identified in a computational genome-wide search for novel sRNAs, as well as in an expression-based search (57, 62). RybB binds the RNA chaperone protein Hfq, suggesting that the RNA acts by pairing with mRNAs. Steady-state levels of RybB could be detected only in stationary phase in rich media (57, 62). However, the conditions leading to the synthesis of RybB and its effects on the cell remained largely uncharacterized until recently.
In characterizing the regulation of RybB synthesis, we found that RpoE (sigma E or σE) was absolutely necessary for its synthesis and was a direct regulator of its transcription. σE is a member of the extracytoplasmic family of alternative sigma factors (31). σE guides the core RNA polymerase in binding to the promoter region and initiating transcription of specific genes in response to extracytoplasmic stress (excess heat shock or misfolded outer membrane proteins [OMPs]) (36, 52). RybB also represses σE synthesis, creating an autoregulatory loop.
(A portion of this work was used in partial fulfillment of the Ph.D. degree to K.M.T. from Howard University [Department of Microbiology].)
MATERIALS AND METHODS
Strains, plasmids, and media.
The strains used are listed in Table S1 in the supplemental material. Most are derivatives of E. coli K-12 MG1655. Strains were grown in Luria-Bertani (LB) medium (KD Medical, Columbia, MD) in an Innova 3100 water bath shaker (New Brunswick Scientific, Edison, NJ) at approximately 200 rpm at 37°C unless otherwise indicated. Unless otherwise indicated, marked mutations were moved into the desired strain background using bacteriophage P1 transduction (49). Transductants were selected on LB plates supplemented with 25 μg/ml kanamycin, 10 μg/ml chloramphenicol, or 25 μg/ml tetracycline (KD Medical, Columbia, MD), depending on the marker used. Tetrazolium (2,3,5-triphenyltetrazolium chloride [TTC]) (Sigma)-lactose (TTC-Lac) plates were prepared as previously described (38). Insertion mutants were screened on MacConkey-lactose plates supplemented with 25 μg/ml kanamycin (KD Medical) or TTC-Lac plates supplemented with 25 μg/ml kanamycin (TTC-Lac-Kan). All plasmids used in this study are listed in Table S2 in the supplemental material. Plasmids were generally introduced into strains by TSS (transformation and storage solution) transformation (10). Transformants were selected on LB agar plates supplemented with 100 μg/ml ampicillin (KD Medical). Genomic DNA transformants were screened on MacConkey-lactose plates supplemented with 50 μg/ml of ampicillin (Mac-Lac-Amp) (KD Medical).
Construction of rybB-lacZ transcriptional fusions.
Single-copy chromosomal rybB-lacZ transcriptional fusions used in this study were first constructed in plasmid pRS1553 (http://www.mimg.ucla.edu/bobs/vectors/Alpha-lac/alphaLac.htm), a derivative of the vectors described in reference 50. To construct the multicopy rybB-lacZ long transcriptional fusion, the region from −77 to +22 of rybB was PCR amplified from MG1655 genomic DNA using primers KT12 and KT14 (see Table S3 in the supplemental material), digested with EcoRI and BamHI, and ligated to a pRS1553 EcoRI/BamHI digest to yield pKMT30. Similarly, in the rybB-lacZ short transcriptional fusion, the region from −36 to +22 of rybB was PCR amplified using primers KT13 and KT14 (Table S3) and ligated to pRS1553 to yield pKMT31. Strains containing these plasmids were then crossed with λRS468; Lac+ recombinants yielded transducing phages λKMT30 and λKMT31. λKMT30 and λKMT31 were used to lysogenize DJ480, yielding strains KMT12000 and KMT11000, respectively.
Construction of an rpoE-lacZ translational fusion.
A region of rpoE from 160 nucleotides (nt) upstream to 270 nt downstream of the ATG was amplified from MG1655 genomic DNA using primers KT132 and KT143 (see Table S3 in the supplemental material), digested with EcoRI and BamHI, and ligated to EcoRI/BamHI-treated pRS1551 to yield pKMT35. Plasmid pKMT35 was then crossed with λRS468 (50) to yield λKMT35. Strain NM6010 was lysogenized with λKMT35, resulting in strain KMT14000.
Construction of pBAD-rybB and pBAD-nsrR plasmids.
To clone the rybB gene under the control of an arabinose-inducible promoter, plasmid pNM12, a derivative of pBAD24 used for expression of sRNAs, was used as the vector (32). Oligonucleotides KT119 and KT120 (see Table S3 in the supplemental material) were annealed, and the resulting product was ligated to an MscI/PstI digest of pNM12, resulting in pKMT5. To clone nsrR under the control of an arabinose-inducible promoter, nsrR was amplified from MG1655 genomic DNA using primers KT191 and KT192, digested with EcoRI and PstI, and ligated to pBAD24, resulting in pKMT8. The constructs were confirmed by DNA sequencing.
Transposon mutant screen.
For screen II, a pool of random insertion mutant derivatives of DJ480 was created by infecting DJ480 with λNK1316 (29) and selecting kanamycin-resistant colonies; a P1 lysate made on this pool was used to deliver the random insertions into KMT12000 by transduction. A total of 4,200 kanamycin-resistant transductants were screened on TTC-Lac-Kan; 22 Lac− colonies were picked and assayed for immunity to phage λimm21. Only two of the Lac− transductants were still immune to the phage and thus retained the chromosomal rybB-lacZ fusion. The two insertions, rrb2::kan and rrb4::kan, were retransduced into KMT12000 to confirm the linkage of the kan insertion to the phenotype.
Insertions were mapped by an inverse PCR method as previously described (9, 43). The first PCR uses a random primer (RandomPrim1) and a transposon-specific primer (TnKm1) (see Table S3 in the supplemental material). In the second PCR, a nested set of primers (Revprim2 and TnKm2) (Table S3) was used. TnKm2 was used to sequence the PCR product to obtain the location of the kan gene. The rrb2::kan insertion mapped to the middle of the rep gene, at nucleotide 3958864, and was renamed rep::kan; the strain carrying this insertion is KMT12002. The rrb4::kan insertion mapped to an intergenic region directly upstream of the dnaK gene, at nucleotide 12109, and was renamed zaa-1000::kan; the strain carrying this insertion is KMT12003.
Genomic DNA library screens.
For screen I, a genomic DNA library was introduced into KMT12000 (long rybB-lacZ fusion) and KMT11000 (short rybB-lacZ fusion) by electroporation (54). Following electroporation, 1 ml of LB medium was added to the electroporation mixtures, the culture was incubated at 37°C for 1 h, and 50-μl aliquots were spread on Mac-Lac-Amp. For KMT12000, approximately 10,500 colonies were screened; for KMT1000, approximately 3,500 colonies were screened.
In addition to screening KMT12000 and KMT11000, the genomic library was also introduced into KMT12002 and KMT12003 (rep::kan and zaa-1000::kan mutant derivatives of KMT12000, Lac− phenotype, from screen II) to screen for multicopy suppressors (screen III). Transformants were screened for a Lac+ phenotype on Mac-Lac-Amp.
Construction of chromosomal rybB and rseA mutants.
Kanamycin-resistant deletion insertions in rseA and rybB were constructed using the mini-λ-based λ Red recombineering system (15). A kan cassette with 50 bp of homology to regions flanking rybB was amplified using primers KT18 and KT19 (Table S3). A kan cassette with 50 bp of homology to rseA was amplified using oligonucleotides KT76 and KT77 (see Table S3 in the supplemental material). A culture of DJ480 containing mini-λ::tet (NM300) was grown at 32°C to mid-log phase and switched to 42°C to induce expression of the λ Red proteins. Aliquots of the uninduced and induced cultures were made electrocompetent, and approximately 100 ng of the respective PCR products was electroporated into a 50-μl aliquot of the electrocompetent cells. Recombinants were selected on LB agar plates containing 50 μg/ml of kanamycin.
RNA isolation and Northern blots.
Total RNA was extracted in mid-exponential or stationary phase using the hot-phenol method (2). RNA samples were run on 8% acrylamide gels, and nonisotopic Northern blotting was performed as previously described (34). The 5′-biotinylated oligonucleotide KT98 was used as a probe for the detection of RybB RNA (see Table S3 in the supplemental material).
In vitro transcription assays.
Single-round in vitro transcription experiments were performed to test the rybB promoter. The rybB promoter DNA template was prepared by PCR from MG1655 genomic DNA and contained sequences from −126 to +112 with respect to the transcription start and also the rybB terminator, using oligonucleotide primers rybB prom and rybB term (see Table S3 in the supplemental material). The control PdegP DNA template was prepared from the vector pUA66, carrying the native PdegP σE promoter sequences from −65 to +20 (46). PCR was used to amplify a degP promoter fragment from −200 to +175 with the primers pMS201-FW200 and pMS201-RV2 terminator (Table S3). Note that the pMS201-RV2 terminator primer contains the rpoC terminator sequence (35) to provide efficient transcription termination at the underlined C in Table S3.
RNA polymerase core enzyme was purified as described in reference 65, and His6-tagged σE was purified using a QIAGEN Ni2+ affinity column per the manufacturer's instructions. The transcription assays were performed as described in reference 45 with the following modifications. Binding reaction mixtures (6 μl) contained 5 nM template DNA, 250 nM core RNA polymerase, 1,250 nM σE, and 1× binding buffer (5% glycerol, 20 mM Tris [pH 8.0], 300 mM potassium acetate, 5 mM magnesium acetate, 0.1 mM EDTA, 1 mM dithiothreitol, 50 μg/ml bovine serum albumin, 0.05% Tween). Single-round transcriptions were initiated with 2 μl of a 2× XTP-heparin mix (to give a final concentration of 200 μM ATP/UTP/GTP, 10 μM CTP, 2.5 μCi [α-32P]CTP [3,000 Ci/mmol], 100 μg/ml heparin in 1× binding buffer), mixtures were incubated for 5 min at 37°C, and then reactions were terminated with 4 μl stop solution (20 mM EDTA, 80% deionized formamide, 0.05% [wt/vol] bromophenol blue, 0.05% [wt/vol] xylene cyanol FF). The RNA transcripts were resolved by electrophoresis after heating at 90°C for 2 min and loading 3 μl on a 6% denaturing polyacrylamide sequencing gel together with DNA sequencing reaction products that functioned as size markers. Transcripts were visualized using a Molecular Dynamics Storm 560 PhosphorImager scanning system.
β-Galactosidase assays.
β-Galactosidase assays were carried out in 96-well plates and read in a SpectraMax 250 (Molecular Devices, Sunnyvale, CA) as previously described (66). Briefly, 50-ml LB cultures (KD Medical, Columbia, MD) were grown at 32°C or 37°C. At various time points, 100-μl aliquots of the cell culture were added to a well containing permeabilization buffer. Specific activity units are 25-fold lower than Miller units.
RESULTS
Genetic screens for regulators of RybB synthesis.
In the study that initially identified RybB, its expression could be detected only in stationary phase (62). This pattern of expression was later confirmed, and the start site of rybB transcription was determined by 5′ rapid amplification of cDNA ends (57), but the regulators of RybB expression were not determined.
Two rybB-lacZ transcriptional fusion strains were initially used to screen for RybB regulators. One fusion, the rybB-lacZ long fusion, contained −79 to +22 of rybB, with respect to the mapped transcription start site; the other, the rybB-lacZ short fusion, contained −36 to +22 of rybB. The rybB-lacZ long-fusion strain was Lac+ on Mac-Lac plates; the rybB-lacZ short-fusion strain was less strongly Lac+. The activity of both fusions increased upon entry of cells into stationary phase, consistent with previous reports where RybB RNA was detected only in stationary phase (57, 62). Expression of the rybB-lacZ fusions was not altered in an rpoS-null mutant, suggesting that stationary-phase induction of rybB expression is RpoS independent (data not shown). The short fusion was used only in the first of the screens described below; tests with this fusion suggest that it is qualitatively similar in its regulation to the long fusion but may be missing some promoter sequences necessary for optimal expression.
The initial screen for regulators of rybB (screen I) was of an E. coli genomic DNA plasmid library in the rybB-lacZ fusion strains (54). A total of 11,000 transformants of the rybB-lacZ long fusion and 3,000 transformants of the rybB-lacZ short fusion were screened for Lac− or Lac++ phenotypes on Mac-Lac-Amp or TTC-Lac-Amp agar plates; candidate plasmids were confirmed by isolation and retransformation. Twelve independent plasmids, representing 10 different genomic regions, that reduced fusion expression were identified (Lac− phenotype) (Table 1). Two independent plasmids carried the rybB gene itself. These plasmids also contained the upstream flanking gene, ybjK, which is a predicted DeoR-type transcriptional regulator. However, a null mutation of ybjK in the chromosome did not affect the expression of rybB-lacZ activity, suggesting that YbjK is not required for expression of rybB (data not shown). One clone gave a Lac++ phenotype for the rybB-lacZ long fusion; it did not carry any known transcriptional regulators (Table 1). No explanation for the phenotypes resulting from these plasmids was immediately obvious; we return to some of them below.
TABLE 1.
Screen I: genomic library clones that modulate rybB-lacZ fusion expression
| Library screen hosta | Plasmid isolate (fragment boundaries) | Lac phenotype | Gene
|
Comments | |
|---|---|---|---|---|---|
| Nameb | Descriptionc | ||||
| rybB-lacZ (long) | pK-09 (886844-890902); pK-235 (884802-887483) | Lac− | >′ybjK | Putative DeoR-type transcriptional regulator | RybB negative regulation of σE; see text |
| <rybB | sRNA | ||||
| <ybjL | Putative transport protein | ||||
| >ybjM | Unknown function | ||||
| <grxA′ | Glutaredoxin I | ||||
| pK-215 (4402773-4406851); | Lac− | >′purA | Adenylosuccinate synthetase | NsrR effect; this work | |
| pK-221 (4402773-4404689)a | >nsrR | Transcriptional repressor | |||
| >rnr′ | Exoribonuclease R | ||||
| pK-222 (2869026-2871368) | Lac− | <truD | tRNA pseudouridine 13 synthase | Cell division and cell surface effects? | |
| <ispF | 2-C-Methyl-d-erythritol-2,4-cyclodiphosphate synthetase, isprenoid biosynthesis | ||||
| <ispD | 4-Diphosphocytidyl-2C-methyl-d-erythritol synthetase | ||||
| <ftsB | Essential cell division protein | ||||
| <ygbE′ | Putative cytochrome oxidase subunit | ||||
| pK-121 (3025059-3026683) | Lac− | >′guaD | Guanine deaminase | Unknown mechanism | |
| >ygfQ | Unknown function (transporter?) | ||||
| <ygfS′ | Unknown function (oxidoreductase?) | ||||
| pK-126 (835278-777897) | Lac− | >′ybiB | Unknown function (transferase/phosphorylase?) | Unknown mechanism | |
| >ybiC | Unknown (dehydrogenase?) | ||||
| <ybiJ | Unknown function | ||||
| <ybiI | Unknown function | ||||
| <ybiX′ | Unknown function | ||||
| pK-220 (3242589-3244529) | Lac− | <′uxaA | Altronate dehydratase | Unknown mechanism | |
| >exuT | Hexuronate transporter | ||||
| >exuR′ | Negative transcriptional regulator of the Exu operon | ||||
| pK-212 (982071-986106) | Lac− | >′ycbB | Unknown function (amidase?) | σE regulated (45, 46) | |
| >ycbK | Unknown function; σE regulated | ||||
| >ycbL | Unknown function | ||||
| <aspC | Aspartate aminotransferase | ||||
| <ompF′ | Outer membrane protein | ||||
| rybB-lacZ (short) | pK-304 (4102714-4104832) | Lac− | <′cpxA | Membrane sensor kinase in CpxAR two-component signal transduction system | Regulator of periplasmic functions, may down-regulate σE stress |
| <cpxR | Response regulator in CpxAR two-component signal transduction system | ||||
| >cpxP | Periplasmic protein activated by CpxAR, induced in alkaline pH, suppresses toxic envelope protein effects | ||||
| pK-407 (3341506-3345714) | Lac− | >′yhbN | ABC transporter subunit | σE -regulated operon σE competition? (45, 46) | |
| >yhbG | ABC transporter subunit | ||||
| >rpoN | σ54, nitrogen starvation sigma factor | ||||
| >yhbH | Probable σ54 modulation protein | ||||
| >ptsN | Pts, regulates N metabolism | ||||
| >yhbJ′ | ATPase | ||||
| pK-402 (1646935-1649118) | Lac− | ′rzpQ | Unknown function (cryptic prophage) | Cell division defect? | |
| >dicF | sRNA, inhibits ftsZ | ||||
| >dicB | Control of cell division, activates MinC | ||||
| >ydfD | Unknown function (cryptic prophage) | ||||
| >ydfE | Unknown function (cryptic prophage) | ||||
| >insD′ | Cryptic prophage, transposase? | ||||
| rybB-lacZ (long) | pK-601 (2037829-2040601) | Lac++ | >yedY | Unknown function | Unknown mechanism; increase membrane stress? |
| >yedZ | Unknown function (inner membrane protein) | ||||
| >yodA | Cadmium-induced metal binding protein; SoxS, Fur regulated | ||||
| >yodB | Unknown function (cytochrome?) | ||||
Because the rybB-lacZ long fusion resulted in strains that were relatively Lac+, it proved challenging to identify plasmids leading to higher levels of expression (positive regulators); only one was identified in the previous screen. In order to identify possible positive regulators of rybB synthesis, a second screen for random insertion mutations that modulated expression of the rybB-lacZ long fusion was carried out. Approximately 25,000 transposon insertions into the rybB-lacZ strain were screened for changes in the Lac phenotype (screen II). Only two insertions that changed expression in a reproducible fashion were isolated; both reduced rybB expression. Unexpectedly, one was in rep, encoding an ATP-dependent DNA helicase; the second was upstream of dnaK (zaa-1000::kan). The strain carrying the rep::kan insertion grew slowly and was somewhat cold sensitive. The zaa-1000::kan insertion is presumed to be polar on dnaK expression. However, the effect of zaa-1000::kan mutation on DnaK levels was not determined.
While it seemed unlikely that either Rep or DnaK was a direct regulator of rybB transcription, the Lac− phenotype of the strains carrying these insertions allowed an easier screen for multicopy plasmids that increased expression from the rybB-lacZ long fusion. Using strains carrying the rybB-lacZ long fusion containing either the rep::kan (KMT12002) or zaa-1000::kan (KMT12003) insertion, we once again introduced the plasmid genomic library and screened for suppression of the Lac− phenotype on Mac-Lac-Amp plates (screen III). A total of 14,000 transformants were screened for each strain. Four plasmids that increased expression of the fusions were identified; one plasmid was found twice, once in each mutant strain (Table 2). None of the plasmids carried either the rep or dnaK genes. Of the plasmids identified in this screen, only two carried known regulators, emrR and ompR.
TABLE 2.
Genomic library clones isolated as multicopy suppressors; increased expression of rybB-lacZ (screen III)
| Plasmid isolate (fragment boundaries) | Multicopy suppression
|
Gene
|
||
|---|---|---|---|---|
| Original mutation | Modulatory effect in ompR−? | Name | Descriptiona | |
| pK4-14 (1862417-1865705) | rrb2 (rep::kan) and rrb4 | Yes; stimulation | >yeaD | Unknown function, conserved |
| (zaa-1000::kan) | <yeaE | Methylglyoxal reductase | ||
| <mipA | Scaffolding protein for murein polymerase and murein hydrolase | |||
| >yeaG | Unknown function, conserved | |||
| pK4-29 (1403515-1405770) | rrb4 (zaa-1000::kan) | No | >abgR | Putative LysR-type transcriptional regulators |
| <isrA | sRNA | |||
| >ydaL | Unknown function | |||
| <ydaM′ | Unknown function, conserved | |||
| pK4-55 (3531018-3535617) | rrb4 (zaa-1000::kan) | Yes; stimulation | >pckA | Phosphoenolpyruvate carboxykinase |
| (complementation) | <envZ | Inner membrane osomosenser protein, regulates activity of OmpR | ||
| <ompR | Transcriptional regulator involved in osmoregulation of OmpC and OmpF | |||
| >greB | Transcriptional elongation factor | |||
| >yhgF | Unknown function | |||
| pK2-8 (2805651-2809436) | rrb2 (rep::kan) | No | >proX | High-affinity transport for glycine, betaine, and proline |
| >ygaY | Uncharacterized member of the major facilitator superfamily of transporters | |||
| >ygaZ | Predicted transporter | |||
| >ygaH | Predicted inner membrane protein | |||
| >emrR | Negatively regulates transcription of the emrRAB operon encoding subunits of a multidrug efflux pump; acts here via OmpR | |||
EmrR is a negative regulator of genes involved in exporting drugs from the cell (30, 63). While an emrR plasmid stimulated expression of both the long and short rybB-lacZ fusions modestly, a mutation in emrR also had a modest stimulatory effect, suggesting both positive and negative effects of EmrR (data not shown). Epistasis experiments (see below and Fig. 1B) suggested that the EmrR effect was likely to be indirect, and it was not further pursued.
FIG. 1.
OmpR and MipA-YeaE regulation of rybB. (A) β-Galactosidase activity of wild-type (KMT12000) and ΔompR (KMT12005) rybB-lacZ fusion strains. Cells were grown in LB medium at 37°C and assayed as described in Materials and Methods. Numbers on the y axis are machine units as described in Materials and Methods. (B) A ΔompR rybB-lacZ fusion strain (KMT12005) was transformed with plasmid pHDB3 (vector) or library clone pK2-8, pK4-14, pK4-29, or pK4-55 (Table 2), plated on Mac-Lac-Amp plates, and grown overnight at 37°C. (C) Wild-type rybB-lacZ fusion strains carrying either vector control or pK4-14 and ΔompR rybB-lacZ fusion strains carrying either vector control or pK4-14 were grown to an optical density at 600 nm of approximately 0.5 and assayed for β-galactosidase activity. The values are means for three independent experiments. (D) A degP-lacZ (PND254) transcriptional fusion strain carrying either pHDB3 (a vector control) or pK4-14 (mipA-yeaE plasmid) was grown to an optical density at 600 nm of approximately 2.5 to 3.0 and assayed for β-galactosidase activity as described above. Note that PND254 is mutant for cpxA (see Table S1 in the supplemental material), and therefore activation likely reflects only the σE-dependent promoter.
OmpR is the response regulator component of the EnvZ-OmpR two-component signal transduction system. OmpR acts as a transcriptional regulator of OMPs, most notably OmpC and OmpF, in response to changes in osmolarity of the environment (41, 64). OmpR is also a transcriptional activator of two sRNAs, OmrA and OmrB, involved in the posttranscriptional regulation of several OMPs (22). To assess the role of OmpR in the transcription of rybB, a ΔompR derivative of the rybB-lacZ strain was examined. Consistent with multicopy OmpR increasing expression of RybB (Fig. 1B and Table 2), the specific activity of the long rybB-lacZ was reduced approximately fivefold in the ΔompR mutant in stationary phase (Fig. 1A), suggesting that OmpR is necessary for the full activity of the rybB-lacZ fusion.
If OmpR is the sole direct regulator of rybB transcription, we would expect the ompR mutant to be epistatic to all of the stimulatory plasmids from screen III, except for those that carry ompR itself. The four plasmids listed in Table 2 were each transformed into the ΔompR derivative of the long rybB-lacZ fusion, and the Lac phenotype was assessed (Fig. 1B; Table 2). As expected, the plasmid carrying the ompR gene produced a strong Lac+ phenotype (Fig. 1B) in the otherwise Lac− ΔompR background. Two other plasmids, including the one carrying emrR, failed to increase expression in the ompR mutant (Fig. 1B), suggesting that the stimulatory activity of these plasmids was upstream of OmpR and depends upon OmpR. However, the plasmid carrying the mipA (yeaF) and yeaE genes was able to stimulate the rybB-lacZ fusion in both wild-type and ΔompR strains, in both exponential phase (Fig. 1C) and stationary phase (data not shown). Parallel results were found with the short fusion: the mipA plasmid and the ompR plasmid increased activity in an ompR mutant derivative (data not shown).
The finding that mipA-yeaE was epistatic to ompR suggested that OmpR was not the sole primary regulator of rybB. While the function of YeaE is unknown, MipA has been shown to interact with murein synthase and murein hydrolase in vitro (59). MipA is therefore thought to be a scaffolding protein for murein synthesis (59).
These results suggested that possibly neither OmpR nor MipA-YeaE was a direct regulator of rybB. If so, and if they act in a similar way, the stimulation of the rybB promoter upon overproduction of either of these proteins should reflect changes in the activity or synthesis of a transcription factor that acts directly at the rybB promoter. A clue to the identity of this factor came from the realization that ΔompR strains have about a fivefold reduction in both rybB expression and the activity of σE (Fig. 1A) (36). Given the role of MipA in murein synthesis, it seemed possible that these plasmids both act to activate σE, the alternative sigma factor that is activated by periplasmic protein misfolding (4, 36, 37, 48), and that σE promotes transcription of rybB.
An examination of the promoter region of rybB suggested that this was likely to be the case (Fig. 2). The −35 and −10 consensus sequences for E. coli σE-dependent promoters are GGAACTT and TCAAA, respectively, separated by a 16-nt spacer region (46). The rybB upstream region contains highly conserved sequences characteristic of the σE −10 and −35 sequences beginning at −11 and −33 nucleotides upstream from the RybB +1, respectively (Fig. 2). The putative rybB −35 sequence is identical to the −35 sequence for other well-characterized strong σE-dependent promoters in E coli, including the degP, rpoHP3, and rpoEP2 promoters (Fig. 2). The putative rybB −10 sequence has a high degree of similarity to these same promoters, with only one nucleotide change.
FIG. 2.
Alignment of the rybB promoter. The E. coli rybB region from −1 to −39 was aligned with the corresponding sequences from Salmonella, Klebsiella, Yersinia, Photorhabdus, and Erwinia. Below the rybB sequences are the promoter sequences for other well-known E. coli σE-dependent promoters (46). Below the alignment is the consensus sequence previously defined for σE-dependent promoters (46).
Overexpression of OmpR, by increasing the expression of the major OMPs, would be expected to increase σE activity (3) and therefore rybB expression. Does MipA overexpression also increase σE activity? We tested this by determining whether multicopy mipA-yeaE induced transcription of a known σE-dependent promoter, degP. In the presence of the mipA-yeaE plasmid, there is a clear induction of a degP-lacZ transcriptional fusion, compared to the vector control (Fig. 1D), consistent with the idea that rybB transcription may be regulated by σE.
σE is necessary for transcriptional induction of rybB.
The results described above were consistent with the idea that rybB expression was dependent upon σE. We further examined this possibility by testing expression of the rybB-lacZ fusion and RybB RNA levels in strains where σE activity or synthesis was either increased or abolished.
In the absence of RseA, the σE-specific anti-sigma factor, the activity of σE-dependent promoters is significantly increased (16); rybB should be constitutively activated if it is transcribed from a σE-dependent promoter. Indeed, in the absence of RseA, the specific activity of the rybB-lacZ fusion was ∼23-fold higher in exponential phase and 7-fold higher in stationary phase (Fig. 3A). The smaller effect in stationary phase is likely to be due to the fact that σE is already partially activated in stationary-phase cells (14). In addition, overexpression of σE from a plasmid carrying rpoE under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter led to an immediate and significant increase in rybB expression, consistent with σE acting as a direct regulator of rybB transcription (Fig. 3B).
FIG. 3.
σE regulation of rybB. (A) β-Galactosidase activity of wild-type (KMT12000) and rseA (KMT12041) rybB-lacZ strains was assayed in both exponential (optical density at 600 nm [OD600], 0.5) and stationary (OD600, approximately 2.5 to 3.0) phases as described above. (B) β-Galactosidase activity of the wild-type rybB-lacZ strain (KMT12000) containing the vector control pTrc99A (dashed lines) or pLC245 (solid lines) was measured in the presence (solid symbols) or absence (open symbols) of IPTG. Cultures were grown to an OD600 of 0.3, IPTG was added to a final concentration of 1 mM, and aliquots of the culture were taken every 5 min for 30 min. (C) β-Galactosidase activity of wild-type (KMT12000) and ΔrpoE (KMT12047) rybB-lacZ strains were assayed in stationary-phase growth as described above. (D) β-Galactosidase activity of wild-type (KMT12000) and ppGpp0 (KMT12031) rybB-lacZ strains throughout growth.
Conversely, eliminating σE prevented expression of rybB. Although rpoE is essential, it can be deleted in the presence of uncharacterized suppressor mutations (4), allowing us to compare the specific activity of the rybB-lacZ fusion and RybB RNA levels in the presence and absence of rpoE. On MacConkey-lactose plates the ΔrpoE rybB-lacZ fusion strain was totally white (Lac−) (data not shown). The stationary-phase activity of rybB-lacZ seen in the wild type was completely abolished in the ΔrpoE background (Fig. 3C). Northern blots confirmed that RybB RNA was not detected in a ΔrpoE background in stationary phase (Fig. 4A). In cells carrying the plasmid with an IPTG-inducible rpoE gene, expression of the rybB fusion was restored (data not shown) and RybB RNA could be detected even in exponential phase (Fig. 4B).
FIG. 4.
σE-dependent RybB RNA expression in vivo and in vitro. (A) Northern blot of RybB RNA isolated from wild-type (KMT12000), ΔrpoE (ΚΜΤ12047), and ΔrybB (KMT12054) strains. Total RNA was isolated from exponential (optical density at 600 nm [OD600], 0.5 to 0.7) and stationary (OD600, 2.5 to 3.0) phases of growth and processed as described in Materials and Methods. (B) Northern blot for RybB RNA isolated in exponential phase (OD600, 0.5 to 0.7). The wild-type rybB-lacZ strain (KMT12000) containing the vector control pTrc99A or pLC245 was grown to an OD600 of 0.3 and treated with 100 μM IPTG for 60 min, and total RNA was isolated from each sample and corresponding no-IPTG control and processed as described in Materials and Methods. (C) In vitro transcription assay with purified core RNA polymerase, purified σE, and the rybB DNA template. Lanes 1 to 3, purified core RNA polymerase only; lanes 4 to 6, purified core RNA polymerase plus purified σE with the respective templates: PdegP, a σE-dependent promoter (lanes 1 and 4); PrybB (lanes 2 and 5); and PyadF, a σ70-dependent promoter (lanes 3 and 6).
It was recently shown that the activity of σE-dependent promoters increases upon entry into stationary phase (14), which is consistent with our observations of rybB-lacZ activity (Fig. 1A). This stationary-phase induction of σE-dependent promoters was shown to be dependent on the alarmone guanosine 3′,5′-bispyrophosphate (ppGpp) (14). If the stationary-phase induction of the rybB promoter is ppGpp dependent, then the stationary-phase induction of rybB-lacZ should be abolished in a ppGpp0 (ΔrelA ΔspoT) genetic background. In order to determine if the rybB promoter was ppGpp dependent, a ΔrelA ΔspoT double mutant of the rybB-lacZ transcriptional fusion was constructed and assayed for β-galactosidase activity throughout growth. Consistent with the observations of Costanzo and Ades (14), the stationary-phase induction of rybB-lacZ is defective in a ppGpp0 genetic background (ΔrelA ΔspoT) (Fig. 3D). Altogether, these results strongly support the idea that transcriptional induction of rybB is completely σE dependent. The short rybB-lacZ fusion gave parallel results, showing full dependence on rpoE on plates and increased synthesis in an rseA mutant (data not shown).
As final evidence that σE directly regulates rybB, in vitro transcription reactions were carried out to determine if there was a biochemical interaction between σE and the rybB promoter. In these experiments, the rybB transcript was expressed 10-fold more than the degP transcript (Fig. 4C), generally considered a good σE-dependent promoter (20). In a control experiment using the vegetative sigma factor, σ70, no transcript was detected for the rybB gene (data not shown). The transcript start determined here is consistent with that predicted from the promoter and previously determined in vivo (Fig. 2) (57). Taken together, the in vivo and in vitro data show that rybB is completely σE dependent and that in the presence of σE, the rybB promoter is highly transcribed.
RybB modulation of σE activity.
Plasmids containing rybB were isolated in screen I as negative regulators of the rybB-lacZ fusion (Table 1), suggesting that RybB may negatively regulate itself. This was tested further by expressing rybB under the control of the pBAD promoter. The activity of rybB-lacZ was decreased 2-fold in exponential phase and 10-fold in stationary phase when RybB was overexpressed by induction with arabinose (Fig. 5A). Thus, overexpression of RybB is sufficient to down-regulate the σE-dependent rybB promoter. The short rybB-lacZ fusion gave similar results; RybB overexpression decreased expression of the fusion 10-fold (data not shown).
FIG. 5.
Regulation of σE by sRNAs. Aliquots of the cultures were assayed for β-galactosidase activity as described in Materials and Methods. The values are means from three independent experiments. Error bars represent standard deviations. Numbers on the y axis are machine units as described in Materials and Methods. (A and B) β-Galactosidase activities of a Δara rybB::kan rybB-lacZ long-transcriptional-fusion strain (KMT12120) containing the vector control pNM12 or pKMT5 (pBAD-rybB+) (A) and the wild-type rpoE-lacZ fusion strain (KMT14000) containing the vector control pNM12 or pKMT5 (B) in the presence of arabinose (0.02%) were assayed in both exponential and stationary phases of growth. (C and D) β-Galactosidase activities of wild-type (KMT12000), ΔrybB (KMT12054), and Δhfq (KMT12055) rybB-lacZ strains (C) and wild-type (KMT14000), ΔrybB (KMT14002), and Δhfq (KMT14003) rpoE-lacZ translational fusion strains (D) were assayed during growth in LB medium.
As discussed above, σE is the direct regulator of rybB transcription. Therefore, it is possible that RybB autorepression is due to a RybB regulatory effect on σE synthesis or activity. This was tested by assaying an rpoE-lacZ translational fusion in the presence of rybB expressed from the pBAD promoter. This fusion contains the σE-dependent promoter for rpoE but apparently not a functional σ70 promoter; it is fully σE dependent (data not shown). It should reflect both transcriptional and translational regulation of rpoE. The activity of rpoE-lacZ decreased approximately 3-fold in exponential and approximately 11-fold in stationary phase when RybB was overexpressed (Fig. 5B). These results parallel those of the rybB-lacZ fusion, indicating that RybB overexpression is sufficient to inhibit σE synthesis and/or activity.
Introduction of a rybB-null mutation into the rybB-lacZ transcriptional fusion strain led to a modest increase (less than twofold) in fusion expression (Fig. 5C). Not surprisingly, this effect was detectable only in stationary phase, when RybB is normally expressed. Since RybB was reported to bind to the RNA chaperone Hfq, and most Hfq-binding sRNAs require Hfq for their activity (55), mutating hfq should also increase the activity of the rybB-lacZ fusion. In fact, a null mutation in hfq increased expression of rybB-lacZ dramatically, in both exponential and stationary phase (Fig. 5C). Parallel effects were seen with the rpoE-lacZ fusion (Fig. 5D). Deletion of the rybB gene led to a twofold increase in the activity of the rpoE-lacZ fusion in stationary phase, and the absence of hfq increased the activity of the rpoE-lacZ fusion throughout growth about 5- to 10-fold (Fig. 5D). Thus, it seems likely that, in addition to RybB, other Hfq-dependent RNAs also negatively regulate σE synthesis or activity. Furthermore, the high level of expression of these σE reporters in an hfq mutant under otherwise unperturbed growth conditions suggests that the σE stress response is significantly activated under these conditions. It is interesting that the activity of both rybB-lacZ and rpoE-lacZ fusions still increases in stationary phase, even in the absence of hfq. Other recent reports of Hfq-dependent negative regulation of σE in E. coli (19, 23, 42) (see Discussion), in Salmonella (21), and in Vibrio cholerae (18) are consistent with our findings.
Characterization of sRNA regulation of σE.
One model for the RybB negative regulation of σE activity by sRNAs is by negative regulation of OMP synthesis, leading to a decrease in periplasmic stress. OMPs, including the major outer membrane porin OmpC, contain a C-terminal Y-X-F tripeptide motif. This region has been shown to interact with the PDZ domain of the periplasmic protease DegS, leading to cleavage of the RseA anti-sigma factor (1, 61). As noted above, a mutation in ompR that reduces the synthesis of the major outer membrane porins OmpC and OmpF leads to decreased expression of σE-dependent reporters (Fig. 1A). This suggests that decreasing the flux of proteins to the cell surface is sufficient to decrease periplasmic stress. A number of sRNAs that negatively regulate OMPs with this C-terminal motif have been identified (reviewed in references 22 and 58). This set of sRNAs that target OMPs may provide an explanation for the strong effect of hfq mutants on σE activity.
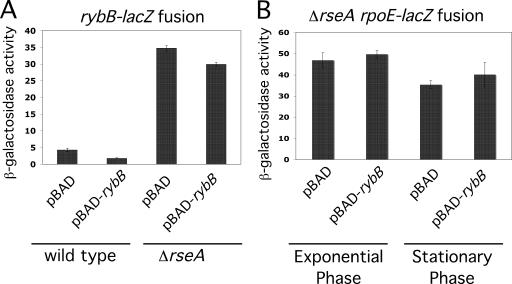
If RybB autorepression and RpoE repression are indirect, via down-regulating OMP synthesis, we would expect the effect on σE-dependent promoters to depend upon RseA. This was tested with three different σE-dependent reporters, rybB-lacZ, rpoE-lacZ (Fig. 6), and rpoHP3-lacZ (data not shown). A wild type and a ΔrseA derivative of each reporter were tested for the effect of overexpressing RybB from an arabinose-inducible promoter. The rseA mutation significantly increased the basal level of expression of the rybB-lacZ, rpoEP2-lacZ (Fig. 6; compare Fig. 6B to Fig. 5B) and rpoHP3-lacZ (data not shown) fusions, as expected. RybB was unable to down-regulate the expression of any of these fusions in the ΔrseA strain background (Fig. 6 and data not shown).
FIG. 6.
RybB autorepression is RseA independent. β-Galactosidase activities were assayed as for Fig. 5. (A) β-Galactosidase activity of the Δara rybB::kan rybB-lacZ transcriptional fusion strain (KMT12120) containing the vector control pNM12 or pKMT5 and the ΔrseA rybB::kan rybB-lacZ transcriptional fusion strain (KMT12119) containing the vector control pNM12 or pKMT5 (pBAD-rybB+) grown with arabinose (0.02%); results for exponential growth are shown. (B) β-Galactosidase activity of the ΔrseA rpoE-lacZ fusion strain (KMT14001) containing the vector control pNM12 or pKMT5 grown with arabinose and assayed in both exponential and stationary phases of growth.
Reexamination of plasmids from screens.
The identification of σE as an essential regulator of rybB-lacZ suggested that plasmids and mutations that modulated rybB-lacZ expression in our screens should also act through effects on σE synthesis or activity, as RybB itself seems to. We reexamined the plasmids isolated in these screens to see whether they provided any further insights into σE signaling. Regulation of expression of the σE-dependent reporter might occur in various ways: (i) by increasing or decreasing periplasmic stress by changes in synthesis of OMPs mediated through RseA, the anti-sigma factor for σE (1, 4, 5, 61); (ii) by titration of σE by the presence of σE promoter on a multicopy plasmid; or (iii) by affecting σE translation or activity. In addition, it is possible that a given σE reporter might be sensitive to an additional regulator that is not active on other σE promoters.
The stimulatory plasmids described in Table 2 all either directly or indirectly affect the synthesis of OMPs that signal to the RseA protease. Overexpression of OmpR presumably leads to the overexpression of OmpC or OmpF, both known to induce σE activity. Similarly, MipA, an OMP involved in murein synthesis (39, 59), carries the DegS-activating sequence in its C terminus (61), and activates not only the rybB-lacZ fusion but also another σE-dependent fusion, degP-lacZ (Fig. 1D). The other two plasmids isolated in screen III (Table 2) acted upstream of OmpR and therefore, we believe, act by signaling to OmpR, indirectly increasing Omp synthesis. It seems likely that overexpression of many genes perturbs the cell envelope and/or osmolarity control sufficiently to cause induction of the EnvZ/OmpR system and therefore stimulate σE expression.
In a reciprocal manner, we expect that the plasmids in screen I (Table 1) that decreased expression of the rybB-lacZ fusion decrease the synthesis and/or activity of σE. Three of the plasmids contain the regions previously identified either as carrying σE promoters or showing regulation by σE in genome-wide expression arrays (Table 1) (46). Titration of σE is one possible explanation for the down-regulation of the rybB promoter by these plasmids, but it is also likely that the products are made in a σE-dependent fashion because they contribute to decreasing periplasmic stress. One plasmid carries cpxR and part of cpxA; cpxR is the response regulator for a set of genes also involved in periplasmic stress relief (44). Presumably, overexpressing CpxR turns on genes that reduce the basal level signaling to σE. Consistent with this idea, overexpression of CpxR is known to functionally compensate for lack of σE at high temperatures (13).
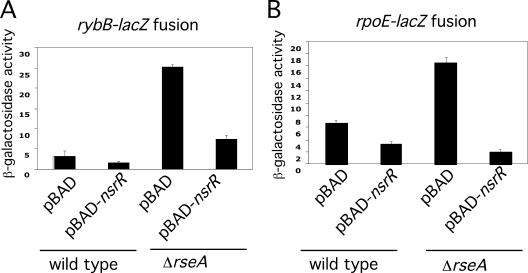
One plasmid identified in screen I carried nsrR, a transcriptional repressor that responds to nitric oxide and helps the cell respond to the stress of nitric oxide exposure (7). Overexpression of nsrR from the pBAD promoter decreased expression of the rybB-lacZ fusion, as well as expression of rpoE-lacZ (Fig. 7A and B, respectively). Therefore, the activity of the original plasmid appears to be due to NsrR. The overexpression of nsrR inhibited these two σE reporters even in a ΔrseA strain (Fig. 7). This identifies NsrR as a novel, RseA-independent regulator of σE-dependent genes, although whether regulation is direct or indirect remains to be determined. This suggests that the nitrosative stress response may somehow communicate with the envelope stress response. None of the other plasmids from screen I were tested in this way.
FIG. 7.
NsrR regulation of rybB and rpoE expression (A) β-Galactosidase activities of the Δara714 leu::Tn10 (Tetr) rybB-lacZ transcriptional fusion strain (KMT12069) containing the vector control pNM12 or pKMT8 (pBAD-nsrR+) and the Δara714 leu::Tn10 (Tetr) ΔrseA rybB-lacZ transcriptional fusion strain (KMT12078) containing the vector control pNM12 or pKMT8 assayed in the presence of arabinose (0.02%) in late exponential phase. (B) β-Galactosidase activities of the wild type (KMT14000) and the ΔrseA rpoE-lacZ fusion strain (KMT14001) containing the vector control pNM12 or pKMT8 assayed in late exponential phase.
DISCUSSION
The σE regulon of E. coli controls a set of periplasmic proteases and chaperones, as well as proteins that act to properly direct proteins to the outer membrane (46). The work described here, along with recent publications and reports from other groups, extends the regulon to noncoding regulatory RNAs and demonstrates that these regulatory RNAs have an important role in reducing the basal level of induction of the σE system, presumably by reducing the flux of OMPs to the cell surface (24, 42).
RybB, the sRNA studied here, is a conserved 81-nt RNA that binds Hfq and is expressed during stationary-phase growth (57, 62). In the present study, we examined the regulation and function of RybB. We found that rybB was fully dependent upon σE for expression in vivo, was transcribed efficiently by σE in vitro, and has a promoter that fits the previously described consensus for σE promoters (46). These findings indicate that rybB is an sRNA member of the σE regulon. We further found that RybB function is to negatively regulate the σE regulon: RybB overexpression decreases expression of several σE promoters, including rybB itself; deletion of rybB has the converse effect.
While the manuscript was in preparation, two groups working with E. coli and Salmonella reported that RybB and another regulatory RNA, MicA, require σE for their transcription, down-regulate σE activity, and have as their direct targets several OMPs (24, 42, 53). The only known target of MicA is the major OMP OmpA; RybB down-regulates transcripts for at least two porins in E. coli and 14 transcripts for OMPs in Salmonella. Some targets down-regulated after overexpression of σE, for instance, ompX, have no known sRNA regulators, suggesting that there may be other σE-regulated sRNAs as well (23, 46).
The primary role of σE has been thought to be responding to envelope stress that results from environmental change. The emerging evidence that the σE regulon encodes multiple sRNAs that negatively regulate porins and other envelope proteins changes this view in a very important way. It has long been known that cells have mechanisms to balance production of OMPs. For example, upon overexpression of some OMPs, the expression of other OMPs is lowered (11, 12, 17). Therefore, there must be a mechanism for coordination of OMP expression that functions above the level of individual transcription factors such as OmpR. We propose that σE, through its sRNA arm, performs this coordination function. It appears that during normal cell growth, some, if not most, OMP genes are transcribed and would be translated in excess, beyond the level needed for function. The relative levels of many of these proteins are determined by sRNAs of the regulon utilizing two mechanisms: (i) sRNAs directly target certain mRNAs for destruction, and (ii) relative levels of these targets in turn alter σE activity, either directly by activating DegS to initiate RseA cleavage (OmpC, OmpW) or indirectly by altering the envelope folding environment (OmpA). Together, these mechanisms allow the cell to set the relative levels of many envelope constituents. Moreover, additional features enhance the reach of this integrative function of σE. Many sRNAs not encoded in the σE regulon have targets that affect σE activity (e.g., MicF and MicC), thereby allowing their effects to be coordinated with those of σE. Taken together, these results suggest that a primary function of σE is to coordinate expression of envelope constituents, especially porins, under all growth conditions.
The importance of this coordination can be best appreciated when it is disrupted. Overexpression of the σE regulon causes cell growth problems and lysis in E. coli (25, 40). In Salmonella, a strain with an rseA deletion rapidly acquired secondary mutations that decreased σE activity, and an rseA hfq double mutation proved to be lethal (21). Presumably under these conditions, the σE regulon is fully induced and there is abundant machinery for folding OMPs and inserting them into the outer membrane. Thus, the tendency for these cells to lyse may instead reflect too high a protein load in the outer membrane, rather than the accumulation of unfolded OMPs in the periplasm. The limitation of OMP synthesis in a σE-dependent fashion thus appears to reflect a critical role in regulating both the flux of OMPs (to allow proper folding and insertion) and the capacity of the outer membrane itself.
Other levels of regulation of the RybB promoter.
In this picture of the σE network, RybB and MicA are made in response to σE stress and down-regulate that stress, either directly or indirectly by down-regulating the synthesis of OMPs that are the substrates for the σE functions. The strong phenotype of an hfq mutant in expressing σE genes even in exponential growth suggests that the down-regulation of other OMPs by sRNAs, probably not all under σE control, is operating under all growth conditions. RybB overexpression down-regulates three different σE-dependent fusions tested, rybB-lacZ, rpoHP3-lacZ, and rpoEP2-lacZ, all in an RseA-dependent fashion. We expect all OMP signaling to be via RseA cleavage of this anti-sigma factor. The observation that the three fusions are not down-regulated by RybB in an rseA mutant strongly suggests that the RybB effect on σE activity and/or synthesis is RseA dependent (i.e., via OMPs). This is consistent with the observations of others that RybB down-regulates OMPs (24, 42).
The rybB-lacZ fusion proved to be a sensitive reporter for examining the σE signaling pathway. As noted above, one of the plasmids isolated in screen I which down-regulated the rybB-lacZ fusion (Table 1) contained nsrR, which proved to be an RseA-independent regulator of two σE-dependent fusions, rybB-lacZ and rpoE-lacZ (Fig. 7). The identification of NsrR as a regulator of σE-dependent genes suggests a connection between periplasmic stress and nitric oxide stress that warrants further investigation.
Upstream signals that affect σE.
The approach used in this study to investigate rybB regulation identified both multicopy plasmids and mutations that affected the rybB-lacZ fusion, now known to reflect σE function. Among the plasmids that reduced function were some that may titrate σE or reduce stress that signals to σE; some stimulatory plasmids identified in screen III act by activating OmpR, presumably increasing the synthesis of outer membrane proteins. Previous plasmid screens for stimulators of a σE-dependent promoter gave a nonoverlapping list of candidates (36), suggesting that there are significant numbers of genes and pathways that can lead to induction.
Most puzzling of the genetic loci identified in our screens was the insertion mutation in the rep gene, encoding a DNA helicase. This insertion significantly decreases expression of the fusion, and therefore presumably σE activity. The Rep helicase is involved in the restart of damaged replication forks, and in cells devoid of rep, there may be accumulation of DNA for early regions of the chromosome (in front of possible replication errors) compared to later regions. The genes for the major outer membrane porins, ompC, ompF, and ompA, are all relatively far from oriC. Their location may cause a lower level of OMP synthesis in the rep mutant. If this is the explanation for the lower activity in the rep mutant, it may be serendipitous, or it may reflect a mechanism for ensuring that the dosage of these genes does not increase until near the end of the replication cycle, when cells, and therefore the available cell surface, are larger.
Our investigation of rybB regulation, combined with work by others, points to a critical role for Hfq-regulated sRNAs in the control of the cell surface and the transport of OMPs to the cell surface. At the core of this response are MicA and RybB, made in large amounts only when the export machinery is stressed, and capable of down-regulating major outer membrane porins. The combination of multiple sRNAs, regulated in a variety of ways and targeting a variety of different OMP transcripts, provides the flexibility and capability of responding to many signals that the cell is likely to need.
Supplementary Material
Acknowledgments
We thank members of the Gottesman lab for their help at various stages of this project and Carol Gross for her support and comments on the work and the manuscript. In particular, we thank Nadim Majdalani for advice and assistance with various strain constructions. Also, we thank Carin Vanderpool and Mitsuoki Kawano for help with sequencing of the transposon insertions and for providing the pool of random kanamycin insertions. We thank Gisela Storz, Nadim Majdalani, Carin Vanderpool, Colleen McCullen, Maude Guillier, Alexander Bougdour, Pierre Mandin, and Kyung Moon for their comments and critical reading of the manuscript.
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 6 April 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-σ factor. Genes Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 3.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 4.Alba, B. M., H. J. Ji Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σE activity. Mol. Microbiol. 40:1323-1333. [DOI] [PubMed] [Google Scholar]
- 5.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 7.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S., E. A. Lesnik, T. A. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65:157-177. [DOI] [PubMed] [Google Scholar]
- 9.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 10.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Click, E. M., G. A. McDonald, and C. A. Schnaitman. 1988. Translational control of exported proteins that results from OmpC porin overexpression. J. Bacteriol. 170:2005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Click, E. M., and C. A. Schnaitman. 1989. Export-defective LamB protein is a target for translational control caused by OmpC porin overexpression. J. Bacteriol. 171:616-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly, L., A. De Las Penas, B. M. Alba, and C. A. Gross. 1997. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 11:2012-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costanzo, A., and S. E. Ades. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigma E, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Court, D. L., S. Swaminathan, D. Yu, H. Wilson, T. Baker, M. Bubunenko, J. Sawitzke, and S. K. Sharan. 2003. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene 315:63-69. [DOI] [PubMed] [Google Scholar]
- 16.De Las Peñas, A., L. Connolly, and C. A. Gross. 1997. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 17.Diedrich, D. L., and J. A. Fralick. 1982. Relationship between the OmpC and LamB proteins of Escherichia coli and its influence on the protein mass of the outer membrane. J. Bacteriol. 149:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53:345-354. [DOI] [PubMed] [Google Scholar]
- 19.Douchin, V., C. Bohn, and P. Bouloc. 2006. Down-regulation by porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 281:12253-12259. [DOI] [PubMed] [Google Scholar]
- 20.Erickson, J. W., V. Vaughn, W. A. Walter, F. C. Neidhardt, and C. A. Gross. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419-432. [DOI] [PubMed] [Google Scholar]
- 21.Figueroa-Bossi, N., S. Lemire, D. Maloriol, R. Balbontin, J. Casadesus, and L. Bossi. 2006. Loss of Hfq activates the sigma E-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 62:838-852. [DOI] [PubMed] [Google Scholar]
- 22.Guillier, M., and S. Gottesman. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 59:231-247. [DOI] [PubMed] [Google Scholar]
- 23.Guisbert, E., V. A. Rhodius, N. Ahuja, E. Witkin, and C. A. Gross. 2007. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 189:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 364:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Kabir, M. S., D. Yamashita, S. Koyama, T. Oshima, K. Kurokawa, M. Maeda, R. Tsunedomi, M. Murata, C. Wada, H. Mori, and M. Yamada. 2005. Cell lysis directed by σE in early stationary phase and effect of induction of the rpoE gene on global gene expression in Escherichia coli. Microbiology 151:2721-2735. [DOI] [PubMed] [Google Scholar]
- 26.Kawamoto, H., T. Morita, A. Shimizu, T. Inada, and H. Aiba. 2005. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 19:328-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawano, M., A. A. Reynolds, J. Miranda-Rios, and G. Storz. 2005. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 33:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 30.Lomovskaya, O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDowell, J. C., J. W. Roberts, D. J. Jin, and C. Gross. 1994. Determination of intrinsic transcription initiation by RNA polymerase elongation rate. Science 266:822-825. [DOI] [PubMed] [Google Scholar]
- 36.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of σE, an Escherichia coli heat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 37.Mecsas, J., R. Welch, J. W. Erickson, and C. A. Gross. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J. Bacteriol. 177:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, and K. L. Williams. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 40.Nitta, T., H. Nagamitsu, M. Murata, H. Izu, and M. Yamada. 2000. Function of the σE regulon in dead-cell lysis in stationary phase Escherichia coli. J. Bacteriol. 182:5231-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norioka, S., K. Ramakrishnan, K. Ikenaka, and M. Inouye. 1986. Interaction of a transcriptional activator, OmpR, with reciprocally osmoregulated genes, ompF and ompC, of Escherichia coli. J. Biol. Chem. 261:17113-17119. [PubMed] [Google Scholar]
- 42.Papenfort, K., V. Pfeiffer, F. Mika, S. Lucchini, J. C. D. Hinton, and J. Vogel. 2006. σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters, J. E., and N. L. Craig. 2000. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell 6:573-582. [DOI] [PubMed] [Google Scholar]
- 44.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhodius, V., N. Savery, A. Kolb, and S. Busby. 2001. Assays for transcription factor activity. Methods Mol. Biol. 148:451-464. [DOI] [PubMed] [Google Scholar]
- 46.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:0043-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rivas, E., R. J. Klein, T. A. Jones, and S. R. Eddy. 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 11:1369-1373. [DOI] [PubMed] [Google Scholar]
- 48.Rouvière, P., A. De Las Penas, J. Mescas, C. Zen Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, σE, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 51.Sledjeski, D. D., and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 52.Straus, D. B., W. A. Walter, and C. A. Gross. 1989. The activity of σ32 is reduced under conditions of excess heat shock protein production in Escherichia coli. Genes Dev. 3:2003-2010. [DOI] [PubMed] [Google Scholar]
- 53.Udekwu, K. I., and E. G. Wagner. 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 35:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulbrandt, N. D., J. A. Newitt, and H. D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187-196. [DOI] [PubMed] [Google Scholar]
- 55.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 51:1525-1533. [DOI] [PubMed] [Google Scholar]
- 56.Vanderpool, C. K., and S. Gottesman. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076-1089. [DOI] [PubMed] [Google Scholar]
- 57.Vogel, J., V. Bartels, H. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. H. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel, J., and K. Papenfort. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9:605-611. [DOI] [PubMed] [Google Scholar]
- 59.Vollmer, W., M. von Rechenberg, and J.-V. Holtje. 1999. Demonstration of molecular interaction between the murein polymerase PBP1, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 60.Wagner, E. G. H., and R. W. Simons. 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 48:713-742. [DOI] [PubMed] [Google Scholar]
- 61.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 62.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong, A., A. Gottman, C. Park, M. Baetens, S. Pandza, and A. Matin. 2000. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 44:2905-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida, M., L. Qin, L. A. Egger, and M. Inouye. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114-17123. [DOI] [PubMed] [Google Scholar]
- 65.Young, B. A., L. C. Anthony, T. M. Gruber, T. M. Arthur, E. Heyduk, C. Z. Lu, M. M. Sharp, A. Heydorn, R. R. Burgess, and C. A. Gross. 2001. A coiled-coil from the RNA polymerase β′ subunit allosterically induces selective nontemplate strand binding by σ70. Cell 105:935-944. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, Y.-N., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.