Abstract
Along with methane, methanol and methylated amines represent important biogenic atmospheric constituents; thus, not only methanotrophs but also nonmethanotrophic methylotrophs play a significant role in global carbon cycling. The complete genome of a model obligate methanol and methylamine utilizer, Methylobacillus flagellatus (strain KT) was sequenced. The genome is represented by a single circular chromosome of approximately 3 Mbp, potentially encoding a total of 2,766 proteins. Based on genome analysis as well as the results from previous genetic and mutational analyses, methylotrophy is enabled by methanol and methylamine dehydrogenases and their specific electron transport chain components, the tetrahydromethanopterin-linked formaldehyde oxidation pathway and the assimilatory and dissimilatory ribulose monophosphate cycles, and by a formate dehydrogenase. Some of the methylotrophy genes are present in more than one (identical or nonidentical) copy. The obligate dependence on single-carbon compounds appears to be due to the incomplete tricarboxylic acid cycle, as no genes potentially encoding alpha-ketoglutarate, malate, or succinate dehydrogenases are identifiable. The genome of M. flagellatus was compared in terms of methylotrophy functions to the previously sequenced genomes of three methylotrophs, Methylobacterium extorquens (an alphaproteobacterium, 7 Mbp), Methylibium petroleiphilum (a betaproteobacterium, 4 Mbp), and Methylococcus capsulatus (a gammaproteobacterium, 3.3 Mbp). Strikingly, metabolically and/or phylogenetically, the methylotrophy functions in M. flagellatus were more similar to those in M. capsulatus and M. extorquens than to the ones in the more closely related M. petroleiphilum species, providing the first genomic evidence for the polyphyletic origin of methylotrophy in Betaproteobacteria.
Methylotrophy is the metabolic capacity to grow on reduced carbon compounds containing no C—C bonds, such as methane, methanol, methylated amines, etc. (1, 39). While the role of methanotrophs in the reduction of global emissions of methane has been well recognized (27), less attention has been paid to nonmethanotrophic methylotrophs as participants in the global carbon cycle. However, recent models estimate methanol emissions into the atmosphere at 82 to 273 teragrams (Tg) year−1 (with living plants as the major source [19, 25]), putting methanol emission on a scale similar to that of methane emissions (approximately 600 Tg year−1 [36]) and pointing toward the global role of nonmethanotrophic methanol utilizers. While no global modeling has been attempted to characterize the production of methylated amines, they are known to be abundant in marine and freshwater environments and represent dynamic constituents of not only carbon but also nitrogen global cycles (44). So far, only nonmethanotrophic methylotrophs have been implicated in utilizing methylated amines (1, 39).
Methylobacillus flagellatus strain KT utilizes methanol and methylated amines as the sole sources of carbon and energy and is classified as an obligate methylotroph (24). The strain was isolated in the early 1980s from a metropolitan sewer system (24) and selected as a prospective industrial strain due to its high growth rates on methanol, high tolerance to methanol and formaldehyde, high biomass yield, and high coefficient of conversion of methanol into biomass (2, 3, 7, 24). Derivatives of the strain have been successfully generated, aimed at commercial production of value-added compounds (6, 21, 41). In addition to its commercial potential, the strain has become one of the most prominent models for studying biochemistry of methylotrophy, as it presents a facile genetic system with a variety of tools for manipulation, such as suicide vectors for site-directed mutagenesis, expression vectors, promoter probe vectors, etc. (8, 17, 18, 33). Thus, the genome-based analysis of methylotrophy in this organism is complemented by a body of previous genetic and biochemical data. Based on 16S rRNA sequence, M. flagellatus belongs to the Betaproteobacteria class and is most closely related to other members of the family Methylophilaceae (24). The genomic sequence of M. flagellatus reported here presents an excellent case study for the comparative analysis of the molecular basis of methylotrophy in alpha-, beta-, and gammaproteobacteria.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations were used in the text: CRISPR, clustered regularly interspaced short palindromic repeats; H4MPT, tetrahydromethanopterin; PQQ, pyrroloquinoline quinone; EPS, exopolysaccharide; MDH, methanol dehydrogenase; MADH, methylamine dehydrogenase; RuMP cycle, ribulose monophosphate cycle; GND, 6-phosphogluconate dehydrogenase; FDH, formate dehydrogenase; PEP, phosphoenolpyruvate; OAA, oxaloacetate; GAP, glyceraldehyde phosphate; TCA cycle, tricarboxylic acid cycle.
Methylobacillus flagellatus strain KT (ATCC 51484) was obtained from the laboratory collection. For the isolation of genomic DNA, cultures were grown in 100 ml of minimal medium (28) supplemented with 2% (wt/vol) methanol. Genomic DNA was isolated from late-exponential-phase cultures in accordance with recommendations of the Department of Energy's Joint Genome Institute (DOE-JGI; Walnut Creek, CA). The genome was sequenced using the whole-genome shotgun method (16) and assembled using standard tools as described on the JGI website (http://www.jgi.doe.gov/). Gaps remaining in the assembled sequence were closed by primer walking or by sequencing specifically amplified PCR fragments. The sequence was finished and polished at the JGI Los Alamos National Laboratory facility. The assembled genome was computationally annotated, and automated functional assignments were manually curated.
Nucleotide sequence accession number.
The sequence of the complete M. flagellatus strain KT genome is available under GenBank accession number CP000284 and also from the JGI website (http://genome.jgi-psf.org/finished_microbes/metfl/metfl.home.html).
RESULTS
General genome features and basic functions.
The genome consists of a single circular chromosome of 2,971,517 base pairs (55.7% GC content), of which 143,032 base pairs represent a direct identical repeat (see below). A total of 2,766 coding regions are recognized in the genome, of which 144 are identical doubles, as these are parts of the extended repeat. Of the total translatable open reading frames, 233 are unique to M. flagellatus, 2,520 have top BLAST hits with bacterial genes, 10 have top hits with archaeal genes, and 3 have top hits with eukaryotic genes. Based on protein identity scores, the closest relatives of M. flagellatus whose complete genome sequences are available are the betaproteobacteria Thiobacillus denitrificans (492 of a total of 1,681 betaproteobacterial top hits) and Dechloromonas aromatica (299 top hits). Of 568 gammaproteobacterial top hits, 108 are with the proteins translated from the chromosome of Methylococcus capsulatus, an obligate methane utilizer whose genome has been recently sequenced (54). Some of these are well-characterized methylotrophy genes (see below), while most of the remaining genes are hypothetical genes, and some of these may be involved in methylotrophy as well. The chromosome contains two ribosomal (16S-23S-5S) operons and all the genes encoding ribosomal proteins. There are a total of 46 tRNA genes corresponding to 38 tRNA acceptors for recognizing all 20 amino acids. Only one complete (Tn3 type) transposase gene is present in the genome (Mfla1495); a partial gene is present nearby (Mfla1488), and the two surround a group of genes predicted to be involved in arsenate resistance. A number of phage-related genes were identified in the genome, and most of these appear to have homologs in related betaproteobacteria, with a few exceptions that are unique to M. flagellatus (see below). The genome of M. flagellatus, like many microbial genomes (40), contains a region of CRISPR. It is organized as 93 identical sequences of 32 nucleotides interspaced by nonidentical sequences of 33 to 39 nucleotides (nucleotides 632591 to 638803), preceded by six genes encoding CRISPR-associated proteins (Mfla601 to Mfla607). The role of CRISPR is not yet well understood, but they have been implicated recently in DNA rearrangements, lateral gene transfers, host cell defense, replication, and regulation (13, 22, 26, 46). They have also been used in evolutionary studies and strain typing (40). So far, the CRISPR structure in the chromosome of M. flagellatus is not homologous to any known CRISPR.
Standard sets of genes are present for DNA replication, transcription, and translation, and complete pathways are apparently present to synthesize all the amino acids and nucleotides and all the essential vitamins and cofactors [biotin, thiamine, riboflavin, pyridoxal phosphate, cyanocobalamin, folic acid, heme, coenzyme A (CoA), NAD(P), tetrahydrofolate, H4MPT, and PQQ]. Few secondary metabolite synthesis pathways are predicted in the genome, for example, a pathway for terpenoid precursor (geranylgeranyl) biosynthesis. A single large gene cluster (Mfla1940 to Mfla1987) is predicted to encode the flagellum functions.
Two major types of terminal oxidases have been previously detected in M. flagellatus, an o-type oxidase similar to the Escherichia coli bo-type oxidase and a bb-type oxidase sharing some properties in common with the E. coli bd-type oxidase (42, 48). Two clusters were recognized in the genome potentially encoding cytochrome c oxidases (complex IV, Mfla629 to Mfla631 and Mfla1292 to 1295, respectively). Based on protein identities, the former cluster is most likely responsible for the bb-type terminal oxidase, while the latter is responsible for the o-type oxidase. In addition, a complete set of genes for NADH quinone oxidoreductase (complex I) was identified, all in one cluster (Mfla2048 to Mfla2061).
A relatively large number of two-component signal transduction proteins predicted in the genome (31 histidine kinases and 31 response regulators), recently conceptualized as “bacterial IQ” (20), point toward a rather high potential for environmental adaptability. In addition, numerous single-component regulatory proteins (predominantly encoded by the lysR and tetR type) are predicted. However, little is known about the regulation of either methylotrophy or the general metabolic functions of M. flagellatus. Thus, the specific functions of the predicted regulators will need to be tested via mutation and/or expression analyses in the future. A large number of genes encoding putative transporters were identified in the genome. At least some of the 31 TonB-dependent siderophore receptor gene homologs are likely involved in iron uptake. Other putative transporters are predicted to be involved in transport of other metals or in nitrate, ammonium, or sulfate metabolism, as well as in EPS transport. Type I, II, and IV secretion systems are also predicted. A cluster of genes encoding parts of a phosphotransferase-type sugar transport system was identified, similar to the clusters previously characterized in Nitrosomonas europaea (31) and Nitrosococcus oceani (38). However, the functions of these genes remain enigmatic (31).
Methylotrophy.
(i) Primary oxidation of methanol and methylamine.
M. flagellatus exhibits high growth rates on methanol or methylamine (up to 0.73 h−1 [2, 3, 7]) and possesses high activities of MDH and MADH, respectively (7, 37). The genome analysis revealed the presence of a gene cluster predicted to encode MDH and accessory proteins (mxaFJGIRSACKLD; Mfla2034 to Mfla2044) similar to the clusters characterized in other methylotrophs (9, 52, 54). In Methylobacterium extorquens, this cluster contains two additional genes, mxaEH, whose functions remain unknown (9), but no homologs were found in the mxa gene cluster of M. flagellatus. In addition to the bona fide MDH gene cluster, four additional gene clusters were identified in the genome predicted to encode homologs of the large subunit of MDH (mxaF; Mfla344, Mfla1451, Mfla1717, and Mfla2314), three of them linked to genes predicted to encode cytochrome c (Mfla342, Mfla1450, and Mfla2312, respectively), the former sharing no sequence identity with MxaG, the cytochrome that accepts electrons from MDH, and the latter two sharing less than 35% with MxaG. Only one of the gene clusters also contained a homolog of mxaJ (Mfla2313), but none contained homologs of mxaI that encode the small subunit of MDH (9). Two additional clusters were identified that were predicted to encode functions essential for the synthesis of active MDH (mxaRSACKL; Mfla687 to Mfla692 and Mfla1895 to Mfla1900), but these were not linked to any genes potentially encoding PQQ-linked dehydrogenases. An additional cluster containing homologs of mxaED (Mfla2124 and Mfla2125) was also identified. Homologs of the genes for MDH subunits as well as other MDH functions are often found in the genomes of both methylotrophs and nonmethylotrophs (10, 15, 29, 49, 54). However, mutation analysis suggests that these homologs are not involved in methanol oxidation, and a MDH proper, composed of the small and the large subunits, is required (10, 29). Genes for biosynthesis of PQQ, the cofactor of MDH, were found in two separate clusters, pqqABCDE (Mfla1680 to Mfla1683 [23]) and pqqFG (Mfla734 to Mfla735), similar to the clusters previously characterized for other methylotrophs (9, 29, 54).
All the genes for MADH synthesis were partially identified previously, and all are present in a single cluster on the chromosome (mauFBEDAGLMNazu; Mfla547 to Mfla556 [17, 18]). No genes with high homologies to the regulatory genes involved in methanol or methylamine oxidation functions that have been characterized in M. extorquens (9) or Paracoccus denitrificans (14, 29, 30) are identifiable in the M. flagellatus chromosome, pointing to either a lack of regulatory systems or to the existence of nonhomologous regulatory systems.
(ii) Formaldehyde oxidation.
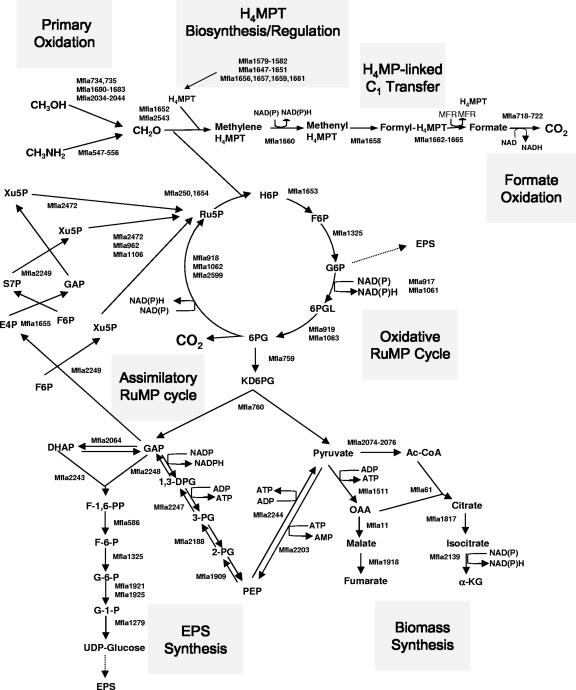
Genes for the two known pathways for formaldehyde oxidation are present, the H4MPT-linked formaldehyde oxidation pathway and the oxidative RuMP cycle (Fig. 1). Two enzymes are specific to the RuMP cycle, hexulosephosphate synthase (HPS) and hexulosephosphate isomerase (HPI [1]). Two copies of hps were identified (Mfla250 and Mfla1654); one is part of a gene cluster previously identified in “Aminomonas aminofaciens” (an uncharacterized Methylobacillus species) and containing genes for histidine biosynthesis (50), while the other is part of the previously characterized large methylotrophy gene cluster containing most of the genes for the H4MPT-linked formaldehyde oxidation pathway, the so-called “archaeal-like” gene cluster (33). It should be noted that the term “archaeal-like” is applicable to methylotroph genetics only as a historical reference, commemorating the time when the genes in question, first discovered in M. extorquens, had homologs only in the Archaea domain (11). Genes predicted to encode HPI (Mfla1653) and transaldolase (Mfla1655) are also found in this cluster. Physical linking on the chromosome of RuMP cycle genes and the H4MPT-linked pathway genes is so far unique to Methylophilaceae (33). Like HPS, the first enzyme of the RuMP cycle, Fae, the first enzyme of the H4MPT-linked pathway (responsible for condensation of formaldehyde with H4MPT [53]) is also predicted to be encoded by two different genes. The first is part of the main “archaeal” gene cluster (Mfla1652 [33]), while the second (Mfla2543) does not appear to be linked to any recognizable methylotrophy genes. The proteins translated from the two genes are 83% identical. In addition to the two bona fide fae genes, two fae homologs are present, previously designated as fae2 (Mfla2524) and fae3 (Mfla2364), and the functions of these genes remain unknown (33). Four additional “archaeal-like” genes involved in H4MPT-linked formaldehyde oxidation pathway (afp, orf20, orf19, orf22, and Mfla1579 to 1582) form a separate gene cluster (33).
FIG. 1.
Central metabolism of M. flagellatus as deduced from the genome sequence and prior genetic/physiological studies. Gray boxes indicate specific methylotrophy metabolic modules. Enzymes predicted to be responsible for specific reactions are represented by Mf1a numbers of open reading frames as translated from the genome sequence (GenBank accession number NC 007947). CH3OH, methanol; CH3NH2, methylamine; CH2O, formaldehyde; H6P, hexulose 6-phosphate; F6F, fructose 6-phosphate; G6P, glucose 6-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose 5-phosphate; Xu5P, xylulose 5-phosphate; E4P, eritrose 4-phosphate; S7P, sedoheptulose 7-phosphate; DHAP, dihydroxyacetone phosphate; F1,6PP, fructose 1,6-bisphosphate; G1P, glucose 1-phosphate; 1,3DPG, 1,3 diphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; α-KG, alpha-ketoglutarate. For more details on methylotrophy genes, see Table S1 in the supplemental material.
The product of HPI, fructose 6-phosphate (6-P), is converted to glucose 6-P by phosphoglucoisomerase (1). A single pgi gene is predicted in the genome (Mfla1325), not linked to any other C1 genes. The genes predicted for glucose 6-P dehydrogenase, 6-P-gluconolactonase and GND form a gene cluster (zwf-gndA-pgl; Mfla917 to Mfla919 and Mfla1061 to Mfla1063) in which gndA and zwf are transcribed in the same direction while pgl is transcribed in the opposite direction, and this cluster is present in two copies, one of which is part of the extended identical repeat. An additional copy of gnd (gndB; Mfla2599) is present in the genome. This latter gene encodes an enzyme 70% identical to GND translated from the genome of M. capsulatus, while it is only 28% identical to gndA. We have previously demonstrated that overexpressing gndA leads to an increase in GND activity linked to NAD, while the NAD(P)-linked activity remains unaffected (8). It is likely that gndB is responsible for the latter activity.
(iii) Formate oxidation.
Genes for a FDH (Mfla718 to Mfla722) homologous to the one encoded in the genome of M. capsulatus (54) were identified in the genome.
(iv) Formaldehyde assimilation.
The assimilatory RuMP cycle branches from the dissimilatory RuMP cycle at the level of 6-P-gluconate (1). Previous enzyme evidence suggested that the 6-P-gluconate dehydratase (KDPG)/ketodeoxy-P-gluconate aldolase/transaldolase version of the RuMP cycle must be operational (37). Indeed, genes for all the enzymes involved are identifiable in the genome (Fig. 1). A total of three genes were identified that were predicted to encode pentose phosphate isomerase (Mfla129, Mfla962, and Mfla1106), of which the latter two are identical copies (as a result of the extended repeat) having 41% amino acid identity with the former. Functioning of the alternative cleavage/regeneration versions of the RuMP cycle (fructose bisphosphate aldolase/sedoheptulose bisphosphatase, fructose bisphosphate aldolase/TA, or KDPG/sedoheptulose bisphosphatase [1]) is not supported by genome analysis, as genes for neither phosphofructokinase nor sedoheptulose bisphosphatase are identifiable.
Pyruvate is the “end product” of the RuMP cycle in M. flagellatus (Fig. 1) (1). To provide necessary cell constituent precursors, reactions converting pyruvate or acetyl-CoA into PEP and OAA are necessary (1). We were able to identify putative genes for PEP synthase (Mfla2203) and pyruvate kinase (Mfla2244) that are likely responsible for interconverting pyruvate and PEP. We also identified a putative gene for pyruvate carboxylase (Mfla1511) that may be responsible for converting pyruvate into OAA. In addition, a gene was identified encoding a product homologous to the alpha subunit of OAA decarboxylase (Mfla1512), another enzyme potentially capable of converting pyruvate into OAA. However, we were not able to identify genes for the beta and gamma subunits associated with this activity (12). An alternative way of synthesizing PEP would be from GAP as shown in Fig. 1. This proposed metabolic loop could serve to balance the levels of pyruvate, GAP, and PEP in the cell.
(v) TCA cycle deficiency as a cause of obligate methylotrophy.
Some methylotrophs (classified as facultative) can grow on multicarbon substrates in addition to C1 substrates, while others (classified as obligate) can grow only on C1 substrates (1, 39). The lack of a complete TCA cycle has been suggested as one of the main causes for obligate methylotrophy (1), based on the experimental evidence that a loss of alpha-ketoglutarate caused a facultative methylotroph to become an obligate methylotroph (4). However, the recent sequencing of an obligate methane utilizer, M. capsulatus, revealed the presence of all the genes for the TCA cycle (54), but it should be noted that the enzymes are not all present at measurable levels (55). This is also the case with chemolithoautotrophic ammonia-oxidizing bacteria (31, 38). On the contrary, analysis of the M. flagellatus genome revealed that genes for three enzymes of the TCA cycle were not identifiable (those encoding malate, alpha-ketoglutarate, and succinate dehydrogenases). Thus, in the case of M. flagellatus, the obligate methylotrophy may be explained by the lack of the main energy-generating pathway for multicarbon substrate metabolism. Gene candidates for the reactions leading to the formation of alpha-ketoglutarate were identified (Mfla61, Mfla1817, Mfla2074 to Mfla2076, and Mfla2139; Fig. 1), suggesting a biosynthetic function for the partial TCA cycle. A gene predicted to encode an alternative enzyme for converting OAA into malate, malate:quinone oxidoreductase (Mfla11), is present in the genome, likely providing a source of malate for cell biosyntheses. Genes are also predicted for interconverting succinate and succinyl-CoA (Mfla1888 and Mfla1889). However, we were unable to predict how succinate or succinyl-CoA could become parts of central metabolism.
(vi) Exopolysaccharide synthesis.
Methylotrophs employing the RuMP cycle for formaldehyde assimilation are known to produce large amounts of EPS (51, 56, 57). The polysaccharide may be a means to balance carbon assimilation and energy generation under specific conditions, a means for detoxifying formaldehyde (47), or an agent essential for the existence of these microbes in the environment (for example, to function in biofilm formation [5]). This metabolic peculiarity of methylotrophs has been explored in terms of commercial production of EPS as a food additive (56, 57). A Methylobacillus strain closely related to M. flagellatus, Methylobacillus sp. strain 12S, has been employed in studies aiming to define the set of genes involved in EPS synthesis and their specific functions (56, 57). As a result, a cluster of 21 genes has been characterized, and chemical properties of the EPS, named methanolan, have been studied. The latter has been found to be a heteropolymer composed of glucosyl, galactosyl, and mannosyl residues (3:1:1 [57]). Interestingly, a large gene cluster was detected in the genome of M. flagellatus (Mfla2007 to Mfla2029) that revealed significant gene syntheny (gene order conservation) with the cluster in Methylobacillus sp. strain 12S (56). However, similarity between the polypeptide counterparts was very low, not exceeding 51% identity. As homologs of most of the genes involved in EPS biosynthesis in Methylobacillus sp. strain 12S were present, similar chemical properties could be predicted for the EPS excreted by M. flagellatus. However, significant divergence in gene sequence suggests separate histories for the respective gene clusters in M. flagellatus and Methylobacillus sp. strain 12S. An additional gene cluster predicted to be involved in EPS biosynthesis was identified in the genome (Mfla1268 to Mfla1280) containing a number of genes with (distant) homologs in the former gene cluster. This gene cluster may be involved in the biosynthesis of a different EPS.
As sugar phosphates are central intermediates in the metabolism of C1 compounds by M. flagellatus, theoretically, precursors for EPS biosynthesis could be drawn straight from the RuMP cycle. Alternatively, sugar phosphate precursors may be synthesized de novo, via the reactions of gluconeogenesis, as shown in Fig. 1. There are two arguments in favor of the enzymes in question being involved in gluconeogenesis as opposed to glycolysis, the latter theoretically allowing growth on glucose and fructose: (i) the apparent lack of sugar phosphorylation enzymes and (ii) the apparent lack of a gene for phosphofructokinase.
The identical repeat as a sign of genome evolution.
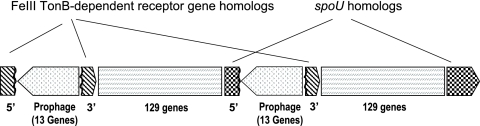
A direct repeat of 143,032 base pairs was identified in the genome. The analysis of the repeated sequence and its flanking regions has shown that part of the repeat is made up of a group of genes unique to M. flagellatus and most probably represents a prophage (Mfla820 to Mfla832 and Mfla964 to Mfla976), based on predictions that some of these genes encode phage-related functions. The sequence analysis also revealed that one copy of the putative prophage interrupts a TonB-dependent receptor gene homolog (Mfla818 and Mfla833), while the entire sequence of the repeat interrupts a spoU gene homolog (Mfla963 and Mfla1107). The structure of the repeat schematically represented in Fig. 2 points toward a likely possibility that the duplication event occurred as a result of phage integration and suggests both the potential existence of free-living phages that infect Methylophilaceae and a mechanism by which methylotroph genomes may evolve. At this time we are not in a position to speculate whether the integration and/or duplication sites (TonB-dependent receptor gene homolog and a spoU homolog) were random or specific sites for recombination.
FIG. 2.
Schematic representation of the chromosomal region containing a direct identical repeat. 3′ and 5′ indicate 3′ and 5′ partial genes, respectively.
The polyphyletic nature of methylotrophy as deduced from genome comparisons.
M. flagellatus is the fourth methylotroph whose genomic determinants for methylotrophy are being reported. The organisms previously characterized in these terms include M. extorquens, an alphaproteobacterial facultative methylotroph (9), M. capsulatus, a gammaproteobacterial obligate methylotroph (54), and the recently described Methylibium petroleiphilum, a betaproteobacterial facultative methylotroph (34, 43). Methylotrophy has been characterized before in terms of functional metabolic modules, which encompass enzymes and factors involved in a single metabolic goal, such as methanol oxidation, formaldehyde oxidation, or C1 assimilation (9). The major metabolic modules involved in methylotrophy in M. flagellatus, as described above, include oxidation systems for methanol and methylamine, the RuMP cycle for formaldehyde oxidation that overlaps to a large degree with the assimilatory RuMP cycle, and the H4MPT-linked formaldehyde oxidation pathway. While at least one putative formate dehydrogenase is encoded in the genome, its contribution to methylotrophy is predicted to be minor, based on the previous experiments demonstrating low levels of FDH activity during growth on C1 compounds and the predominant role of cyclic oxidation of formaldehyde that does not involve formate as an intermediate (7, 37). In terms of primary C1 oxidation functions, genome comparisons revealed that gene clusters encoding the methanol dehydrogenase function in M. flagellatus were similar to those of the respective gene clusters in M. extorquens and M. capsulatus (9, 54), while no major methanol oxidation cluster encoding the large and the small subunits of MDH and an associated cytochrome (MxaG) were identified in M. petroleiphilum (34). The nature of the enzyme responsible for methanol oxidation in this organism remains unknown. The mxaF homolog, xoxF, that has been identified in the genome of M. petroleiphilum, if active, would represent a different module, along with xoxGJ. Homologs of xoxFJG are also present in M. flagellatus, M. capsulatus, and M. extorquens, and in the latter organism, mutation analysis failed to establish a function for this module in methanol oxidation (10). The methylamine utilization gene cluster in M. flagellatus was found to be similar to the one in M. extorquens, except that the gene for amicyanin, a natural electron acceptor for MADH in M. extorquens, was missing from the M. flagellatus cluster. Instead, a gene for a similar blue copper electron acceptor protein (azurin) was present (17, 18). No genes encoding methylamine oxidation were detected in the M. petroleiphilum or M. capsulatus genome. For formaldehyde assimilation, M. flagellatus employs the KDPG/TA version of the RuMP cycle, and the same module is employed by M. capsulatus (37, 54). In contrast, M. petroleiphilum does not encode key functions of the RuMP cycle. Instead, its genome contains a complete set of genes for the serine cycle, the C1 assimilatory pathway also employed by M. extorquens (Table 1) (9). Different gene clustering patterns and low gene similarity between the two organisms do not imply a recent transfer from an alphaproteobacterial methylotroph into M. petroleiphilum. The only methylotrophy module shared by all the organisms involved in comparisons, besides formate dehydrogenases that are ubiquitous, was the H4MPT-linked C1 transfer module. We have previously conducted comparative analyses of gene clusters encoding H4MPT-linked C1 transfer reactions in methylotrophs (33). These analyses have revealed that the cluster in M. flagellatus was more similar, in terms of gene syntheny, to the clusters in gammaproteobacterial methanotrophs than to the clusters in two other betaproteobacteria, M. petroleiphilum and Burkholderia xenovorans. Phylogenetic analyses further supported the finding that in terms of H4MPT-linked C1 transfer functions, M. flagellatus is more closely related to M. capsulatus than to betaproteobacteria of the order Burkholderiales (33). These analyses suggest that M. flagellatus (and other Methylophilaceae members) and M. petroleiphilum (and other Burkholderiales members) have acquired genes for H4MPT-linked C1 transfers as results of at least two independent events. Considering the lack of other overlapping methylotrophy modules in Methylophilaceae and Burkholderiales, we propose that methylotrophy as a metabolic capability evolved at least twice in betaproteobacteria.
TABLE 1.
Methylotrophy metabolic modules in M. flagellatus compared to those of other methylotrophs
| Methylotrophy module | Strains
|
|||
|---|---|---|---|---|
| M. flagellatus | M. petroleiphilum | M. capsulatus | M. extorquens | |
| Methane monooxygenase | − | − | − | + |
| Methanol dehydrogenase | + | − | + | + |
| Methylamine dehydrogenase | + | − | − | + |
| H4MPT-linked C1 transfer | + | + | + | + |
| Ribulose monophosphate cycle | + | − | + | − |
| Serine cycle | − | + | + | + |
| Calvin-Benson-Bassham cycle | − | + | + | − |
DISCUSSION
We described here the findings from the genome analysis of an obligate methanol and methylamine utilizer, M. flagellatus strain KT, that represents a large and environmentally abundant group of methylotrophs belonging to the family Methylophilaceae (45). In terms of methylotrophy functions, genome analysis revealed few surprises. Sets of genes encoding methylotrophy pathways previously predicted based on biochemical and genetic analyses (8, 18, 23, 37) were identified. Some of the methylotrophy genes were found in more than one identical or nonidentical copy. In addition, genes for enzymes converting pyruvate, the “end product” of the assimilatory RuMP cycle into PEP and OAA, were identified. M. flagellatus excretes large amounts of EPS during growth, equaling up to 20% of total biomass (51). All the genes encoding gluconeogenesis enzymes are present in the genome, and these may be implicated in EPS biosynthesis. On the contrary, the operation of the Embden-Meyerhof-Parnas pathway is unlikely, as no gene for phosphofructokinase is identifiable in the genome. As expected, no genes for known sugar transporters were identified in the genome. While a partial set of genes homologous to a fructose transport system were identified, they are not predicted to encode a functional transporter (31, 38). However, previous experiments of the stimulation of biomass yield on methanol by the addition of glucose (37), likely due to enhanced EPS production, indicate that M. flagellatus is able to take up sugars, even if nonspecifically. The main cause for obligate methylotrophy of M. flagellatus must be the incomplete TCA cycle, as the genome is lacking three enzymes essential to its operation. While the function of malate dehydrogenase may be replaced by malate:quinone oxidoreductase (35), no enzymes that would functionally replace alpha-ketoglutarate or succinate dehydrogenases are predicted in the genome.
The M. flagellatus genome is predicted to encode the biosynthesis of all the amino acids and nucleotides and all the essential vitamins and cofactors. Transport systems involved in essential metal (iron and molybdenum) homeostasis are identifiable, while few transporters predicted to take up complex organic compounds (such as amino acids) are present. No secondary metabolite biosynthesis pathways, such as antibiotic biosynthesis, and no known xenobiotic degradation pathways are encoded. Overall, M. flagellatus appears to possess a streamlined, compact genome encoding few metabolic capacities in excess of the ones devoted to the efficient growth on C1 compounds, possibly pointing to the unique environmental function of M. flagellatus and likely other Methylophilaceae in consuming C1 compounds. However, the relatively large number of signal transduction proteins encoded in the genome point toward the existence of sophisticated adaptive mechanisms the organism must possess, despite the narrow range of growth substrates.
The availability of the genomic sequence of M. flagellatus allowed comparisons with other methylotroph genomes in terms of methylotrophy functions. It is remarkable that in terms of methylotrophy metabolic modules, M. flagellatus has more in common with M. capsulatus, a gammaproteobacterium, than with M. petroleiphilum, a betaproteobacterium. While biochemistry of methylotrophy in M. petroleiphilum is not nearly as well studied as in M. flagellatus, some of the essential methylotrophy modules are clearly missing from its genome, such as a gene cluster encoding a bona fide methanol dehydrogenase or key genes for the RuMP cycle (34). Instead, a complete serine cycle for formaldehyde assimilation that was believed until recently to be characteristic of alphaproteobacterial methylotrophs (1, 39) is encoded in the M. petroleiphilum genome (34). The only methylotrophy module shared by M. flagellatus and M. petroleiphilum is the H4MPT-linked formaldehyde oxidation pathway. However, previous phylogenetic analyses argued that even in terms of this module, M. flagellatus is more closely related to gammaproteobacterial methylotrophs than to M. petroleiphilum (33). While the questions of evolution of methylotrophy as a metabolic capability are far from being answered, it is clear that at least in betaproteobacteria, methylotrophy has evolved more than once. It is worth noting that M. petroleiphilum is not an isolated case of an organism possessing “noncanonical” methylotrophy metabolic modules. We have recently characterized a group of strains, classified as Methyloversatilis universalis of the family Rhodocyclaceae, which like M. petroleiphilum, do not appear to possess classical dehydrogenases for methanol or methylamine and utilize serine cycle for formaldehyde assimilation (32). The relatedness of Methylophilales, Rhodocyclales, and Burkholderiales suggests recent evolution for one of the two and possibly both distinct modes of methylotrophy within betaproteobacteria.
Supplementary Material
Acknowledgments
Sequencing was funded by the U.S. Department of Energy's (DOE) Office of Biological and Environmental Research and was carried out at the Joint Genome Institute and completed at the Los Alamos National Laboratory (JGI-LANL). Computational annotation was carried out at the Oak Ridge National Laboratory.
L.C. was supported in part by NSF grants MCB-0131957 and MCB-0604269 and NIH grant 5R01 GM 58933.
Footnotes
Published ahead of print on 6 April 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom.
- 2.Baev, M. V., N. L. Schklyar, L. V. Chistoserdova, A. Y. Chistoserdov, B. M. Polanuer, Y. D. Tsygankov, and V. E. Sterkin. 1992. Growth of the obligate methylotroph Methylobacillus flagellatum under stationary and nonstationary conditions during continuous cultivation. Biotechnol. Bioeng. 39:688-695. [DOI] [PubMed] [Google Scholar]
- 3.Baev, M. V., L. V. Chistoserdova, B. M. Polanuer, V. E. Sterkin, M. Y. Kiriukhin, and Y. D. Tsygankov. 1992. Effect of formaldehyde on growth of obligate methylotroph Methylobacillus flagellatum in a substrate non-limited continuous culture. Arch. Microbiol. 158:145-148. [Google Scholar]
- 4.Bolbot, J. A., and C. Anthony. 1980. The metabolism of pyruvate by the facultative methylotroph Pseudomonas AM1. J. Gen. Microbiol. 120:233-244. [Google Scholar]
- 5.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdov, A. Y., M. R. Eremashvili, S. V. Mashko, A. L. Lapidus, and M. A. Skvortsova. 1987. Expression of the human interferon alpha F gene in the obligate methylotroph Methylobacillus flagellatum KT and Pseudomonas putida. Mol. Gen. Mikrobiol. Virusol. 8:36-41. (In Russian.) [PubMed] [Google Scholar]
- 7.Chistoserdova, L. V., A. Y. Chistoserdov, N. L. Schklyar, M. V. Baev, and Y. D. Tsygankov. 1991. Oxidative and assimilative enzyme activities in continuous cultures of the obligate methylotroph Methylobacillus flagellatum. Antonie Leeuwenhoek 60:101-107. [DOI] [PubMed] [Google Scholar]
- 8.Chistoserdova, L., L. Gomelsky, J. A. Vorholt, M. Gomelsky, Y. D. Tsygankov, R. K. Thauer, and M. E. Lidstrom. 2000. Analysis of two formaldehyde oxidation pathways in Methylobacillus flagellatus KT, a ribulose monophosphate cycle methylotroph. Microbiology 146:233-238. [DOI] [PubMed] [Google Scholar]
- 9.Chistoserdova, L., S. W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova, L., and M. E. Lidstrom. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 143:1729-1736. [DOI] [PubMed] [Google Scholar]
- 11.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 12.Dahinden, P., Y. Auchli, T. Granjon, M. Taralczak, M. Wild, and P. Dimroth. 2005. Oxaloacetate decarboxylase of Vibrio cholerae: purification, characterization, and expression of the genes in Escherichia coli. Arch. Microbiol. 183:121-129. [DOI] [PubMed] [Google Scholar]
- 13.DeBoy, R. T., E. F. Mongodin, J. B. Emerson, and K. E. Nelson. 2006. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 188:2364-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delorme, C., T. T. Huisman, W. N. Reijnders, Y. L. Chan, N. Harms, A. H. Stouthamer, and R. J. van Spanning. 1997. Expression of the mau gene cluster of Paracoccus denitrificans is controlled by MauR and a second transcription regulator. Microbiology 143:793-801. [DOI] [PubMed] [Google Scholar]
- 15.Denef, V. J., M. A. Patrauchan, C. Florizone, J. Park, T. V. Tsoi, W. Verstraete, J. M. Tiedje, and L. D. Eltis. 2005. Growth substrate- and phase-specific expression of biphenyl, benzoate, and C1 metabolic pathways in Burkholderia xenovorans LB400. J. Bacteriol. 187:7996-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleischmann, R. D., M. D. Adams, O. White, R. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spiggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-498. [DOI] [PubMed] [Google Scholar]
- 17.Gak, E. R., A. Y. Chistoserdov, and M. E. Lidstrom. 1995. Cloning, sequencing, and mutation of a gene for azurin in Methylobacillus flagellatum KT. J. Bacteriol. 177:4575-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gak, E. R., Y. D. Tsygankov, and A. Y. Chistoserdov. 1997. Organization of methylamine utilization genes (mau) in “Methylobacillus flagellatum” KT and analysis of mau mutants. Microbiology 143:1827-1835. [DOI] [PubMed] [Google Scholar]
- 19.Galbally, I. E., and W. Kirstine. 2002. The production of methanol by flowering plants and the global cycle of methanol. J. Atmos. Chem. 43:195-229. [Google Scholar]
- 20.Galperin, M. Y. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganushkina, L. A., R. R. Azizbekian, T. G. Grigor'eva, V. I. Iakubovich, I. V. Chernov, and V. P. Sergiev. 1999. Use of the recombinant bacterial strain to control blood-sucking mosquito larvae. Med. Parazitol. (Moskva) 4:46-50. (In Russian.) [PubMed] [Google Scholar]
- 22.Godde, J. S., and A. Bickerton. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62:718-729. [DOI] [PubMed] [Google Scholar]
- 23.Gomelsky, M., F. Biville, F. Gasser, and Y. D. Tsygankov. 1996. Identification and characterization of the pqqDGC gene cluster involved in pyrroloquinoline quinone production in an obligate methylotroph Methylobacillus flagellatum. FEMS Microbiol. Lett. 141:169-176. [DOI] [PubMed] [Google Scholar]
- 24.Govorukhina, N. I., L. V. Kletsova, Y. D. Tsygankov, Y. A. Trotsenko, and A. I. Netrusov. 1987. Characteristics of a new obligate methylotroph. Microbiologiya 56:849-854. [Google Scholar]
- 25.Guenther, A. 2002. The contribution of reactive carbon emissions from vegetation to the carbon balance of terrestrial ecosystems. Chemosphere 49:837-844. [DOI] [PubMed] [Google Scholar]
- 26.Haft, D. H., J. Selengut, E. F. Mongodin, and K. E. Nelson. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harder, W., M. Attwood, and J. R. Quayle. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 29.Harms, N., J. Ras, S. Koning, W. N. M. Reijnders, A. H. Stouthamer, and R. J. M. van Spanning. 1996. Genetics of C1 metabolism regulation in Paracoccus denitrificans, p. 126-132. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 30.Harms, N., W. N. Reijnders, S. Koning, and R. J. van Spanning. 2001. Two-component system that regulates methanol and formaldehyde oxidation in Paracoccus denitrificans. J. Bacteriol. 183:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hommes, N. G., L. A. Sayavedra-Soto, and D. J. Arp. 2003. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J. Bacteriol. 185:6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalyuzhnaya, M. G., P. De Marco, S. Bowerman, C. C. Pacheco, J. C. Lara, M. E. Lidstrom, and L. Chistoserdova. 2006. Methyloversatilis universalis gen. nov., sp. nov., a novel taxon within the betaproteobacteria represented by three methylotrophic isolates. Int. J. Syst. Evol. Microbiol. 56:2517-2522. [DOI] [PubMed] [Google Scholar]
- 33.Kalyuzhnaya, M. G., N. Korotkova, Crowther, C. J. Marx, M. E. Lidstrom, and L. Chistoserdova. 2005. Analysis of gene islands involved in methanopterin-linked C1 transfer reactions reveals new functions and provides evolutionary insights. J. Bacteriol. 187:4607-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kane, S. R., A. Y. Chakicherla, P. S. Chain, R. Schmidt, M. W. Shin, T. C. Legler, K. M. Scow, F. W. Larimer, S. M. Lucas, P. M. Richardson, and K. R. Hristova. 2006. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J. Bacteriol. 189:1931-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kather, B., K. Stingl, M. E. van der Rest, K. Altendorf, and D. Molenaar. 2000. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J. Bacteriol. 182:3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keppler, F., J. T. G. Hamilton, M. Bras, and T. Rockmann. 2006. Methane emissions from terrestrial plants under aerobic conditions. Nature 439:187-191. [DOI] [PubMed] [Google Scholar]
- 37.Kletsova, L. V., N. I. Govorukhina, Y. D. Tsygankov, and Y. A. Trosenko. 1987. Metabolism of the obligate methylotroph Methylobacillus flagellatum. Microbiologiya 56:901-906. [Google Scholar]
- 38.Klotz, M. G., D. J. Arp, P. S. G. Chain, A. F. El-Sheikh, L. J. Hauser, N. G. Hommes, F. W. Larimer, S. A. Malfatti, J. M. Norton, A. T. Poret-Peterson, L. M. Vergez, and B. B. Ward. 2006. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl. Environ. Microbiol. 72:6299-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lidstrom, M. E. 2001. Aerobic methylotrophic prokaryotes. In E. Stackebrandt (ed.), The prokaryotes, 3rd ed. Springer-Verlag, New York, NY.
- 40.Makarova, K., N. V. Grishin, S. A. Shabalina, Y. Wolf, and E. V. Koonin. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishina, I. M., A. B. Pshenichnikova, V. I. Shvets, D. A. Skladnev, and Y. D. Tsygankov. 2002. Use of methylotrophic bacteria Methylobacillus flagellatum KT for isolation of deuterated exogenous carbohydrates. Prikl. Biokhim. Mikrobiol. 38:393-400. (In Russian.) [PubMed] [Google Scholar]
- 42.Muntyan, M. S., T. Y. Dinarieva, M. Baev, and A. I. Netrusov. 2002. Effect of growth conditions on the synthesis of terminal oxidases in Methylobacillus flagellatus KT. Arch. Biochem. Biophys. 398:118-124. [DOI] [PubMed] [Google Scholar]
- 43.Nakatsu, C. H., K. Hristova, S. Hanada, X.-Y. Meng, J. R. Hanson, K. M. Scow, and Y. Kamagata. 2005. Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the “betaproteobacteria.” Int. J. Syst. Evol. Microbiol. 56:983-989. [DOI] [PubMed] [Google Scholar]
- 44.Naqvi, S. W. A., H. W. Bange, S. W. Gibb, C. Goyet, A. D. Hatton, and R. C. Upstill-Goddard. 2005. Biogeochemical ocean-atmosphere transfers in the Arabian Sea. Prog. Oceanogr. 65:116-144. [Google Scholar]
- 45.Nercessian, O., E. Noyes, M. G. Kalyuzhnaya, M. E. Lidstrom, and L. Chistoserdova. 2005. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 71:6885-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653-663. [DOI] [PubMed] [Google Scholar]
- 47.Southgate, G., and P. M. Goodwin. 1989. The regulation of exopolysaccharide production and of enzymes involved in C1 assimilation in Methylophilus methylotrophus. J. Gen. Microbiol. 135:2859-2867. [Google Scholar]
- 48.Strom, E. V., T. Y Dinarieva, and A. I. Netrusov. 2001. Methylobacillus flagellatus KT contains a novel cbo-type cytochrome. FEBS Lett. 515:109-112. [DOI] [PubMed] [Google Scholar]
- 49.Sy, A., E. Giraud, P. Jourand, N. Garsia, A. Willems, P. de Lajudie, Y. Prin, M. Nevra, M. Gillis, C. Boivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, E. J., N. L. Smith, J. Colby, S. J. Charnock, and G. W. Black. 2004. The gene encoding the ribulose monophosphate pathway enzyme, 3-hexulose-6-phosphate synthase, from Aminomonas aminovorus C2A1 is adjacent to coding sequences that exhibit similarity to histidine biosynthesis enzymes. Antonie Leeuwenhoek 86:167-172. [DOI] [PubMed] [Google Scholar]
- 51.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 52.Van Spanning, R. J. M., C. W. Wansell, T. De Boer, M. J. Hazelaar, H. Anazawa, N. Harms, L. F. Oltmann, and A. H. Stouthamer. 1991. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J. Bacteriol. 173:6948-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward, N., O. Larsen, J. Sakwa, L. Bruseth, H. Khouri, A. S. Durkin, G. Dimitrov, L. Jiang, D. Scanlan, K. H. Kang, M. Lewis, K. E. Nelson, B. Methé, M. Wu, J. F. Heidelberg, I. T. Paulsen, D. Fouts, J. Ravel, H. Tettelin, Q. Ren, T. Read, R. T. DeBoy, R. Seshadri, S. L. Salzberg, H. B. Jensen, N. Kåre Birkeland, W. C. Nelson, R. J. Dodson, S. H. Grindhaug, I. Holt, I. Eidhammer, I. Jonasen, S. Vanaken, T. Utterback, T. V. Feldblyum, C. M Fraser, J. R. Lillehaug, and J. A. Eisen. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood, A. P., J. P. Aurikko, and D. P. Kelly. 2004. A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol. Rev. 28:335-352. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, T., Y. Ayabe, M. Yasunaga, Y. Usami, H. Habe, H. Nojiri, and T. Omori. 2003. Genes involved in the synthesis of the exopolysaccharide methanolan by the obligate methylotroph Methylobacillus sp. strain 12S. Microbiology 149:431-444. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, T., M. Horinouchi, Y. Ayabe, T. Yamaguchi, N. Shibuya, H. Habe, H. Nojiri, H. Yamane, and T. Omori. 2000. Saccharide production from methanol by transposon 5 mutants derived from the extracellular polysaccharide-producing bacterium Methylobacillus sp. strain 12S. Appl. Microbiol. Biotechnol. 54:341-347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.