Abstract
Two theories have been proposed to explain the evolution of introns within eukaryotic genes. The introns early theory, or “exon theory of genes,” proposes that introns are ancient and that recombination within introns provided new exon structure, and thus new genes. The introns late theory, or “insertional theory of introns,” proposes that ancient genes existed as uninterrupted exons and that introns have been introduced during the course of evolution. There is still controversy as to how intron–exon structure evolved and whether the majority of introns are ancient or novel. Although there is extensive evidence in support of the introns early theory, phylogenetic comparisons of several genes indicate recent gain and loss of introns within these genes. However, no example has been shown of a protein coding gene, intronless in its ancestral form, which has acquired an intron in a derived form. The mammalian sex determining gene, SRY, is intronless in all mammals studied to date, as is the gene from which it recently evolved. However, we report here comparisons of genomic and cDNA sequences that now provide evidence of a de novo insertion of an intron into the SRY gene of dasyurid marsupials. This recently (approximately 45 million years ago) inserted sequence is not homologous with known transposable elements. Our data demonstrate that introns may be inserted as spliced units within a developmentally crucial gene without disrupting its function.
Since their discovery in 1977 (1), introns have been the subject of considerable debate (2, 3). The two theories that have been advanced to explain the evolution of introns in the eukaryotic genome represent opposing viewpoints about intron evolution. The introns early theory proposes that introns are remnants of ancient DNA that facilitate the shuffling of exons and provide new gene and protein structures as the raw material for the evolution of new gene functions (4, 5). Alternatively, the introns late theory proposes that ancient genomes carried few, if any, introns and that those observed in most eukaryotic genes have been introduced during evolution (1, 6, 7). Both theories have met with opposition; introns in the eukaryotic genome have probably arisen both by exon shuffling and insertional mechanisms.
Support for the introns early theory has come from the correlation between DNA exonic regions and protein structural units (4, 8–10), in which intron positions are related to protein structure. For example, it has been demonstrated statistically that the triosephosphate isomerase gene was probably constructed out of exons (4), which act as the units of gene structure. The conservation of intron position between plant and animal genes (4, 11, 12), and between nuclear and mitochondrial genes that diverged in the progenote (13, 14), provides evidence that introns antedate the plant/animal and the nuclear/mitochondrial divergence and are therefore ancient elements. Most recently, support has been provided for the ancient origin of introns by statistical analyses of intron-phase bias and correlation (15, 16), which showed that most introns are in phase zero (i.e., they lie between codons). However, it has been argued that the introns early theory does not take into account the catalytic function of RNA, and the theory has been challenged by phylogenetic evidence that suggests that nuclear pre-mRNA introns may have spread late in eukaryotic lineages (17).
The identification of novel intron positions in the triosephosphate isomerase gene (18, 19), statistical and phylogenetic analyses of aldehyde dehydrogenase (20), and a large scale analysis of 10 different protein families (21) offer support for the introns late theory. The variable number and random distribution of intron sites within these genes are best explained by an insertional model of intron insertion evolution. The introns late theory is also supported by the existence of self-splicing introns in rRNA and tRNA, including archael introns, group I and group II introns. In addition, the intron of U6 small nuclear RNA in fungal species has been shown to have been recently inserted by a series of events including reverse splicing, reverse transcription, and gene conversion (22, 23).
Although the discovery of introns with self-splicing ability suggested a mechanism for the late insertion of intervening sequences, only a few intron sequences bear any resemblance to known transposable elements (24, 25). Moreover, the introns late theory implies an insertional mechanism for the placement of introns in the genome, so it must address the problem that insertion of any new element, other than those elements capable of self-splicing, would likely disrupt gene function by altering ORFs or regulatory regions of transcripts.
Here we present evidence of a recent de novo insertion of an intron, bearing no resemblance to known transposable elements, into a well-characterized mammalian gene with a critical function in reproduction. The SRY gene lies on the Y chromosome of all therian mammals and has been shown to be the mammalian testis determining factor in at least human and mouse (26, 27). SRY is a member of the SOX gene family, which code for proteins containing a DNA-binding domain [high mobility group (HMG) box] capable of binding and bending DNA (28). Several SOX genes in addition to SRY have been shown to have important roles in development (29, 30).
It has been proposed that SRY evolved from the X-borne SOX-3 gene (31), its closest relative in the SOX family. As the mammalian sex chromosomes differentiated, SRY and SOX3 diverged, leaving SRY isolated on the nonrecombinant Y chromosome. Other vertebrate groups including monotremes (P. Western and A. Pask, personal communication), birds, and reptiles, retain a sequence homologous to SOX3 on an autosome pair and have no evidence of a sex-specific SRY gene (32). SRY is intronless in the more than 65 mammalian species studied to date (33) (GenBank, July 1997). SOX-3, like most SOX genes, also lacks introns in all eutherian and marsupial mammals studied to date, as well as in monotremes (P. Western and A. Pask, personal communication) and chickens (unpublished results), implying that the ancestral SRY/SOX3 gene was also intronless (31). Here we report that the SRY gene in one marsupial family is interrupted by an intron, implying the recent insertion of a spliceosomal element in this gene and providing evidence for the introns late theory.
MATERIALS AND METHODS
Species.
The following were used in this study. Macropodid marsupials: Macropus eugenii (tammar wallaby), Wallabia bicolor (swamp wallaby), Petrogale sp. (rock wallaby); Dasyurid marsupials: Sminthopsis macroura (stripe-faced dunnart), Planigale gilesi (paucident planigale), Antechinus swainsonii (dusky antechinus); Phalangerid marsupials: Trichosurus vulpecula (brush-tailed possum); and other mammals: Tachyglossus aculeatus (short-beaked echidna), Homo sapiens (human), Ovis aries (sheep), Mus musculus domesticus (mouse).
Reverse Transcriptase (RT)–PCR Analysis.
Genomic amplification of SRY from S. macroura was performed using 100 ng each of the primers 5′-GGATGAGTGAAATGGTGA-3′ and 5′-GTAAACTTCTGCATGTTTCAG-3′, 0.25 mM dNTPs, and 2 units of Taq polymerase in the recommended buffer containing 1.5 mM MgCl2 (Boehringer Mannheim). The cycling conditions on 100 ng of total genomic DNA were: 94°C, 1′; 60°C, 1′; 72°C, 1′) ×40. The fMOL Cycle Sequencing kit (Promega) was used for direct sequencing according to the manufacturer’s instructions. RNA was extracted from testis of S. macroura and M. eugenii by using the Tri-Pure Isolation Reagent (Boehringer Mannheim) according to the manufacturer’s protocol. Total RNA (2 μg) was added with 0.625 mM each dATP, dCTP, dGTP, and dTTP, 80 ng oligo-dT15–18 primer, 20 units RNase inhibitor, and 40 units avian myeloblastosis virus–RT according to the manufacturer’s instructions (Boehringer Mannheim), and the reaction was allowed to proceed at 42°C for 1 hr. One-fifth volumes of the total first strand synthesis was used in subsequent PCR reactions. RT–PCR was performed on S. macroura as described above. Genomic and RT–PCR amplification of SRY from M. eugenii were performed by using primers 5′-GCTATGTATGGCTTCTTGAATG-3′ and 5′-CTGTCATTCGTTTCAGGTTTAAC-3′ under the same conditions as described above.
Southern Blot Analysis.
TaqI and EcoRI digests of 10 μg each of male and female S. macroura genomic DNA and EcoRI digests of genomic DNA from male and female M. eugenii, T. vulpecula, T. aculeatus, human, O. aries, and M. musculus domesticus were carried out by using the standard buffer conditions (Boehringer Mannheim) overnight at 37°C. Digests were then electrophoresed on a 1% agarose gel in 0.5 × TBE buffer and subsequently blotted onto Hybond N+ (Amersham) membrane according to the manufacturer’s instructions. Two hundred and fifty nanograms each of the RT–PCR product and the intron from S. macroura were purified by passage through a Microcon 100 concentrator and labeled with 32P-dCTP by priming with random 10-mers and 2 units Klenow enzyme at room temperature for 1 hr. Unincorporated nucleotides were removed by spinning the probe through a Sepharose column. The blots were prehybridized for 1 hr and hybridized overnight with the probe at 65°C in a modified Church and Gilbert buffer (0.25 M NaHPO4, pH 7.0/5 mM EDTA/7% SDS) containing 100 μg/ml of denatured salmon sperm DNA. The filter was subsequently washed at 65°C in 2 × SSC/0.1% SDS and exposed to film overnight.
Cross-Species PCR.
The region surrounding the intron in SRY was amplified from 100 ng each of S. macroura, P. gilesi, and A. swainsonii genomic DNA using 100 ng each of the primers 5′-GTAAACTTCTGCATGTTTCAG-3′ and 5′-GCCGCCTGAACAAGAAAA-3′, using conditions described previously.
Sequencing.
Each PCR product was ligated into pGEM T-Vector (Promega) and sequence analysis was carried out by cycle sequencing by using the fMOL Sequencing kit (Promega) and the primers listed above.
RESULTS
Identification of a Novel Intron in SRY.
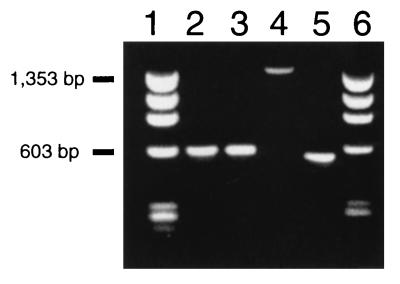
Initial studies of the evolution of SRY involved the characterization of the Y-specific gene in species representing two Australian marsupial families, Macropodidae (kangaroos and wallabies) and Dasyuridae (small carnivores and insectivores), which diverged about 45 million years ago (34). Initially, a cDNA clone was obtained from a S. macroura testis cDNA library. To confirm the integrity of this clone, primers were constructed from the cDNA sequence and PCR was performed on genomic DNA. Surprisingly, the 1.5-kb genomic PCR product was longer than the corresponding 0.6-kb PCR product from the cDNA (Fig. 1). This suggested either the presence of an intron in SRY of S. macroura or a second copy containing an insertion.
Figure 1.
Amplification of marsupial SRY using RT–PCR and genomic PCR separated on a 1.5% agarose gel. Lanes: 1, Phi X-174/HaeIII marker; 2, M. eugenii genomic PCR; 3, M. eugenii RT–PCR; 4, S. macroura genomic PCR; 5, S. macroura RT–PCR; 6, Phi X-174/HaeIII marker.
Characterization of the Intron.
To investigate this anomaly, we subjected the 1.5-kb genomic PCR fragment to direct sequencing. The same primers were then used for RT–PCR of testis RNA, and direct sequencing confirmed the presence of a single 0.6-kb transcript. Sequence analysis showed perfect identity between the S. macroura genomic and RT–PCR fragments, with the exception of an insertion of 825 bp in the genomic sequence in the region 3′ to the HMG box (Fig. 2a). This insertion lies within the coding sequence 550 bp from the start codon and is inserted in phase one (i.e., between the first and second bases of the codon) (Fig. 2b).
Figure 2.
SRY structure and sequence in S. macroura. (a) Schematic diagram of SRY structure. Vertical arrow indicates the point of insertion of the 825 bp intron 3′ to the HMG box. Arrows indicate the direction and position of primers used in PCR. Splice consensus sequences are indicated both in the coding sequence flanking the insertion and at the ends of the intron. (b) Sequence of S. macroura SRY. The coding sequence is presented in uppercase letters and the intron sequence in lowercase letters. TaqI restriction sites are boxed and the splice consensus sequences are underlined.
To eliminate the remote possibility that the RT–PCR product was a transcript from an intronless copy of SRY which, for some reason, was not amplified from genomic DNA, we performed Southern blot analysis by using the RT–PCR product as a probe. Sequence data showed that the intron contains a single TaqI site (Fig. 2b). TaqI digestion could therefore be used to distinguish a genomic copy of SRY containing the intron, which would produce an ≈500 bp band, from an intronless copy, which would produce a band >2 kb. The Southern blot showed only the 500 bp band expected for a TaqI fragment, which extends from the 5′ coding region to within the intron (Fig. 2b), indicating that the copy containing the intron is the only genomic copy of SRY (Fig. 3).
Figure 3.
Southern blot of S. macroura genomic DNA digested with TaqI and probed with S. macroura SRY RT–PCR product. Lane 1, male; lane 2, female. Markers are shown on the right in base pairs (bp) and the arrow indicates the male specific band representing the single copy of SRY present in the male genome.
The absence of the 825 bp insertion in the RT–PCR product implies that it is actively spliced out of the primary transcript. This is consistent with the finding that sequences at the junctions of the insertion and the coding regions correspond to the known consensus sequences for the splice junctions of introns (35, 36) (Fig. 2b).
A search of GenBank (December 1996) using the sequence for the 825 bp intron found no significant evidence of homology to any known sequence.
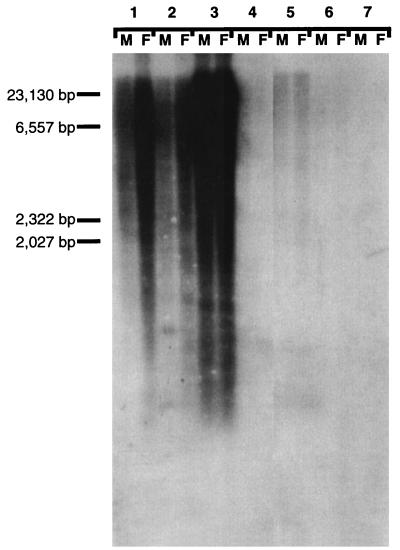
To determine the frequency and distribution of the intron sequence in the dasyurid genome, the cloned intronic sequence was hybridized to a Noah’s Ark blot containing DNA from males and females of several marsupial species, including a macropod in which SRY is intronless, S. macroura, the brush-tailed possum, in which the 3′ sequence of SRY is unknown, the short-beaked echidna, which does not carry an SRY gene, human, sheep, and mouse. No signal was obtained in eutherian or monotreme DNA. However, a smear was obtained in all marsupial lanes, indicating that the intron contains a highly repetitive element ubiquitous to marsupials (Fig. 4). The intron was then digested with RsaI and HinfI, which dissected it into five fragments. Each fragment was individually probed to genomic DNA from a male S. macroura (data not shown). Only the last two fragments, comprising nearly one-half of the intron sequence, were highly repeated. The remaining fractions hybridized only to a single band representing the original intron.
Figure 4.
Noah’s Ark blot representing several mammalian orders probed with the intron from S. macroura SRY. Lanes: 1, M. eugenii male and female; 2, S. macroura male and female; 3, T. vulpecula male and female; 4, T. aculeatus male and female; 5, H. sapiens male and female; 6, O. aries male and female; 7, M. musculus domesticus male and female.
Evolution of SRY Gene Structure in Marsupials.
To compare SRY gene structure among different marsupials with a view to dating the insertion event, SRY sequence was PCR amplified from genomic DNA and RNA from several marsupials. We analyzed SRY gene structure in two other dasyurid genera, A. swainsonii and P. gilesi. By using primers positioned on either side of the intron in the S. macroura gene, we amplified and sequenced an approximately 900 bp fragment from the genomic DNA of P. gilesi and an approximately 700 bp fragment from the genomic DNA of A. swainsonii (Fig. 5a). The position of the intron, as well as the splice junction motif (Fig. 5b), are well conserved in these other dasyurids.
Figure 5.
SRY intron in other dasyurid marsupials. (a) PCR products of SRY using primers flanking the intron shown on a 1.5% agarose gel. Lanes: 1, Phi X-174/HaeIII marker; 2, S. macroura; 3, P. gilesi; 4, A. swainsonii; 5, Phi X-174/HaeIII marker. (b) Sequence alignment of the intron isolated from S. macroura, Antechinus stuartii, and P. gilesi using Clustal V. Asterisks denote identity and dashes denote gaps. Intron boundaries are indicated by arrows. The primer sequence has been excluded from this alignment.
Amplification of SRY from the macropodid marsupials M. eugenii (Fig. 1), Wallabia bicolor (data not shown), and 12 Petrogale species and subspecies (37) yielded a single 0.6 kb product in both the genomic and RT–PCR. The absence of the intron in macropodids and its presence in dasyurids suggests that the intron was inserted into the SRY of a dasyurid ancestor after the divergence of macropodids and dasyurids.
DISCUSSION
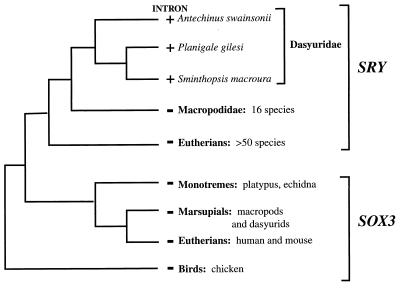
The presence of an intron in the dasyurid SRY gene, and its absence from SRY in all other eutherian and marsupial clades examined, is most parsimoniously explained by de novo insertion after the divergence of macropodids and dasyurids about 45 million years ago (Fig. 6). The alternative hypothesis of intron loss in macropodid and eutherian mammals requires two independent events. Moreover, the loss of an intron in SOX3, the ancestor of SRY, would also be required. This would necessitate a loss of an intron in SOX3 after the divergence of the mammalian sex chromosomes in the X-borne SOX3 of the mammalian lineages, and independently in the autosomal SOX3 in the lineage leading to birds and reptiles (Fig. 6). The simplest conclusion, therefore, is that this intron has been recently inserted into SRY in the dasyurid lineage. The presence of the intron in all three dasyurid genera implies that it was inserted prior to the dasyurid radiation and has been retained in the lineage.
Figure 6.
Representation of phylogeny of the SRY gene indicating the presence (+) or absence (−) of the intron within each lineage. Only animals for which SRY has been included in the SRY phylogeny are shown. Birds and monotremes lack the gene yet contain the ancestral SOX3.
The SRY gene is critical for sex determination in mammals, and disruption of its function causes aberrant sexual development and sterility in humans and mice (26, 27). It is therefore highly unlikely that SRY could have been inactivated by an out-of-phase insertion within the coding sequence and subsequently reactivated by a favorable change into a spliced intron. The presence of the intron in such an essential gene is more likely to have resulted from insertion of a sequence that included splice-junction motifs.
The imprecise positioning of splice junctions leading to small changes in the size and sequence of the final transcript, a characteristic of many transposable elements that act as introns (25), does not appear to have occurred in the dasyurid SRY, because alignment to M. eugenii SRY reveals no significant addition of bases flanking the splice site in S. macroura SRY. The recent identification of a precisely excised intron that was formed by transposable element insertion has demonstrated that mobile element insertion can occur without disrupting the mRNA coding sequence (24). Therefore, evolution of the intron in dasyurid SRY from a transposable element cannot be dismissed.
The origin of this intron and the mechanism of de novo insertion of a functionally spliced intron remain ambiguous. It may be that the intron represents a remnant transposable element or a new family of insertional sequences that carry their own splice junction motifs, ensuring proper splicing from the coding sequences they interrupt. Alternatively, the mechanism of insertion itself may ensure that the sequence is effectively spliced. There has also been recent evidence that introns are not “junk DNA,” but may contain functional elements (38), which convey selective advantage to the gene itself.
The intron in dasyurid SRY provides the first evidence of de novo insertion of a sequence into a mammalian protein coding gene that is correctly spliced out of the primary transcript. The presence of this intron in the single copy of SRY in dasyurid marsupials, its consensus splicing sequences, and its insertion out of phase strongly support the introns late theory.
Acknowledgments
S. macroura material was kindly provided by Dr. Lynne Selwood (Zoology Department, La Trobe University). We would like to acknowledge Mike O’Neill, Jodie Painter, Amanda Spurdle, and Rab Kusmierski for comments on the manuscript. Marsupial tissue was held under permit RP-97–015. This work was supported by grants to J.A.M.G. from the Australian Research Council and the National Health and Medical Research Council. R.J.W.O.’s postgraduate studies were supported by an Overseas Postgraduate Research Award and a La Trobe University Postgraduate Research Scholarship. M.L.D.’s postgraduate studies were supported by a La Trobe University Postgraduate Research Scholarship.
Footnotes
References
- 1.Williamson B. Nature (London) 1977;270:295–297. [Google Scholar]
- 2.Gilbert W. Nature (London) 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle W F. Nature (London) 1978;272:581–582. [Google Scholar]
- 4.Gilbert W, Glynias M. Gene. 1993;135:137–144. doi: 10.1016/0378-1119(93)90058-b. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert W. Cold Spring Harbor Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 6.Rogers J. Trends Genet. 1989;5:213–216. doi: 10.1016/0168-9525(89)90084-x. [DOI] [PubMed] [Google Scholar]
- 7.Palmer J D, Logsdon J M. Curr Opin Genet. 1991;1:470–477. doi: 10.1016/s0959-437x(05)80194-7. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert W, Marchionni M, McKnight G. Cell. 1986;46:151–154. doi: 10.1016/0092-8674(86)90730-0. [DOI] [PubMed] [Google Scholar]
- 9.Go M. Nature (London) 1981;291:90–93. doi: 10.1038/291090a0. [DOI] [PubMed] [Google Scholar]
- 10.De Souza S J, Long M, Schoenbach L, Roy S W, Gilbert W. Proc Natl Acad Sci USA. 1996;93:14632–14636. doi: 10.1073/pnas.93.25.14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchionni M, Gilbert W. Cell. 1986;46:133–141. doi: 10.1016/0092-8674(86)90867-6. [DOI] [PubMed] [Google Scholar]
- 12.Shah D M, Hightower R C, Meagher R B. J Mol Appl Genet. 1983;2:111–126. [PubMed] [Google Scholar]
- 13.Kersanach R. Nature (London) 1994;367:387–389. doi: 10.1038/367387a0. [DOI] [PubMed] [Google Scholar]
- 14.Seioyama C, Joh T, Tsuzuk T, Shimada K. J Mol Biol. 1988;202:355–364. doi: 10.1016/0022-2836(88)90270-7. [DOI] [PubMed] [Google Scholar]
- 15.Long M, Rosenberg C, Gilbert W. Proc Natl Acad Sci USA. 1995;92:12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long M, De Souza S J, Gilbert W. Curr Opin Genet. 1995;5:774–778. doi: 10.1016/0959-437x(95)80010-3. [DOI] [PubMed] [Google Scholar]
- 17.Mattick J S. Curr Opin Genet. 1994;4:823–831. doi: 10.1016/0959-437x(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 18.Kwiatowski J, Krawczyk M, Kornacki M, Bailey K, Ayala F J. Proc Natl Acad Sci USA. 1995;92:8503–8506. doi: 10.1073/pnas.92.18.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logsdon J M, Tyshenko M G, Dixon C, D-, Jafari J, Walker V K, Palmer J D. Proc Natl Acad Sci USA. 1995;92:8507–8511. doi: 10.1073/pnas.92.18.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rzhetsky A, Ayala F J, Hsu L C, Chang C, Yoshida A. Proc Natl Acad Sci USA. 1997;94:6820–6825. doi: 10.1073/pnas.94.13.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho G, Doolittle R F. J Mol Evol. 1997;44:573–584. doi: 10.1007/pl00006180. [DOI] [PubMed] [Google Scholar]
- 22.Tani T, Oshima Y. Nature (London) 1989;337:87–90. doi: 10.1038/337087a0. [DOI] [PubMed] [Google Scholar]
- 23.Tani T, Oshima Y. Genes Dev. 1989;5:1022–1031. doi: 10.1101/gad.5.6.1022. [DOI] [PubMed] [Google Scholar]
- 24.Giroux M J, Clancy M, Baier J, Ingham L, McCarty D, Hannah C. Proc Natl Acad Sci USA. 1994;91:12150–12154. doi: 10.1073/pnas.91.25.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puruganann M D. Trends Ecol Evol. 1993;8:239–243. doi: 10.1016/0169-5347(93)90198-X. [DOI] [PubMed] [Google Scholar]
- 26.Koopman P, Munserberg A, Capel B, Vivian N, Lovell-Badge R. Nature (London) 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- 27.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B, Smith M, Foster J, Frischauf A-M, Lovell-Badge R, Goodfellow P N. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 28.Rimini R, Pontiggia A, Spada F, Ferrari S, Harley V R, Goodfellow P N, Bianchi M E. Philos Trans R Soc London B. 1995;350:215–220. doi: 10.1098/rstb.1995.0154. [DOI] [PubMed] [Google Scholar]
- 29.Foster J W, Brennan F E, Hampikian G K, Goodfellow P N, Sinclair A H, Lovell-Badge R, Selwood L, Renfree M B, Cooper D W, Graves J A M. Nature (London) 1992;359:531–533. doi: 10.1038/359531a0. [DOI] [PubMed] [Google Scholar]
- 30.Wright E M, Snopek B, Koopman P. Nucleic Acids Res. 1993;21:744. doi: 10.1093/nar/21.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster J W, Graves J A M. Proc Natl Acad Sci USA. 1994;91:1927–1931. doi: 10.1073/pnas.91.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graves J A M. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 33.Tucker P K, Lundrigan B L. Philos Trans R Soc London B. 1995;350:221–227. doi: 10.1098/rstb.1995.0155. [DOI] [PubMed] [Google Scholar]
- 34.Archer M. In: Vertebrate Zoogeography and Evolution in Australasia. Archer M, Clayton G, editors. W. A.: Hesperian; 1984. pp. 633–809. [Google Scholar]
- 35.Watson J D, Gilman M, Witkowski J, Zoller M, editors. Recombinant DNA. San Francisco: Freeman; 1992. [Google Scholar]
- 36.Li W-H, Graur D. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer; 1991. [Google Scholar]
- 37.O’Neill R J W, Eldridge M D B, Crozier R H, Graves J A M. Mol Biol Evol. 1997;14:350–353. doi: 10.1093/oxfordjournals.molbev.a025769. [DOI] [PubMed] [Google Scholar]
- 38.Tycowski K T, Shu M-D, Steltz J A. Nature (London) 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]