Abstract
In various bacteria, Zur, a zinc-specific regulator of the Fur family, regulates genes for zinc transport systems to maintain zinc homeostasis. It has also been suggested that Zur controls zinc mobilization by regulating some ribosomal proteins. The antibiotic-producing soil bacterium Streptomyces coelicolor contains four genes for Fur family regulators, and one (named zur) is located downstream of the znuACB operon encoding a putative zinc uptake transporter. We found that zinc specifically repressed the level of znuA transcripts and that this level was derepressed in a Δzur mutant. Purified Zur existing as homodimers bound to the znuA promoter region in the presence of zinc, confirming the role of Zur as a zinc-responsive repressor. We analyzed transcripts for paralogous forms of ribosomal proteins L31 (RpmE1 and RpmE2) and L33 (RpmG2 and RpmG3) for their dependence on Zur and found that RpmE2 and RpmG2 with no zinc-binding motif of conserved cysteines (C's) were negatively regulated by Zur. C-negative RpmG3 and C-positive RpmE1 were not regulated by Zur. Instead, they were regulated by the sigma factor σR as predicted from their promoter sequences. The rpmE1 and rpmG3 genes were partially induced by EDTA in a manner dependent on σR, suggesting that zinc depletion may stimulate the σR regulatory system. This finding reflects a link between thiol-oxidizing stress and zinc depletion. We determined the Zur-binding sites within znuA and rpmG2 promoter regions by footprinting analyses and identified a consensus inverted repeat sequence (TGaaAatgatTttCA, where uppercase letters represent the nucleotides common to all sites analyzed). This sequence closely matches that for mycobacterial Zur and allows the prediction of more genes in the Zur regulon.
Zinc is an essential element for a large number of enzymes and proteins. It supports various physiological activities by maintaining both the structural stability and functional activities of several proteins, including regulatory factors (10, 14). Even though it is a redox-inert metal and does not participate in oxidation-reduction reactions, it can serve as an antioxidant, partly through protecting sulfhydryl groups of proteins from free radical attack and through antagonizing free radical formation by competing with redox-active transition metals such as iron and copper (7). At higher concentrations, it inhibits various physiological functions by blocking important thiols or competing with other functional metal ions for binding sites in proteins, as demonstrated with cytochrome c oxidase (1, 34). Therefore, cells need to maintain zinc homeostasis for optimal survival.
Fur (ferric uptake regulator) family proteins regulate metal homeostasis and oxidative stress responses, as first demonstrated for iron transport in Escherichia coli (5, 22). In a wide range of bacteria, Fur orthologues serve as global regulators of iron homeostasis, antioxidative responses, and in some cases, virulence (23, 42). Other than iron, the list of specific metals that bind to Fur family regulators and hence specifically regulate their own homeostasis is increasing. Examples of the metal-specific Fur family regulators are Zur for zinc (39), Mur for manganese (13), and Nur for nickel (2). In E. coli, Zur is known to exhibit sensitivity to femtomolar levels of free intracellular zinc (36) and regulates the high-affinity zinc uptake system ZnuACB (40). In Bacillus subtilis, Zur regulates not only zinc uptake (17) but also the mobilization of zinc through ribosomal proteins (3, 35, 38). In B. subtilis, it has been shown that among the two forms of ribosomal protein L31 that either contain a zinc-binding motif with conserved cysteines (C+; RpmE) or do not (C−; YtiA) (30), the C− form YtiA liberates zinc-containing RpmE from the ribosome under zinc-deficient conditions, and this action was proposed to provide the needed metal (3). The induction of YtiA under zinc-deficient conditions is mediated through Zur (35) as predicted by in silico analyses (38). The regulation of ribosomal proteins and metal transport systems by Zur in Mycobacterium tuberculosis was also observed recently (29).
The genome information for Streptomyces coelicolor, a model organism for differentiation and antibiotic production, predicts the presence of four Fur homologues (6). Three of them have already been characterized as FurA (the product of gene SCO0561 in the S. coelicolor database ScoDB [http://Streptomyces.org.uk]), CatR (a B. subtilis PerR orthologue and the product of gene SCO5206), and Nur (the product of gene SCO4180), which regulate catalase-peroxidase (20), catalase A (21), and superoxide dismutases (2, 9), respectively. The fourth Fur homologue (the product of gene SCO2508) has not yet been characterized. We report here the characterization of this homologue as Zur, a zinc uptake regulator that controls both zinc uptake and ribosomal proteins predicted to be involved in zinc mobilization.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The S. coelicolor A3(2) M145 strain was used as a wild type and routinely grown in YEME liquid medium containing 10.3% sucrose (25). To prepare metal-chelated medium, we modified NMMP medium without trace metal elements (25) by chelating it with Chelex-100 resin (Bio-Rad) for 2 h before the addition of phosphate buffer and glucose, followed by filtration to remove resin. For PCR-targeted mutagenesis, E. coli BW25113 carrying the pIJ790 plasmid was used as recommended in reference 19. E. coli ET12567 carrying the pUZ8002 plasmid was used for conjugal transfer (32).
Construction of Δzur mutants.
Two Δzur mutants were constructed by replacing most (S701) or all (S700) of the zur coding sequence with the apramycin resistance cassette [aac(3)IV] by using PCR-targeted mutagenesis (19). The upstream forward primer used to create strain S701 contains a 39-nucleotide (nt)-long zur sequence immediately upstream of nt 100 (G; in bold) relative to the start codon linked to the aac(3)IV sequence (underlined; 5′-CCG CCC TTC AGG AGG TCG AGG AGT TCC GCA GCG CGC AGG ATT CCG GGG ATC CGT CGA CC-3′). The upstream primer for S700 contains the zur sequence up to the start codon (in bold; 5′-GGC CCG GCA AGC CGC GAA GAC GTG AGG AGG AAT CCA GTG ATT CCG GGG ATC CGT CGA CC-3′). The same downstream reverse primers were used (5′-TAC AGG GGC CCG GGG CAC AGC CCC GGC CCC ACC GGC TCA TGT AGG CTG GAG CTG CTT C-3′; the zur stop codon is in bold, and the aac(3)IV sequence is underlined). The purified PCR product was introduced by electroporation into the E. coli BW25113 strain (11), which harbors the λred recombination plasmid pIJ790 and cosmid SCC121 (a gift from K. Chater at John Innes Centre) carrying the zur gene. The gene structure of the resulting cosmid (SCC121Δzur::apr) recovered from the selected transformants was verified, and the cosmid was introduced into E. coli ET12567 carrying pUZ8002 and then transferred into S. coelicolor M145 by conjugation. Apramycin-resistant and kanamycin-sensitive exconjugants were selected, and the expected gene structures in Δzur mutants (S700 and S701) were confirmed by PCR and Southern hybridization.
Overexpression and purification of S. coelicolor Zur from E. coli.
The coding region of the zur gene was amplified from the S. coelicolor cosmid SCC121 by using mutagenic primers Zur-up (5′-GGA GGA ATC ATA TGA CCA CCG CTG-3′; the NdeI site is underlined) and Zur-down (5′-CCG TTC AGG CGG ATC CAG GGG C-3′; the BamHI site underlined). The 470-bp PCR product was digested with NdeI and BamHI and inserted into pET3a (Novagen) digested with the same enzymes. E. coli BL21(DE3) was transformed with the resulting recombinant plasmid (pET3aZur). For the purification of Zur, an overnight culture from a single colony was used to inoculate 1 liter of Luria-Bertani medium. Cells were grown with vigorous shaking at 37°C to an optical density at 600 nm (OD600) of 0.5 and were induced with 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h at 30°C. Harvested cells were resuspended with binding buffer (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, and 5 mM imidazole), and cell extracts were prepared. Cell extracts were loaded onto a nickel-charged Chelex-100 column washed with 6 volumes of binding buffer followed by 6 volumes of washing buffer (20 mM Tris-HCl [pH 7.9], 0.5 M NaCl, and 60 mM imidazole). Zur was eluted with 10 volumes of elution buffer (20 mM Tris-HCl [pH 7.9] and 0.5 M NaCl) containing linear imidazole gradients from 100 to 500 mM. Fractions containing Zur were pooled and dialyzed against buffer A (20 mM Tris-HCl [pH 7.8], 100 mM NaCl, 5% [vol/vol] glycerol, and 5 mM EDTA) to remove imidazole and nickel, buffer B (20 mM Tris-HCl [pH 7.8], 50 mM NaCl, 10% glycerol, and 0.1 mM dithiothreitol [DTT]), and finally storage buffer (20 mM Tris-HCl [pH 7.8], 50 mM NaCl, 30% glycerol, and 2 mM DTT). The concentration of purified Zur was determined by the Bradford method. The protein was stored at −80°C.
Gel mobility shift assay for DNA-Zur binding.
The znuA promoter DNA fragment from nt −107 to +35 relative to the transcription start site was amplified by PCR by using the forward (5′-GCT TCA GGT TAC CGG CGT GG-3′) and reverse (5′-GCT ATG CCG GAT ATG CGG CG-3′) primers. The purified DNA was labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Binding reactions were performed with approximately 5.5 fmol of labeled znuA DNA fragments and 6.8 to 6,800 fmol of purified Zur in 20 μl of the reaction buffer [20 mM Tris-HCl (pH 7.80), 50 mM KCl, 1 mM DTT, 0.1 mg of bovine serum albumin/ml, 5% glycerol, and 0.1 μg of poly(dI-dC), with or without 25 μM ZnSO4]. Following incubation at room temperature for 20 min, the binding mixture was subjected to electrophoresis at 4°C on a 5% polyacrylamide gel at 130 V in TB (89 mM Trizma base, 89 mM boric acid) buffer. After electrophoresis, the gel was dried and exposed to X-ray film or an X-ray screen for quantification with a phosphorimage analyzer (FLA-2000; Fuji).
S1 nuclease mapping analysis.
RNAs were isolated from the wild-type (M145) and Δzur mutant (S700 and S701) strains grown to an OD600 of 0.3 to ∼0.4 in YEME or chelated minimal medium. Probes for znuA, zur, rpmE1, rpmE2, rpmG2, and rpmG3 transcripts were generated by PCR by using S. coelicolor cosmids (SCC121, znuA and zur; SCE9, rpmE2 and rpmG2; 2SC6G5, rpmE1; and SC5G5, rpmG3) as templates. Primer pairs were as follows (the nucleotide positions corresponding to the 5′ ends are given in parentheses): znuA, 5′-CCA AGC AGG ATC CTG TCG GCG GCC ACG G-3′ (186 nt upstream from the start codon) and 5′-GCT GGA GCA GGC CGA GAG GG-3′ (87 nt downstream from the start codon); zur, 5′-AAC GCG TTG AGC CCG AGG ATC CGG CGG AG-3′ (139 nt upstream from the start codon) and 5′-CCT GAA GGG CGG CCG ACA CGG C-3′ (87 nt downstream from the start codon); rpmE2, 5′-GTC CCT CGG ATC CAC CTA CGT CAC CCG-3′ (124 nt upstream from the start codon) and 5′-GGT GAG GAA GGC GTA GTC CG-3′ (75 nt downstream from the start codon); rpmE1, 5′-CGT CGT CGG ATC CTG GTC GAG AAG G-3′ (262 nt upstream from the start codon) and 5′-GTG GTG AAC GAC GCG CCA CAG-3′ (71 nt downstream from the start codon); rpmG2, 5′-GTC CTG CGG ATC CCC TTC GTG CTC-3′ (248 nt upstream from the start codon) and 5′-GAC GTA GGT GTA GCC GGT CC-3′ (69 nt downstream from the start codon); and rpmG3, 5′-GCT GTG GGG ATC CGC TGC TTC GGC-3′ (258 nt upstream from the start codon) and 5′-GTG ACG TAG GTG ACG CCG GTG-3′ (71 nt downstream from the start codon). The mutagenized 5′ nucleotides (underlined) in the forward primers were designed to inhibit antisense hybridization and to create BamHI sites (in bold). The probe DNA fragments were labeled with [γ-32P]ATP and T4 polynucleotide kinase. Hybridization and S1 nuclease mapping were carried out according to standard procedures. For high-resolution mapping, the protected DNA fragments were loaded onto 6% (wt/vol) polyacrylamide gel containing 7 M urea, along with sequencing ladders generated from the same probe DNA.
Analytical ultracentrifugation.
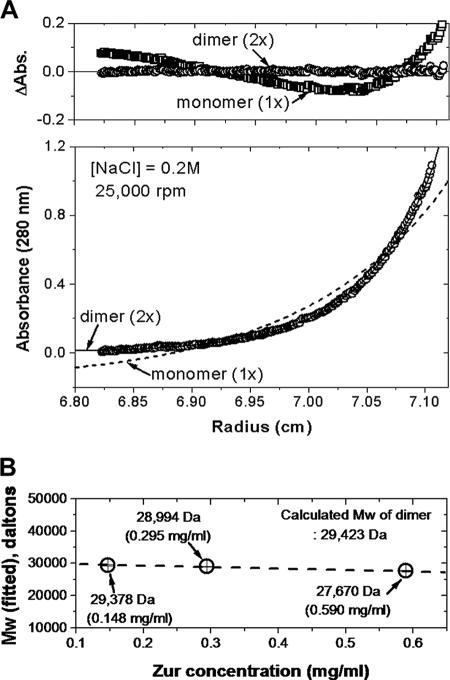
Equilibrium sedimentation analyses were performed using a Beckman ProteomeLab XL-A analytical ultracentrifuge at the Center for Common Facilities at Mokpo National University, Muan, Korea. Zur proteins dissolved in 20 mM Tris-HCl (pH 8.0) buffer containing 2 mM β-mercaptoethanol were measured in six-sector cells at two rotor speeds (25,000 and 35,000 rpm), three concentrations of Zur (10.1 μM [0.148 mg/ml], 20.1 μM [0.295 mg/ml], and 40.2 μM [0.590 mg/ml]), and three salt concentrations (0.05, 0.1, and 0.2 M NaCl) with the loading volume of 130 μl. Five scans were collected and averaged to give the final data for the analysis. All measured data fit well into a dimeric model (for a representative result obtained for 20.1 μM Zur in 0.2 M NaCl, see Fig. 4). The partial specific volume of Zur and the buffer density were calculated using Sednterp (27). The calculated partial specific volume at 20°C was 0.7219 cm3/g. The densities of the buffers containing 0.05, 0.1, and 0.2 M NaCl at 20°C were 1.00087 g/cm3 (0.05 M), 1.00293 g/cm3 (0.1 M), and 1.00704 g/cm3 (0.2 M), respectively.
FIG. 4.
Determination of the oligomeric state of Zur by analytical ultracentrifugation. (A) Sedimentation equilibrium of Zur at 25,000 rpm in 20 mM Tris-HCl buffer containing 0.2 M NaCl at 20°C. (Top panel) Distributions of the residuals as a function of the radial positions for monomer and dimer fits. The random distribution of residuals for the dimer fit indicates that Zur is a homogeneous dimer in solution. (Bottom panel) Distributions of absorbance values as a function of the radial position at a protein concentration of 20.1 μM (0.295 mg/ml). The points (circles) represent experimental data obtained at 280 nm, and the fitting lines correspond to a thermodynamically ideal monomer (dotted line) and dimer (solid line). (B) Experimentally determined molecular masses (Mw) of Zur at different protein concentrations: 10.1 μM (0.148 mg/ml), 20.1 μM (0.295 mg/ml), and 40.2 μM (0.590 mg/ml). Standard deviations were smaller than the sizes of the symbols.
For data analysis by mathematical modeling using nonlinear least-squares curve fitting, the fitting function was as follows: Cr = Cb × exp[ApMp(r2 − rb2)] + ɛ, where Cr is the total concentration at the radial position r, Cb is the concentration of protein at the cell bottom (b), Ap is (1 − υρ)ω2/2RT, Mp is the molecular mass of the protein, ɛ is a baseline error term, υ and ρ are the partial specific volume and the solution density, respectively, ω is the rotor angular velocity, R is the gas constant, and T is the temperature. The selection of the model was made by examining the numbers of the weighted sum or square values and weighted root mean square error values. Further data manipulation and data analysis by mathematical modeling were performed using MLAB (26).
DNase I footprinting.
The probe DNA was prepared by PCR by using a 5′-32P-labeled primer (either forward or reverse) and a nonlabeled primer. For the znuA probe, the forward primer (znuA-up S1, 5′-GCT TCA GGT TAC CGG CGT GG-3′) and the reverse primer (znuA-down S1, 5′-GCT GGA GCA GGC CGA GAG GG-3′) generated a 194-bp probe encompassing a region from nt −107 to +87 relative to the transcription start site. For the rpmG2 probe, the forward primer (rpmG2-up S1, 5′-CAA GCT CGA CAT CCG CTG CTT CGG C-3′) and the reverse primer (rpmG2-down S1, 5′-GTG ACG TAG GTG ACG CCG GTG-3′) generated a 310-bp probe encompassing nt −241 to +69. The amplified products were purified by nondenaturing polyacrylamide gel electrophoresis and eluted by using the standard crush and soak method. Binding reactions were performed as described for the gel mobility shift assay by using an amount of the purified probe corresponding to 18,000 cpm in a final volume of 40 μl per reaction. After 20 min of incubation at room temperature, 40 μl of 5 mM CaCl2-10 mM MgCl2 was added, followed by the addition of 0.06 U of RQI RNase-free DNase (Promega) for 90 s at room temperature. The cleavage reaction was stopped by adding 90 μl of stop solution (200 mM NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate, 125 μg of glycogen/ml), followed by DNA extraction and precipitation. DNA resuspended in formamide loading buffer was separated on a 6% sequencing gel along with G+A sequencing ladders. Autoradiograms were developed on an image plate with a phosphorimage analyzer (FLA-2000; Fuji).
RESULTS
The uncharacterized Fur homologue encoded downstream of the znuACB operon encoding a putative zinc uptake system may be zinc uptake regulator Zur.
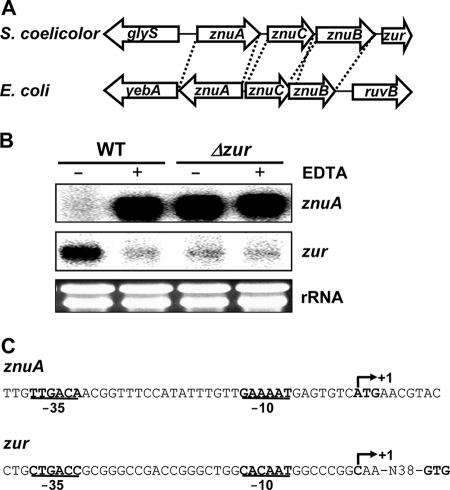
Among the four Fur homologues encoded in the S. coelicolor genome (6), one uncharacterized homologue is encoded by a gene downstream of the putative genes for the high-affinity zinc uptake system (znuACB) (Fig. 1A). The S. coelicolor znu genes show high levels of similarity to previously characterized genes corresponding to high-affinity zinc uptake transporter components in E. coli. ZnuA (the SCO2505 gene product, a probable metal-binding lipoprotein of 328 amino acids [aa]), ZnuC (the SCO2506 gene product, a probable ATPase of 256 aa), and ZnuB (the SCO2507 gene product, a probable membrane permease of 300 aa) show 26.5, 34.8, and 27.3% identity, respectively, to their E. coli counterparts. Unlike those in E. coli and other proteobacteria, an S. coelicolor Fur homologue gene (SCO2508) is located downstream of the znuACB genes. It encodes a Fur family protein of 139 aa with a molecular mass of 14,711 Da that shows 36% identity and 52% similarity to E. coli Fur. A lower level of sequence similarity to E. coli Zur was found, with 29% identity and 39% similarity. We named this gene zur for zinc uptake regulator and characterized it further.
FIG. 1.
Gene organization and regulation of znuACB and zur loci in S. coelicolor. (A) Comparison of gene organization patterns of znuACB loci in S. coelicolor and E. coli. (B) Regulation of znuA and zur genes by Zur. Transcripts from znuA and zur genes were analyzed by S1 nuclease mapping. RNAs were prepared from the wild-type (WT) and Δzur (S701) strains grown in YEME complex medium. To chelate metals, EDTA at a 2 mM final concentration was added for 1 h before cell harvesting. For each S1 analysis, the same amount of RNA (50 μg) was used and the amount was monitored by using rRNAs. +, present; −, absent. (C) Transcription start sites and predicted promoter elements for the znuA and zur genes. Transcription start sites (+1) were determined by high-resolution S1 mapping analyses. Predicted −10 and −35 elements are shown in bold characters and underlined.
Zur negatively regulates znuA gene expression and positively regulates the expression of its own gene.
In order to characterize the function of Zur, we made Δzur (SCO2508) mutants that lack the entire coding region (S700) or most of the coding region downstream from nt 100 relative to the start codon (S701) by using PCR-targeted mutagenesis (19). Both mutants grew more slowly and reached lower plateau values (OD600 of about 0.8) than the wild type (OD600 of about 1.4) in YEME liquid medium. On R2YE plates (25), both mutants were significantly retarded, by more than 2 days, in forming aerial mycelia and formed very small numbers of spores and amounts of the blue antibiotic pigment actinorhodin. The retarded growth and differentiation of the mutants were restored to the wild-type levels by introducing a wild-type gene through the pSET152 plasmid (data not shown). These phenotypes of Δzur mutants demonstrate the importance of the genes regulated by Zur, which most likely exert their effects through the control of zinc homeostasis, in the proper growth and differentiation of S. coelicolor.
Using the Δzur strain that lacks most of the coding region but retains the promoter and some of the coding region (S701), we examined the znuA and zur transcripts by S1 nuclease mapping and compared them to those from the wild type (M145). In order to monitor metal-dependent expression, exponentially grown cells (OD600 of 0.3 to ∼0.4 in YEME) were treated with 2 mM EDTA, a metal chelator, for 1 h. As demonstrated in Fig. 1B, the znuA transcript level in the wild type was drastically increased by EDTA, whereas the level of zur transcripts was decreased. This demonstrates that znuA and zur gene expression is tightly controlled by metal in opposite ways. In the Δzur mutant, transcripts from the znuA gene were expressed constitutively regardless of EDTA treatment, whereas zur expression was decreased. These results suggest that Zur acts as a repressor of znuA gene expression and as a positive regulator of its own expression.
The transcription start sites for zur and znuA promoters were determined by high-resolution S1 mapping analysis. As summarized in Fig. 1C, znuA transcription starts from the A residue of the translation start codon (ATG), with predicted promoter elements of GAAAAT (−10) and TTGACA (−35), most likely to be recognized by the housekeeping sigma factor σHrdB. zur transcription starts from the C residue located 44 nt upstream of the translational start codon (GTG), with predicted promoter elements of CACAAT (−10) and CTGACC (−35), which are also likely to be recognized by σHrdB.
Zinc-specific regulation of znuA and zur genes.
In order to define the metal specificity of znu and zur gene expression, we tested the effects of various metals on cells grown in metal-chelated minimal medium. Wild-type cells were grown to exponential phase (OD600 of 0.3 to 0.4) in chelated minimal medium as described in Materials and Methods. Either 2 mM EDTA or 25 μM divalent metal salt (FeSO4, NiSO4, CoCl2, ZnSO4, or MnSO4) was added for 30 min before cell harvesting. S1 mapping results shown in Fig. 2A demonstrate that znuA gene expression was fully derepressed in the chelated medium, as judged by the lack of additional induction by EDTA. znuA expression was inhibited by more than 20-fold only by zinc among the various divalent metals tested, implying that the znuACB genes most likely encode a zinc-specific transporter system as predicted. zur gene expression, on the other hand, was specifically induced by zinc by about fivefold. We then varied the concentration of zinc to observe the sensitivity of gene expression. The results in Fig. 2B show that znuA was half repressed by the addition ZnSO4 to the medium at around 100 nM, whereas zur expression was half induced by the addition of ZnSO4 to the medium at around 75 nM.
FIG. 2.
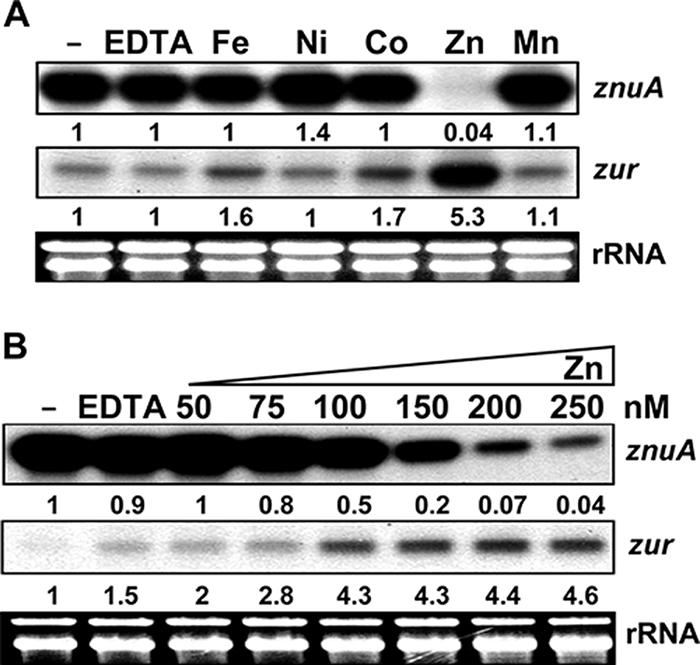
Zinc-specific regulation of znuA and zur genes. (A) Effects of various divalent metals on znuA and zur transcripts. The wild-type and Δzur (S701) cells were grown in chelated minimal medium (NMMP medium incubated with Chelex-100 resin). Either EDTA (2 mM final concentration) or various metals (from Fe to Mn, at 25 μM) were added, and the cultures were incubated for 30 min before harvesting. Following S1 mapping, the amount of protected DNA probe was quantified by a phosphorimage analyzer (FLA-2000; Fuji). Representative results are presented with the quantified signal values. −, control. The signal intensity relative to that of the untreated control was presented. (B) Sensitivity of gene expression to exogenously added zinc. Cells were grown as described in the legend to panel A except that increasing amounts of ZnSO4 (final concentrations, 50 to 250 nM) were added to the culture for 30 min before cell harvesting. Representative results are presented with quantified signal values.
Purified Zur binds to the znuA promoter DNA fragment in a zinc-dependent manner.
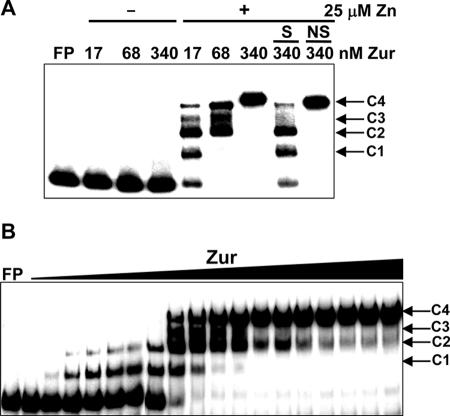
We prepared S. coelicolor Zur protein by overproducing it in E. coli and examined its binding by gel mobility shift assays. The DNA fragment that contains the znuA promoter region from −107 to +35 was generated by PCR and used as a probe. The binding reaction was carried out by incubating approximately 0.3 nM labeled DNA and various amounts of purified Zur in 20 μl of binding buffer with or without 25 μM ZnSO4 for 20 min at room temperature. As demonstrated in Fig. 3A, binding occurred only in the presence of zinc, creating multiple shifted bands presumably representing protein-DNA complexes. At least four complex bands were detected, with the most retarded band observed at the highest concentration of Zur. Competition with excess amounts of either specific or nonspecific competitors demonstrated that these complexes were specific ones. By adding increasing amounts of Zur from 0.34 to 340 nM (in steps corresponding to monomer concentrations) to each binding reaction mixture in the presence of 25 μM zinc, we observed the progressive formation of complexes, from faster-moving (C1) to slower-moving (C2, C3, and C4) ones, in correlation with changing concentrations (Fig. 3B). These multiple complexes most likely reflect multiple bindings of Zur protein to the znuA probe we used.
FIG. 3.
Zinc-dependent binding of purified Zur to znuA promoter DNA. (A) Results from gel mobility shift assays in the presence (+) and absence (−) of zinc. Purified Zur protein at various concentrations (from 17 to 340 nM, with increases corresponding to monomeric units) was incubated with 32P-labeled znuA DNA fragments (0.3 nM) in the absence or presence of 25 μM ZnSO4. To confirm the specificity of binding complexes, either a 100-fold molar excess of specific competitors (S; unlabeled znuA DNA) or a 300-fold molar excess of nonspecific competitors (NS; HpaII digest of pGEM-3Zf plasmid) was added to the binding mixture. FP denotes the lane containing the labeled probe DNA only. Four distinct bands with different mobilities were designated C1 to C4. (B) Progressive formation of multiple complex bands (C1 to C4) by increasing amounts of Zur from 0.34 to 340 nM in the presence of 25 μM ZnSO4.
In order to determine the oligomeric status of Zur in solution, we employed analytical ultracentrifuation. The equilibrium sedimentation results demonstrated in Fig. 4 correlate well with a dimeric status of Zur in solution at three different concentrations (10.1, 20.1, and 40.2 μM). The absence of increase in the molecular mass at higher concentrations confirms that Zur exists as homogeneous dimers, and there is no indication of self-association. This result is in agreement with the oligomeric behavior proposed for other Fur homologues examined so far. Therefore, the multiple complex bands most likely represent multiple bindings of dimeric Zur to the znuA DNA probe.
Zur-dependent regulation of ribosomal protein genes in S. coelicolor.
Recent observations with B. subtilis that a zinc-containing ribosomal protein, L31 (RpmE), can be replaced with a zincless L31 variant (YtiA) under zinc-depleted conditions as a way to mobilize zinc (3, 35) and that ytiA transcription is regulated by Zur (35, 38) led us to examine the regulation of RpmE holomogues in S. coelicolor. Three genes rpmE paralogues were found in the S. coelicolor genome, rpmE1 (SCO5359), rpmE2 (SCO3427), and rpmE3 (SCO1150) (6). The rpmE1 gene contains the zinc-binding motif (the gene product is C+), whereas rpmE2 and rpmE3 do not (the gene products are C−). The rpmE1 gene has a consensus promoter sequence for the sigma factor σR and has previously been demonstrated to be regulated by σR in response to disulfide stress (37). We also looked for rpmG paralogues that encode L33, which is another ribosomal component predicted to participate in zinc mobilization (30). There are three rpmG paralogues in the S. coelicolor genome, rpmG1 (SCO4635), rpmG2 (SCO3428), and rpmG3 (SCO0570). The rpmG1 gene contains a zinc-binding motif, whereas rpmG2 and rpmG3 do not. We identified another σR consensus promoter sequence in rpmG3. The search results are summarized in Table 1. We therefore tested the dependence of the expression of these paralogous genes on metal depletion and disulfide stress. S1 mapping analyses demonstrated single transcript start sites for rpmE1, rpmG2, and rpmG3. The rpmE2 gene lies immediately downstream of rpmG2 and most likely forms an operon with rpmG2. We were not able to determine transcription start sites for rpmE3 and rpmG1, since there were numerous S1-protected bands for rpmG1 and no bands for rpmE3. We therefore analyzed transcripts from rpmE1, rpmE2, rpmG2, and rpmG3 for further details.
TABLE 1.
Paralogous genes encoding L31 and L33 ribosomal proteinsa
| Ribosomal protein | Gene | Gene numberb | Cysteine motifc | Proposed regulator |
|---|---|---|---|---|
| L31 | rpmE1 | SCO5359 | + | σR |
| rpmE2 | SCO3427 | − | Zur | |
| rpmE3 | SCO1150 | − | NDd | |
| L33 | rpmG1 | SCO4635 | + | NDd |
| rpmG2 | SCO3428 | − | Zur | |
| rpmG3 | SCO0570 | − | σR |
For the numbering of paralogues, we used the number 1 for those that contain conserved cysteine motifs, different from the annotations in the S. coelicolor database ScoDB (http://Streptomyces.org.uk).
The rpmG2 (SCO3428) and rpmE2 (SCO3427) genes are cotranscribed from the rpmG2 promoter.
+, present; −, absent.
ND, not determined.
Results presented in Fig. 5 show that levels of rpmE2 and rpmG2 transcripts increased drastically upon metal depletion and that this induction was mediated through the inactivation of Zur activity, as demonstrated by constitutive expression in a Δzur mutant. The expression of rpmE1 and rpmG3, however, was induced only partially by metal depletion compared with the drastic induction by diamide, a thiol oxidant, in a manner independent of Zur. Therefore, the C− forms of L31 (RpmE2) and L33 (RpmG2) in S. coelicolor are regulated by Zur, being induced under zinc-depleted conditions, consistent with a model of zinc mobilization through these ribosomal proteins. Interestingly, another C− form of L33 (RpmG3) is not regulated by Zur. The induction of rpmE1 and rpmG3 by diamide was indeed mediated by σR, as demonstrated by the lack of induction in a Δ(sigR-rsrA) mutant (data not shown).
FIG. 5.
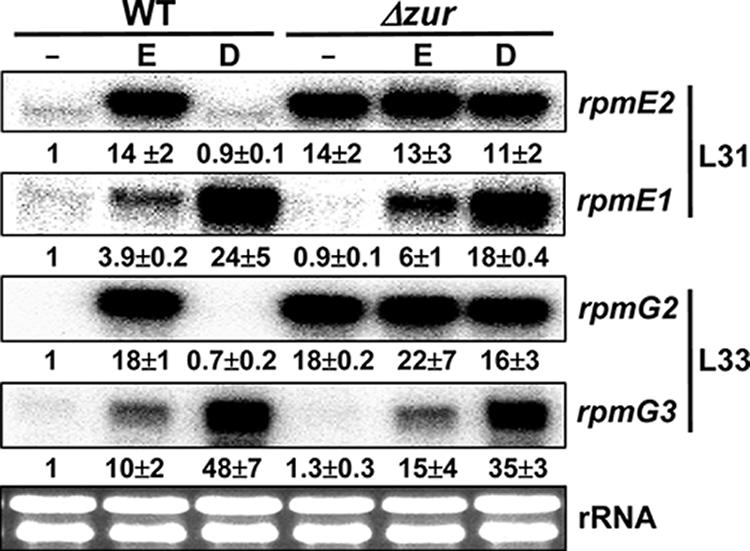
Regulation of ribosomal protein genes encoding paralogous forms of L31 and L33. The transcripts for rpmE1 and rpmE2 encoding L31 with and without a zinc-binding motif, respectively, were examined along with those for rpmG2 and rpmG3, encoding paralogous forms of L33 without zinc-binding motifs. The wild-type (WT) and Δzur (S700) mutant cells were grown in YEME medium and either untreated (−) or treated with 2 mM EDTA (E) or 0.5 mM diamide (D) for 1 h before RNA preparation for S1 analyses. Protected DNA fragments were visualized and quantified by a phosphorimage analyzer (FLA-2000; Fuji). Average values from three independent experiments are presented. The rRNA in each sample (50 μg of total RNA) was monitored by gel electrophoresis as a loading control.
Binding site of Zur.
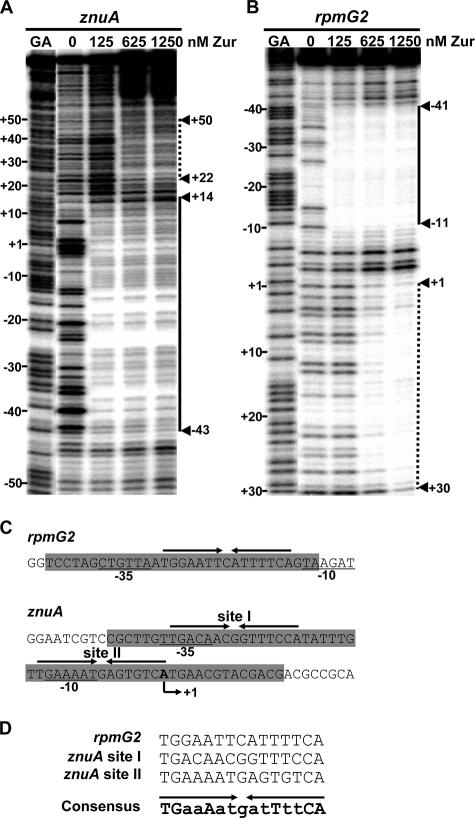
We performed footprinting analyses to delineate Zur-binding sites in znuA and rpmG2 promoter regions. Using 194 bp of the znuA DNA probe, from nt −107 to +87 relative to the transcription start site, we were able to detect a primary protected region from nt −43 to +14 (Fig. 6A). Further secondary protection from nt +22 to +50 was observed at higher concentrations of Zur. By using 310 bp of rpmG2 promoter DNA encompassing the region from nt −241 to +69, a primary protection region from nt −11 to −41 was detected. Further protection from nt +1 to +30 was observed at higher concentrations of Zur. The primary Zur-binding sites overlap entirely with both promoters and thus are consistent with a model of repression by Zur.
FIG. 6.
DNase I footprinting analysis of znuA and rpmG2 promoter regions. (A) Footprint pattern of Zur on znuA promoter DNA (from nt −107 to +87 relative to the transcriptional start site) labeled at the top strand. Increasing amounts of Zur (from 125 to 1,250 nM in steps of monomeric concentrations) were added in the binding reaction. The G+A (GA) sequencing ladders generated from the same DNA probe were run in parallel. The primary protected region (from nt −43 to +14) is indicated with a solid line. The secondary protected region (from nt +22 to +50) at higher Zur concentrations is indicated with a dotted line. (B) Footprint pattern of Zur on rpmG2 promoter DNA (from nt −241 to +69 relative to the transcriptional start site) labeled at the bottom strand. The footprinting assay was done as described in the legend to panel A. The primary (from nt −11 to −41) and secondary (from nt +1 to +30) protected regions are indicated. (C) Location of inverted repeat sequences within the footprinted (shaded) regions of rpmG2 and znuA promoters. The positions of −10 and −35 promoter elements are indicated by underlining. The bent arrow indicates the direction of transcription. (D) Identification of the Zur-binding consensus sequence from the alignment of 15-bp (7-1-7) inverted repeat sequences.
The primary protected region in rpmG2 was found to be much smaller than that in the znuA promoter. We hypothesized that there might be more than one binding site separated by some distance, producing broader protection in znuA. Since Zur exists as a homodimer, its recognition sequence most likely consists of inverted repeats, as proposed for other Fur homologues (16). We looked for such an inverted repeat sequence within the 30-bp protected region in an rpmG2 promoter and identified one such sequence. When the protected region of the znuA promoter was examined for the presence of such motifs, we were able to identify two motifs, one overlapping with the −35 region (site I) and the other overlapping with the −10 region (site II) (Fig. 6C). From an alignment of the three inverted repeat sequences, a provisional consensus recognition motif for S. coelicolor Zur was predicted to be TGaaAatgatTttCA, where uppercase letters represent the nucleotides common to all sites analyzed (Fig. 6D). No obviously similar sequences were found within the secondary protected regions.
DISCUSSION
Our work demonstrated that the uncharacterized Fur homologue (the product of gene SCO2508) in S. coelicolor is zinc uptake regulator Zur that regulates a putative high-affinity zinc uptake system and some ribosomal proteins predicted to be involved in zinc mobilization. The regulation of zinc uptake systems by Zur in a broad range of bacteria, including E. coli (39), B. subtilis (17, 18), and M. tuberculosis (29), has been demonstrated previously. Zur has also been hypothesized to control the mobilization of zinc from several paralogous ribosomal proteins on the basis of comparative genomic analyses of Zur regulons (38). According to this model, the ribosomal proteins that contain a zinc-binding motif (C+) can serve as zinc storage forms and can be replaced with their Zur-regulated paralogues without a zinc-binding motif (C−). This hypothesis was experimentally supported for a ribosomal large-subunit protein, L31 from B. subtilis, where Zur-regulated paralogue YtiA (C−) replaces RpmE (C+) under zinc-deficient conditions (3, 35). Recent observations from microarray and in vitro binding experiments with M. tuberculosis demonstrated that Zur regulates genes for C− paralogues of L33 (RpmG), S14 (RpsN), and S18 (RpsR), supporting this hypothesis (29). Our work adds a convincing proposal that Zur regulates the synthesis of C− paralogues of L31 and L33 in S. coelicolor. Considering the positions of Zur-binding sites within the intergenic regions of rpmG2 (SCO3428) and rpmB2 (SCO3429), it is very likely that rpmB2, encoding a C− paralogue of L28, is also regulated by Zur, along with the downstream gene rpsN2 (SCO3430), encoding a C− paralogue of S14. These observations strengthen the generalization of the proposal that bacteria utilize zinc-specific regulators such as Zur to control paralogous ribosomal proteins without zinc-binding abilities, enhancing zinc storage through ribosomal proteins under conditions of zinc sufficiency and enhancing zinc mobilization from these proteins through replacement with paralogues lacking zinc-binding sites under conditions of zinc deficiency.
An interesting observation that the rpmE1 and rpmG3 genes were regulated by σR was made in this study. RpmE1 is a zinc-containing form, whereas RpmG3 is not. Therefore, a simple correlation between the regulation by σR and the presence or absence of zinc-binding motifs in the paralogous ribosomal proteins could not be made. However, our work reflects some link between thiol-oxidizing stress and the depletion of metals, most likely zinc. In this regard, the partial induction of rpmE1 and rpmG3 by EDTA suggests that zinc depletion may be sensed as a kind of thiol-oxidizing stress in the cell. Since σR is regulated by an anti-sigma factor RsrA, which contains zinc molecules that are released by the formation of disulfide bonds (4, 24, 28), it is possible that zinc depletion may cause the inactivation of RsrA and thus the induction of the σR regulon. Previous observations that zinc is necessary for the stability of RsrA and the binding affinity of RsrA for σR (4, 28) support this proposal. The precise roles of σR and thiol-oxidative stress responses in zinc homeostasis need be investigated in the future.
There is limited information about the autoregulation of Fur homologues. In B. subtilis, PerR has been suggested to repress its own gene and the fur gene (15). In S. coelicolor, on the contrary, we found that Zur is necessary for the zinc-induced expression of its own gene. A similar phenomenon was observed in M. tuberculosis, where zinc enhances zur expression (29). In Mycobacterium, the zinc-dependent induction of the zur gene is mediated through an SmtB/ArsR family regulator that is encoded by the upstream gene cotranscribed with zur (8, 33). In S. coelicolor, we were not able to identify any Zur-binding sequence near the zur promoter nor to observe any specific Zur binding in vitro to the promoter DNA fragment from nt −51 to +39 relative to the transcription start site (data not shown). This result suggests that the positive regulation of zur gene expression by Zur most likely occurs indirectly through an as-yet-unidentified regulator whose synthesis may be controlled by Zur, as exemplified by the positive regulation of iron-utilizing genes by Fur through a small RNA in E. coli (31). Another possible example of positive regulation by Fur is found in Helicobacter pylori, where Fur can mediate its own gene induction under low-iron conditions by shifting its binding sites in the fur promoter region (12). In this respect, further systematic study of the regulation of the zur gene in S. coelicolor is anticipated.
The identification of the proposed Zur-binding consensus sequence in our study depends on the assumption that a Zur dimer recognizes an inverted repeat sequence. A simple sequence alignment of the footprinted regions did not reveal any salient inverted repeat sequences among the conserved nucleotides. The identification of an inverted repeat motif within the small (30-bp) protected region in the rpmG2 gene provided a lead sequence to identify two such motifs within the broader (57-bp) protected region in the znuA gene. It is possible that the binding of one Zur dimer to either of those two sites may create a broad footprint pattern, even though we cannot rule out the possibility that two Zur dimers bind to two sites simultaneously. The presently proposed Zur-binding consensus sequence in a 7-1-7 format (TGaaTat-g-atTttCA) closely resembles the B. subtilis Fur box consensus sequence (TGATAAT-N-ATTATCA) (16) but is distant from the B. subtilis Zur box sequence (TCGTAAT-N-ATTACGA) and the PerR box sequence (TTATAAT-N-ATTATAA). The S. coelicolor Zur box sequence closely matches the proposed Zur box consensus sequence in M. tuberculosis (TGAAAAT-N-ATTTTCA) (29). When the amino acid sequences of the DNA-binding helices in Fur homologues were compared according to the structure model determined for Pseudomonas aeruginosa Fur (41), S. coelicolor Zur shared 13 identical amino acids out of 17 with M. tuberculosis Zur and 9 residues with B. subtilis Fur. On the other hand, it shared only 5 identical residues with B. subtilis Zur or PerR, consistent with the lack of conservation in the binding sequences.
By allowing a 1-nt mismatch and maintaining an AT content of more than 50% within the more variable 11-nt central sequences (TGN11CA), we were able to draw out several candidate target genes of Zur regulation. The list includes rpmF2 (SCO0436), encoding a C− form of ribosomal protein L32; a putative zinc transporter gene (SCO0475); a solute-binding lipoprotein gene (SCO6644); and a putative efflux protein gene (SCO6751). The regulation of rpmF2 as well as rpmB2 by Zur has been demonstrated by Paget and colleagues in their study of the zinc-responsive regulation of alternative ribosomal protein genes (M. Paget, personal communication), supporting our proposal for the consensus Zur box in S. coelicolor. Further analyses are needed to disclose the compositions and the roles of Zur-regulated genes in the growth and differentiation of S. coelicolor.
Acknowledgments
This work was supported by a grant from the Ministry of Science and Technology to J.-H. Roe for the National Research Laboratory of Molecular Microbiology at the Institute of Microbiology, Seoul National University. J.-H. Shin and S.-Y. Oh were recipients of BK21 graduate fellowships from the Ministry of Education and Human Resources.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Aagaard, A., and P. Brzezinski. 2001. Zinc ions inhibit oxidation of cytochrome c oxidase by oxygen. FEBS Lett. 494:157-160. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, B. E., J. Cha, E. J. Lee, A. R. Han, C. J. Thompson, and J. H. Roe. 2006. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59:1848-1858. [DOI] [PubMed] [Google Scholar]
- 3.Akanuma, G., H. Nanamiya, Y. Natori, N. Nomura, and F. Kawamura. 2006. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J. Bacteriol. 188:2715-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, J. B., J. H. Park, M. Y. Hahn, M. S. Kim, and J. H. Roe. 2004. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J. Mol. Biol. 335:425-435. [DOI] [PubMed] [Google Scholar]
- 5.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Bray, T. M., and W. J. Bettger. 1990. The physiological role of zinc as an antioxidant. Free Radic. Biol. Med. 8:281-291. [DOI] [PubMed] [Google Scholar]
- 8.Canneva, F., M. Branzoni, G. Riccardi, R. Provvedi, and A. Milano. 2005. Rv2358 and FurB: two transcriptional regulators from Mycobacterium tuberculosis which respond to zinc. J. Bacteriol. 187:5837-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, H. J., J. H. Choi, E. J. Kim, Y. H. Cho, and J. H. Roe. 1999. Negative regulation of the gene for Fe-containing superoxide dismutase by an Ni-responsive factor in Streptomyces coelicolor. J. Bacteriol. 181:7381-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, J. E. 1998. Zinc enzymes. Curr. Opin. Chem. Biol. 2:222-234. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2003. An anti-repression Fur operator upstream of the promoter is required for iron-mediated transcriptional autoregulation in Helicobacter pylori. Mol. Microbiol. 50:1329-1338. [DOI] [PubMed] [Google Scholar]
- 13.Diaz-Mireles, E., M. Wexler, G. Sawers, D. Bellini, J. D. Todd, and A. W. Johnston. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150:1447-1456. [DOI] [PubMed] [Google Scholar]
- 14.Falchuk, K. H. 1993. Zinc in developmental biology: the role of metal dependent transcription regulation. Prog. Clin. Biol. Res. 380:91-111. [PubMed] [Google Scholar]
- 15.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuangthong, M., and J. D. Helmann. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, J. S., S. Y. Oh, and J. H. Roe. 2000. Regulation of the furA and catC operon, encoding a ferric uptake regulator homologue and catalase-peroxidase, respectively, in Streptomyces coelicolor A3(2). J. Bacteriol. 182:3767-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn, J. S., S. Y. Oh, K. F. Chater, Y. H. Cho, and J. H. Roe. 2000. H2O2-sensitive Fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275:38254-38260. [DOI] [PubMed] [Google Scholar]
- 22.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 23.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 24.Kang, J. G., M. S. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner, and J. H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 26.Knott, G. D. 1979. MLAB: a mathematical modeling tool. Comput. Programs Biomed. 10:271-280. [DOI] [PubMed] [Google Scholar]
- 27.Laue, T. M., B. Shah, T. M. Ridgeway, and S. L. Pelleitier. 1992. Analytical ultracentrifugation in biochemistry and polymer science, p. 90-125. Royal Society of Chemistry, Cambridge, United Kingdom.
- 28.Li, W., A. R. Bottrill, M. J. Bibb, M. J. Buttner, M. S. Paget, and C. Kleanthous. 2003. The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J. Mol. Biol. 333:461-472. [DOI] [PubMed] [Google Scholar]
- 29.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palu, G. Riccardi, and R. Manganelli. 10 November 2006. Global analysis of Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed]
- 30.Makarova, K. S., V. A. Ponomarev, and E. V. Koonin. 2001. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2:RESEARCH 0033. [DOI] [PMC free article] [PubMed]
- 31.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazodier, P., R. Petter, and C. Thompson. 1989. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milano, A., M. Branzoni, F. Canneva, A. Profumo, and G. Riccardi. 2004. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155:192-200. [DOI] [PubMed] [Google Scholar]
- 34.Mills, D. A., B. Schmidt, C. Hiser, E. Westley, and S. Ferguson-Miller. 2002. Membrane potential-controlled inhibition of cytochrome c oxidase by zinc. J. Biol. Chem. 277:14894-14901. [DOI] [PubMed] [Google Scholar]
- 35.Nanamiya, H., G. Akanuma, Y. Natori, R. Murayama, S. Kosono, T. Kudo, K. Kobayashi, N. Ogasawara, S. M. Park, K. Ochi, and F. Kawamura. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52:273-283. [DOI] [PubMed] [Google Scholar]
- 36.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 37.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the óR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 38.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 100:9912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 40.Patzer, S. I., and K. Hantke. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321-24332. [DOI] [PubMed] [Google Scholar]
- 41.Pohl, E., J. C. Haller, A. Mijovilovich, W. Meyer-Klaucke, E. Garman, and M. L. Vasil. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903-915. [DOI] [PubMed] [Google Scholar]
- 42.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]