Abstract
Cutaneous propionibacteria are important commensals of human skin and are implicated in a wide range of opportunistic infections. Propionibacterium acnes is also associated with inflammatory acne vulgaris. Bacteriophage PA6 is the first phage of P. acnes to be sequenced and demonstrates a high degree of similarity to many mycobacteriophages both morphologically and genetically. PA6 possesses an icosahedreal head and long noncontractile tail characteristic of the Siphoviridae. The overall genome organization of PA6 resembled that of the temperate mycobacteriophages, although the genome was much smaller, 29,739 bp (48 predicted genes), compared to, for example, 50,550 bp (86 predicted genes) for the Bxb1 genome. PA6 infected only P. acnes and produced clear plaques with turbid centers, but it lacked any obvious genes for lysogeny. The host range of PA6 was restricted to P. acnes, but the phage was able to infect and lyse all P. acnes isolates tested. Sequencing of the PA6 genome makes an important contribution to the study of phage evolution and propionibacterial genetics.
Propionibacterium acnes is a universal inhabitant of human skin and is found at high population densities on the fat-rich areas of the face, scalp, and upper trunk (16, 23). P. acnes has been implicated in the pathogenesis of inflammatory acne, as treatments that reduce the numbers of P. acnes cells on the skin are therapeutic and the failure of such therapies has been associated with the emergence of resistance in P. acnes (7, 9, 30). Acne vulgaris is the most common disease of skin in adolescents and affects approximately 80% of individuals at some stage in their lives (5). This disease can be extremely painful and can lead to numerous psychological problems. Inflammatory lesions give rise to significant scar formation in 30% of patients (6). In addition to its association with inflammatory acne, P. acnes is an increasingly common opportunistic pathogen, causing a wide range of infections in immunocompromised patients (2, 8, 31, 34).
Bacteriophages that infect P. acnes can be readily isolated from human skin. The study of these bacteriophages has until now been limited to the development of phage typing systems to distinguish the different serotypes of P. acnes (19, 33). Bacteriophages are the most abundant organisms on Earth, and they play an important role in bacterial diversity and pathogenesis. Greater study of P. acnes bacteriophages should make a significant contribution to the study of propionibacterial genetics and to the wider field of phage biology and evolution.
With the sequencing of bacteriophages comes the potential to genetically manipulate the host bacterium. Although the physiology of P. acnes has been well studied in vitro (4, 13, 14), genetic studies have been limited by the lack of genetic transfer systems. The inability to manipulate P. acnes genetically has severely hindered progress toward understanding the role of this organism in acne and other diseases. Similar problems existed several years ago with the study of pathogenic mycobacteria, which are closely related to propionibacteria taxonomically. The development of transformation systems and plasmid vectors for mycobacteria has allowed greater study of the genetics of these organisms (18, 27). In addition, several mycobacteriophages have been characterized in detail, and the genomes of several such phages have been sequenced and analyzed (10, 11, 15, 24, 25). These phages have been proposed to be useful tools for the genetic manipulation of mycobacteria and for understanding mycobacterial gene expression. Studies of them have also made a significant contribution to the study of phage evolution and have demonstrated that there is a high degree of mosaicism among the mycobacteriophages.
Further studies of P. acnes bacteriophages may also lead to the development of phage therapy for acne. Such therapy may overcome the current problems surrounding long-term use of antibiotics and the emergence of resistance in P. acnes and other skin commensals. We present here the first genome sequence of a P. acnes bacteriophage along with an extensive sequence analysis and characterization of the phage morphology and host range.
MATERIALS AND METHODS
Strains of bacteria and phage PA6.
P. acnes bacteriophage PA6 was isolated from a skin scrub wash sample taken from a patient attending the Department of Dermatology at Leeds General Infirmary. A plaque was observed among heavy bacterial growth following plating of this sample on media selective for propionibacteria. A stock was prepared from this plaque, and this stock was used in subsequent experiments and was grown on lawns of P. acnes AT1. P. acnes was grown in TYG broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.25% [wt/vol] glucose) and on reinforced clostridial agar (Oxoid) at 34°C in an N2/CO2/H2 (80:10:10, by volume) atmosphere.
Phage purification and DNA extraction.
Particles of phage PA6 were prepared using a standard plate lysate procedure (28). An aliquot of a stock of PA6 sufficient to produce nearly confluent bacterial lysis was added to 100 μl of late-log-phase P. acnes AT1 resuspended to an optical density at 600 nm of 2.5 in SM buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 60 mM MgSO4, 0.01% [wt/vol] gelatin). Phage were allowed to adsorb for 15 min at 34°C before the mixture was added to 3 ml molten top agarose (0.7% [wt/vol] agarose type VII; Sigma) and poured onto a 90-mm reinforced clostridial agar plate. Ten plates were prepared in this way and incubated anaerobically overnight at 34°C. Phage were harvested by overlaying each plate with 3 ml SM buffer and incubating it for 4 h at 4°C. Buffer was removed from the plates and centrifuged to remove bacteria (8,000 × g, 10 min, 4°C). Phage DNA was subsequently extracted using the Wizard Lambda Preps DNA purification system (Promega) according to the manufacturer's instructions.
DNA sequencing assembly and analysis.
Sequencing of the phage genome was carried out using a shotgun strategy by Lark Technologies Inc. Fragmented phage DNA was cloned into the pUC19 vector, and clones were chosen at random for sequencing. Dye terminator sequencing reactions were performed with ABI3100 and ABI3700 automatic sequencers (Applied Biosystems). DNA sequencing data were assembled and edited using the Phred-Phrap software package (CodonCode Corp.). Single-stranded extensions not identified after shotgun sequencing were isolated by self-ligation of the PA6 genome, PCR over the ligated region, and sequencing of the PCR product. Open reading frames (ORFs) were identified using GeneMark (1) and FramePlot (17). Assignment of an ORF was based on the presence of a possible initiation codon (AUG, GUG, or UUG) preceded by a potential ribosome binding site and the third-letter G+C content of codons within the ORF. Codon usage was determined using the Countcodon program (http://www.kazusa.or.jp/codon/countcodon.html), and protein transmembrane prediction was carried out using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Electron microscopy.
Purified phage from a plate lysate were placed onto carbon-coated copper grids and negatively stained with 1% (wt/vol) uranyl acetate. Particles were visualized by using a Philips CM10 transmission electron microscope.
Nucleotide sequence accession number.
The GenBank accession number for the bacteriophage PA6 genome is DQ431235.
RESULTS
Morphology of PA6 particles.
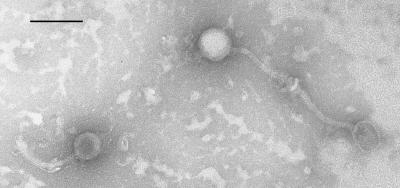
The morphology of PA6 particles was assessed by transmission electron microscopy (Fig. 1). PA6 possessed an icosahedral head with a diameter of approximately 50 nm and a flexible tail that was 165 nm long. PA6 resembles phages of the Siphoviridae family that possess icosahedral heads and long, noncontractile tails (10, 15, 24).
FIG. 1.
Transmission electron micrograph of negatively stained PA6 particles. Bar = 100 nm.
Host range of PA6.
In order to determine the host range of PA6, plaque assays were carried out with 32 different isolates of P. acnes (type I and II), other species of cutaneous propionibacteria, and members of other genera commonly found on human skin. PA6 was able to infect and lyse all 32 isolates of P. acnes tested. The majority of these isolates were clinical isolates from acne patients and included strains that were either sensitive or resistant to erythromycin/clindamycin. Also included were a laboratory strain (P37), the type strain (NCTC737), and the recently sequenced strain KPA171202 (3). PA6 was not able to infect isolates of Propionibacterium granulosum, Propionibacterium avidum, Staphylococcus epidermidis, or Corynebacterium bovis that are also members of the human skin microflora.
DNA sequence and organization of the PA6 genome.
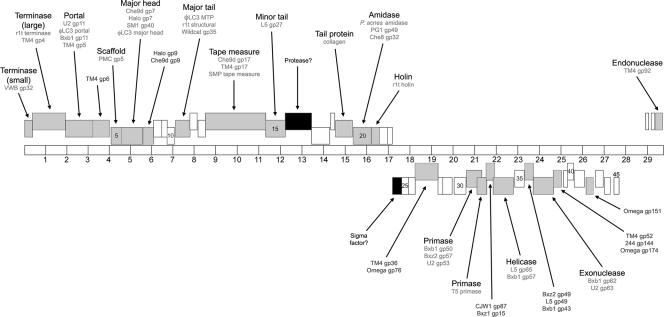
The linear, double-stranded DNA genome of phage PA6 was sequenced using a shotgun strategy. A single contig consisting of 29,739 bp was assembled from 681 sequences and was identified as the full-length phage genome, and >9-fold coverage was achieved. An additional 13-base single-stranded extension (CCTCGTACGGCTT) was identified at each end of the PA6 genome. The overall G+C content of the PA6 genome was 54.0%, which is slightly lower than the G+C content determined for the genome of P. acnes (60%) (3). A total of 48 ORFs were identified in the genome of PA6. Seventeen ORFs possessed potential GUG initiation codons, and the remaining ORFs had AUG initiation codons. All three of the standard termination codons were used, and 29 ORFs had a UGA termination codon. No tRNA genes were found in the genome. The coordinates of the protein-encoding genes identified are shown in Table 1, and a diagram of the PA6 genome is presented in Fig. 2.
TABLE 1.
Coordinates of PA6 genes and molecular masses, putative functions, and homologues of predicted proteins
| Gene | Start site | Stop site | Molecular mass of protein (kDa) | Function | Homologue | E value |
|---|---|---|---|---|---|---|
| 1 | 2 | 373 | 14.1 | Terminase (small) | VWB gp32 | 2e-04 |
| 2 | 378 | 1889 | 53.9 | Terminase (large) | Streptococcus pyogenes phage terminase | 6e-34 |
| Phage r1t putative terminase | 1e-33 | |||||
| Mycobacteriophage TM4 gp4 | 3e-24 | |||||
| 3 | 1886 | 3211 | 48.1 | Portal protein | Mycobacteriophage U2 gp11 | 2e-33 |
| Phage φLC3 putative portal protein | 1e-29 | |||||
| Mycobacteriophage Bxb1 gp11 | 3e-27 | |||||
| Mycobacteriophage TM4 gp5 | 5e-03 | |||||
| 4 | 3218 | 3973 | 27.9 | Mycobacteriophage TM4 gp6 | 2e-04 | |
| 5 | 4084 | 4638 | 19.6 | Scaffold protein | Mycobacteriophage PMC gp5 | 0.85 |
| 6 | 4645 | 5592 | 32.7 | Major head protein | Mycobacteriophage Che9d gp7 | 6e-47 |
| Mycobacteriophage halo gp7 | 2e-36 | |||||
| Streptococcus mitis phage SM1 gp40 | 2e-29 | |||||
| Phage φLC3 major head protein | 2e-28 | |||||
| 7 | 5638 | 6099 | 16.4 | Mycobacteriophage halo gp9 | 3e-25 | |
| Mycobacteriophage Che9d gp9 | 1e-10 | |||||
| 8 | 6101 | 6448 | 13.1 | |||
| 9 | 6455 | 6745 | 10.2 | |||
| 10 | 6766 | 7113 | 12.9 | |||
| 11 | 7172 | 7813 | 22.8 | Major tail protein | Phage φLC3 MTP | 6e-07 |
| Phage r1t structural protein | 8e-07 | |||||
| Mycobacteriophage wildcat gp35 | 4e-04 | |||||
| 12 | 7842 | 8138 | 10.6 | |||
| 13 | 8237 | 8524 | 10.6 | |||
| 14 | 8532 | 11297 | 94.5 | Tape measure | Mycobacteriophage Che9d gp17 | 3e-29 |
| Mycobacteriophage TM4 gp17 | 2e-28 | |||||
| Streptococcus suis phage SMP tape measure | 2e-20 | |||||
| 15 | 11315 | 12262 | 34.9 | Minor tail subunit | Mycobacteriophage L5 gp27 | 0.16 |
| 16 | 12270 | 13427 | 42.8 | Protease? | ||
| 17 | 13444 | 14262 | 28.9 | |||
| 18 | 14331 | 14570 | 8.8 | |||
| 19 | 14630 | 15319 | 22.3 | Tail protein | Human αI(III) collagen chain | 1e-15 |
| 20 | 15376 | 16236 | 31.3 | Amidase | P. acnes putative amidase | 5e-49 |
| Mycobacteriophage PG1 gp49 | 1e-06 | |||||
| Mycobacteriophage Che8 gp32 | 3e-04 | |||||
| 21 | 16249 | 16650 | 13.7 | Holin | Phage r1t holin | 2e-02 |
| 22 | 16663 | 16959 | 11.3 | |||
| 23 | 16984 | 17217 | 8.6 | |||
| 24 | 17637 | 17245 | 14.7 | Sigma factor? | Pfam04545 σ70 | 3e-03 |
| COG1595 σ24 (σE) | 8e-03 | |||||
| 25 | 17916 | 17641 | 10.1 | |||
| 26 | 18258 | 17938 | 12.3 | |||
| 27 | 19315 | 18269 | 39.4 | Mycobacteriophage TM4 gp36 | 1e-28 | |
| Mycobacteriophage omega gp76 | 7e-11 | |||||
| 28 | 19506 | 19312 | 7.1 | |||
| 29 | 19953 | 19513 | 15.6 | |||
| 30 | 20616 | 20053 | 22.5 | |||
| 31 | 21332 | 20661 | 25.3 | DNA primase | Mycobacteriophage Bxb1 gp50 | 3e-20 |
| Mycobacteriophage Bxz2 gp57 | 5e-20 | |||||
| Mycobacteriophage U2 gp53 | 6e-20 | |||||
| 32 | 21570 | 21172 | 15.4 | DNA primase | COG0358 DNA primase | 5e-08 |
| Phage T5 putative DNA primase | 5e-05 | |||||
| 33 | 21886 | 21530 | 13.6 | Mycobacteriophage CJW1 gp87 | 4e-08 | |
| Mycobacteriophage Bxz1 gp15 | 9e-05 | |||||
| 34 | 22749 | 21796 | 35.5 | DNA helicase | COG0305 DNA helicase | 2e-03 |
| Mycobacteriophage L5 gp65 | 9e-15 | |||||
| Mycobacteriophage Bxb1 gp57 | 1e-13 | |||||
| 35 | 23261 | 22791 | 17.3 | |||
| 36 | 23722 | 23312 | 15.1 | Mycobacteriophage Bxz2 gp49 | 0.11 | |
| Mycobacteriophage L5 gp49 | 0.11 | |||||
| Mycobacteriophage Bxb1 gp43 | 0.27 | |||||
| 37 | 24666 | 23719 | 35.8 | Exonuclease | COG2887 RecB exonuclease | 8e-05 |
| Mycobacteriophage Bxb1 gp62 | 2e-12 | |||||
| Mycobacteriophage U2 gp63 | 5e-12 | |||||
| 38 | 25025 | 24666 | 13.3 | Mycobacteriophage TM4 gp52 | 7e-12 | |
| Mycobacteriophage 244 gp114 | 2e-10 | |||||
| Mycobacteriophage omega gp174 | 9e-06 | |||||
| 39 | 25334 | 25131 | 7.2 | |||
| 40 | 25558 | 25331 | 8.4 | |||
| 41 | 26120 | 25581 | 19.1 | |||
| 42 | 26493 | 26206 | 10.8 | Mycobacteriophage omega gp151 | 7e-04 | |
| 43 | 26933 | 26622 | 11.8 | |||
| 44 | 27273 | 27007 | 9.4 | |||
| 45 | 27612 | 27430 | 7.0 | |||
| 46 | 28878 | 29033 | 5.3 | |||
| 47 | 29190 | 29369 | 6.7 | |||
| 48 | 29388 | 29690 | 11.4 | Endonuclease | Pfam01844 HNH endonuclease | 4e-07 |
| Mycobacteriophage TM4 gp92 | 1e-04 |
FIG. 2.
Genome organization of PA6. The 29,739-bp genome is represented by a horizontal bar with 1-kb intervals marked. Putative ORFs are indicated by boxes at three different heights representing the three frames on each strand. Rightward-transcribed genes are above the genome, and leftward-transcribed genes are below the genome. Predicted functions and homologues are indicated. ORFs with phage and nonphage homologues are indicated by gray and black boxes, respectively. ORFs unique to PA6 are indicated by white boxes.
The PA6 genome has an organization similar to that of other Siphoviridae with a left arm containing rightward-transcribed genes (genes 1 to 23) and a right arm containing leftward-transcribed genes (genes 45 to 24). There is little noncoding DNA between these genes, suggesting that there is a single transcript for each set of genes. The final three genes (genes 46 to 48) at the extreme right end of the genome are transcribed rightward and are separated from the other left arm genes by a 1,447-bp segment of noncoding DNA (bases 27431 to 28877). The most notable feature of this region is its much lower G+C content compared to that of the rest of the genome (45%). This right arm gene organization closely resembles that of mycobacteriophage Bxb1, although the noncoding region between the leftward-transcribed genes and three rightward-transcribed genes at the far right of the Bxb1 genome is much smaller, 213 bp. The codon usage of PA6 is somewhat different than that of P. acnes KPA171202 (see the supplemental material), suggesting that in evolutionary terms, PA6 may have only recently become a phage of P. acnes.
Genes encoding structural and assembly proteins.
The left arm of the PA6 genome contains genes encoding proteins of the viral particle. The organization of these genes is similar to that of many of the mycobacteriophages, and as expected, the morphology of the PA6 virion is similar to that observed for these phages (25). Many of the predicted gene products are related to those of mycobacteriophages. Predicted functions have been assigned based on homology to other phage proteins having known or predicted functions and on matches to the COG (http://www.ncbi.nlm.nih.gov/COG/) and Pfam (http://www.sanger.ac.uk/Software/Pfam/) databases.
PA6 gp2 is a putative terminase large subunit and shows a high degree of homology to other members of this protein family (43% similarity to TM4 gp4 and 47% similarity to the lactococcal phage r1t). The smaller gp1 protein shows weak homology to gp32 of phage VWB that is predicted to be the terminase small subunit (32). This homology, along with the size and position of gene 1, makes the gp1 protein the likely terminase small subunit of PA6. This family of proteins is involved in the packaging of phage DNA into the head of the virus particle (12). This process also involves the portal protein (gp3 of PA6) that lies at the head-tail interface and enables passage of genomic DNA into phage heads during packaging and ejection during infection. The gp5 and gp6 proteins are most likely components of the phage head. gp5 is likely to be the scaffold protein that directs assembly of the head proteins, and gp6 is likely to be the major head protein that shows homology to the gp7 head protein of mycobacteriophage Che9d (55% similarity over 312 residues). Three proteins were identified that are proposed components of the phage tail. gp11 exhibits homology with the major tail protein of lactococcal phage φLC3. gp14 is a putative tape measure protein that in bacteriophage λ determines tail length (20, 21). The PA6 tape measure protein does not contain any of the three sequence motifs predicted to have a role in cell wall catabolism, one of which shows homology to the resuscitating-promoting factor family of proteins thought to be involved in stimulating dormant cell growth (22, 26). gp15 is proposed to be a minor tail protein based on homology to the proven tail protein (gp27) of phage L5 (15). One gene of note in this region is ORF16. The predicted product of this gene is a 42.8-kDa protein. The first 80 amino acids of the N terminus of this protein show a low level of homology to a family of protease enzymes that specifically bind to and degrade proteins with nonpolar C termini. It is possible that this protein is involved in some proteolytic process during phage assembly. Alternatively, this protein may be involved in entry of the phage into the host cell through degradation of peptide linkages in the cell wall to allow injection of viral DNA. The location of the gene encoding gp16 suggests that it may be a component of the PA6 tail, making this function not unlikely.
Lysis functions.
ORF20, located near the end of the section of rightward-transcribed genes, encodes a putative N-acetylmuramoyl-l-alanine amidase. This enzyme is most likely involved in cell lysis following phage assembly rather than in phage entry as the gene immediately downstream of ORF20 (ORF21) encodes a putative holin. Following phage assembly, holin proteins assemble to form pores in the cellular membrane, allowing the amidase enzyme access to the surrounding peptidoglycan (35). PA6 gp21 appears to belong to the class II group of holins. Members of this group are usually 65 to 95 residues long and possess two transmembrane domains. Although gp21 is larger (113 residues), it is predicted to contain two transmembrane domains instead of the three transmembrane domains of the class I holins. The PA6 amidase exhibits homology with amidases from the PG1 and Che8 mycobacteriophages. However, the highest degrees of similarity are with an N-acetylmuramoyl-l-alanine amidase and likely an autolysin from its host, P. acnes (67% identity for amino acids 2 to 145). This is perhaps not unexpected considering their shared substrate.
DNA replication functions.
The leftward-transcribed section of the genome contains genes predicted to encode proteins involved in phage DNA replication. PA6 gp31 and gp32 are both putative DNA primases. PA6 gp31 exhibits homology with one of the DNA primases of Bxb1, gp50 (35% identity over 154 residues). The organization of these genes is similar to that of the DNA primase-encoding genes of mycobacteriophages Bxb1, L5, and D29, with an extensive overlap of genes 31 and 32. It has been suggested that in these mycobacteriophages, the primase proteins are produced through a programmed translational frameshift resulting in a single functional protein (24). PA6 gp34 is proposed to be a DNA helicase based on its match to the COG DNA helicase group. This protein contains an ATP-binding domain and exhibits homology to Bxb1 gp57 and L5 gp65, which are also predicted DNA helicases. Other PA6 genes in this region encode proteins with no predicted functions but with homology to proteins of other phages (Table 1).
One significant aspect of the PA6 genome is the absence of any gene encoding an obvious integrase-type enzyme. Plaques of PA6 on lawns of P. acnes are clear with hazy centers and not turbid, as is characteristic of truly lysogenic phages. This plaque morphology has been observed for TM4, which is not temperate but may be capable of forming pseudolysogens in some strains of mycobacteria (11). Also absent from the PA6 genome is any gene encoding a repressor-like protein that would indicate a lysogenic function. Although the presence of such a protein cannot be completely ruled out, the lack of these two lysogeny-related proteins strongly suggests that PA6 is a lytic phage and does not integrate into the host genome. Other PA6 proteins possibly involved in DNA regulation/replication include gp37 and gp48. These proteins are predicted to be an exonuclease and an endonuclease, respectively, due to matches to the COG RecB exonuclease group and Pfam HNH endonuclease family, respectively.
Other genes and features of interest.
Two other predicted ORFs of PA6 are worthy of mention. ORF19 encodes a protein with considerable homology to the human collagen α1(III) chain precursor. In particular, gp19 contains many G-X-Y repeats that are characteristic of many collagens (Fig. 3). The gene encoding gp19 is located in the vicinity of the genes encoding the structural proteins that constitute the phage tail. The presence of collagen-like proteins in phage tails has been noted previously (29). The function of such proteins is unknown, but it has been suggested that they stabilize the tail structure. It is tempting to speculate that in PA6 this protein may have evolved a second function, binding to the host cell and, more specifically, to potential collagen-binding surface proteins of P. acnes. The P. acnes genome contains several genes encoding putative surface adhesins that are likely to be important for the colonization of human skin, one or more of which may bind collagen.
FIG. 3.
Comparison of PA6 gp19 with the human collagen α1(III) chain. Protein sequences are aligned, and identical amino acid residues are marked with asterisks. The glycine residues of the repetitive G-X-Y motifs are in bold type. The numbers indicate the positions of the amino acid sequences in the proteins.
A second gene of interest is ORF24. A BLAST search of the gene product revealed that this protein has a conserved sigma factor domain. The gene encoding this protein is transcribed in the same direction as the genes encoding proteins predicted to be involved in DNA replication. This protein may therefore be a phage-specific sigma factor that directs the host RNA polymerase to transcribe from phage-specific promoters.
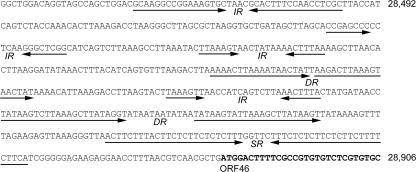
Within the 1.4-kb noncoding region of the PA6 genome there is an approximately 500-bp section that contains numerous direct, inverted, and symmetric repeats (Fig. 4). This 500-bp section has a particularly low G+C content (35%). Such repeat sequences may be binding sites for transcription factors and/or may form secondary structures important for the regulation of gene transcription and DNA replication. The gene organization and direction of transcription indicate that this region is likely to contain promoters for phage gene transcription.
FIG. 4.
Repeat motifs located in the noncoding region of the PA6 genome. The noncoding region between ORF45 and ORF46 contains several direct (DR), inverted (IR), and symmetric (SR) repeats that are indicated by arrows. The numbers indicate the positions in the PA6 genome, and the start of ORF46 is in bold type.
The recent sequencing of the genome of P. acnes strain KPA171202 revealed the presence of a cryptic prophage (3). Very few proteins predicted to be encoded by this prophage show homology to other phage proteins, and there is no great similarity to any PA6 proteins. The only KPA171202 prophage proteins with functions similar to the functions of PA6 proteins are PPA1592, which has similarity to HNH endonucleases and shows weak homology to Bxb1 gp1 and L5 gp4; PPA1596, which appears to be a DNA primase with weak homology to TM4 gp70; and PPA1609, which shows some similarity to exonucleases.
DISCUSSION
Bacteriophages of P. acnes have been isolated previously, but this is the first report of the genomic sequence of a P. acnes phage. The organization of the PA6 genome and the similarity of predicted proteins to the proteins of other phages, especially the mycobacteriophages, provide interesting insights into the phylogenetic relationships of these phages and further highlight the mosaic nature of the phage genomes. Obvious differences between these phages include the apparent lack of genes involved in lysogeny in the PA6 genome. The hazy appearance of the centers of PA6 plaques on lawns of P. acnes suggests that this phage is capable of lysogeny, but genetic and experimental evidence is lacking. Prediction of ORFs in the PA6 genome using a heuristic approach (GeneMark) failed to identify any of the leftward-transcribed genes (ORF24 to ORF45 predicted by FramePlot). Although the protein products of several of these genes exhibit homology to other phage proteins, it is possible that PA6 is a derivative of a larger phage and has lost many genes involved in the regulation of DNA metabolism, including those for lysogeny. The presence of a cryptic prophage within the P. acnes KPA171202 genome indicates that P. acnes phages capable of integration may exist. Genome analysis of other P. acnes bacteriophages could establish phylogenetic relationships and show whether phage PA6 is a derivative of a much larger temperate phage.
The host range of PA6 is restricted to P. acnes. Although it infects and lyses all strains of P. acnes tested to date, PA6 cannot infect other members of the genus Propionibacterium. This restricted range is likely to be dictated by the phage receptor and/or other components of the phage tail involved in entry of the phage into the cell. The PA6 minor tail subunit gp15 may be involved in host specificity. This protein has homology to D29 gp27, L5 gp27, and Bxb1 gp23, which have been proposed to be host specificity determinants (24). Isolation of phages capable of infecting the closely related cutaneous propionibacteria P. granulosum and P. avidum would allow comparisons of these genes to be made. However, phages with the ability to infect these species have not been isolated to date.
The sequencing of PA6 makes a valuable contribution to the study of bacteriophage evolution. The close relationship between mycobacteria and propionibacteria is reflected in the relationships between their respective phages, and PA6 is more closely related to mycobacteriophages than to phages of any other bacterial group. More significantly, PA6 should contribute to the study of propionibacterial genetics. Finally, the sequencing of PA6 should greatly enhance the development of a potential bacteriophage therapy to treat acne and therefore overcome the significant problems associated with long-term antibiotic therapy and bacterial resistance.
Supplementary Material
Acknowledgments
This work was supported by the Leeds Foundation for Dermatological Research and by Sarum Biosciences Ltd.
Footnotes
Published ahead of print on 30 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Borodovsky, M., and J. D. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17:123-133. [Google Scholar]
- 2.Brook, I., and E. H. Frazier. 1991. Infections caused by Propionibacterium species. Rev. Infect. Dis. 13:819-822. [DOI] [PubMed] [Google Scholar]
- 3.Brüggemann, H., A. Henne, F. Hoster, H. Liesegang, A. Wiezer, A. Strittmatter, S. Hujer, P. Dürre, and G. Gottschalk. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305:671-673. [DOI] [PubMed] [Google Scholar]
- 4.Cove, J. H., K. T. Holland, and W. J. Cunliffe. 1983. Effects of oxygen concentration on biomass production, maximum specific growth rate and extracellular enzyme production by three species of cutaneous propionibacteria grown in continuous culture. J. Gen. Microbiol. 129:3327-3334. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe, W. J. 1989. Clinical features of acne, p. 11-75. In R. Marks (ed.), Acne. Martin Dunitz, London, United Kingdom.
- 6.Cunliffe, W. J. 1998. The sebaceous gland and acne—40 years on. Dermatology 196:9-15. [DOI] [PubMed] [Google Scholar]
- 7.Eady, E. A., J. H. Cove, K. T. Holland, and W. J. Cunliffe. 1989. Erythromycin resistant propionibacteria in antibiotic treated acne patients: association with therapeutic failure. Br. J. Dermatol. 121:51-57. [DOI] [PubMed] [Google Scholar]
- 8.Eady, E. A., and E. Ingham. 1994. Propionibacterium acnes—friend or foe? Rev. Med. Microbiol. 5:163-173. [Google Scholar]
- 9.Farrar, M. D., and E. Ingham. 2004. Acne: inflammation. Clin. Dermatol. 22:380-384. [DOI] [PubMed] [Google Scholar]
- 10.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143-164. [DOI] [PubMed] [Google Scholar]
- 11.Ford, M. E., C. Stenstrom, R. W. Hendrix, and G. F. Hatfull. 1998. Mycobacteriophage TM4: genome structure and gene expression. Tubercle Lung Dis. 79:63-73. [DOI] [PubMed] [Google Scholar]
- 12.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 13.Greenman, J., K. T. Holland, and W. J. Cunliffe. 1983. Effects of pH on biomass, maximum specific growth rate and extracellular enzyme production by three species of cutaneous propionibacteria grown in continuous culture. J. Gen. Microbiol. 129:1301-1307. [DOI] [PubMed] [Google Scholar]
- 14.Greenman, J., K. T. Holland, and W. J. Cunliffe. 1981. Effects of glucose concentration on biomass, maximum specific growth rate and extracellular enzyme production by three species of cutaneous propionibacteria grown in continuous culture. J. Gen. Microbiol. 127:371-376. [DOI] [PubMed] [Google Scholar]
- 15.Hatfull, G. F., and G. J. Sarkis. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7:395-405. [DOI] [PubMed] [Google Scholar]
- 16.Holland, K. T. 1989. Microbiology of acne, p. 187-211. In R. Marks (ed.), Acne. Martin Dunitz, London, United Kingdom.
- 17.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the Frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, W. R., M. Tuckman, and B. R. Bloom. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532-535. [DOI] [PubMed] [Google Scholar]
- 19.Jong, E. C., H. L. Ko, and G. Pulverer. 1975. Studies on bacteriophages of Propionibacterium acnes. Med. Microbiol. Immunol. 161:263-271. [DOI] [PubMed] [Google Scholar]
- 20.Katsura, I. 1987. Determination of bacteriophage lambda tail length by a protein ruler. Nature 327:73-75. [DOI] [PubMed] [Google Scholar]
- 21.Katsura, I., and R. W. Hendrix. 1984. Length determination in bacteriophage lambda tails. Cell 39:691-698. [DOI] [PubMed] [Google Scholar]
- 22.Kell, D. B., and M. Young. 2000. Bacterial dormancy and cultivability: the role of autocrine growth factors. Curr. Opin. Microbiol. 3:238-243. [DOI] [PubMed] [Google Scholar]
- 23.Leyden, J. J., K. J. McGinley, and B. Vowels. 1998. Propionibacterium acnes colonization in acne and non-acne. Dermatology 196:55-58. [DOI] [PubMed] [Google Scholar]
- 24.Mediavilla, J., S. Jain, J. Kriakov, M. E. Ford, R. L. Duda, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955-970. [DOI] [PubMed] [Google Scholar]
- 25.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 26.Piuri, M., and G. F. Hatfull. 2006. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62:1569-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranes, M. G., J. Rauzier, M. LaGranderie, M. Gheorghiu, and B. Gicquel. 1990. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a “mini” mycobacterium-Escherichia coli shuttle vector. J. Bacteriol. 172:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Smith, M. C. M., N. Burns, J. R. Sayers, J. A. Sorrell, S. R. Casjens, and R. W. Hendrix. 1998. Bacteriophage collagen. Science 279:1834. [DOI] [PubMed] [Google Scholar]
- 30.Thiboutot, D. M. 1997. Acne: an overview of clinical research findings. Dermatol. Clin. 15:97-109. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, T. P., and A. L. Albright. 1998. Propionibacterium acnes infections of cerebrospinal fluid shunts. Child. Nerv. Syst. 14:378-380. [DOI] [PubMed] [Google Scholar]
- 32.Van Dessel, W., L. Van Mellaert, H. Liesegang, C. Raasch, S. De Keersmaeker, N. Geukens, E. Lammertyn, W. Streit, and J. Anné. 2005. Complete genomic nucleotide sequence and analysis of the temperate bacteriophage VWB. Virology 331:325-337. [DOI] [PubMed] [Google Scholar]
- 33.Webster, G. F., and C. S. Cummins. 1978. Use of bacteriophage typing to distinguish Propionibacterium acnes types I and II. J. Clin. Microbiol. 7:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winward, K. E., S. C. Pflugfelder, H. W. Flynn, T. J. Roussel, and J. L. Davis. 1993. Postoperative Propionibacterium endophthalmitis—treatment strategies and long-term results. Ophthalmology 100:447-451. [DOI] [PubMed] [Google Scholar]
- 35.Young, R., I. N. Wang, and W. D. Roof. 2000. Phages will out: strategies of host cell lysis. Trends Microbiol. 8:120-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.