Abstract
The adaptation of the tubercle bacillus to the host environment is likely to involve a complex set of gene regulatory events and physiological switches in response to environmental signals. In order to deconstruct the physiological state of Mycobacterium tuberculosis in vivo, we used a chemostat model to study a single aspect of the organism's in vivo state, slow growth. Mycobacterium bovis BCG was cultivated at high and low growth rates in a carbon-limited chemostat, and transcriptomic analysis was performed to identify the gene regulation events associated with slow growth. The results demonstrated that slow growth was associated with the induction of expression of several genes of the dormancy survival regulon. There was also a striking overlap between the transcriptomic profile of BCG in the chemostat model and the response of M. tuberculosis to growth in the macrophage, implying that a significant component of the response of the pathogen to the macrophage environment is the response to slow growth in carbon-limited conditions. This demonstrated the importance of adaptation to a low growth rate to the virulence strategy of M. tuberculosis and also the value of the chemostat model for deconstructing components of the in vivo state of this important pathogen.
Despite more than a century of research into tuberculosis (TB), this disease remains the number one killer due to a single infectious agent, making Mycobacterium tuberculosis one of the most successful human pathogens. A key to this bacterium's success is its ability to establish and maintain a latent infection in its human host for many decades (18). The control of TB is severely impeded by the global magnitude of latent TB. One-third of the world's population is estimated to harbor persistent M. tuberculosis primed for reactivation and initiation of clinical disease (7). The bacterial response to the triggers of latency and reactivation are very poorly understood. Determining the mechanisms involved during the establishment, maintenance, and reactivation of latent TB is an important goal for mycobacterial researchers. Such information should lead to the development of novel therapeutics, vaccines, and diagnostic strategies targeted to persistent M. tuberculosis.
In vitro modeling of M. tuberculosis provides simple experimental approaches for studying the physiology and genetic basis of TB. However, the design of adequate models is impeded by the paucity of knowledge about the biological characteristics of both the bacteria and the host environment during human TB, and therefore in vitro modelers must make simplistic assumptions about the environmental variables within the human host. Microaerophilic adaptation, nutrient starvation, drug-persistent, and extended stationary-phase models of persistent TB have been established (9). All these models provide in vitro conditions that are intended to simulate the microenvironments of the host during persistence. While these models have proved to be very valuable for studying persistence, it is unlikely that a single model can recapitulate all the conditions experienced by the bacteria, and the relevance of these models to the in vivo state remains unclear.
An alternative approach is to attempt to deconstruct the physiological state of mycobacteria into its components. It may then be possible to establish molecular fingerprints characterizing the different physiological states of the pathogen that may then be compared with the molecular signature of TB cells in the host. One of the few undisputed characteristics of persistent M. tuberculosis is that the pathogen grows at a lower rate during persistence than during active disease. Studying the adaptation of M. tuberculosis to slow growth is therefore a first step toward deconstructing its physiological state during TB.
Chemostat culture is a technique that was specifically devised to grow microbes under constant, carefully controlled conditions at a single growth rate. We initiated a study of the tubercle bacillus growing in defined media in a chemostat using the Mycobacterium bovis BCG vaccine strain as a model organism (3). Our first step was to establish chemostat culture of the tubercle bacillus growing at two different rates in carbon-limited conditions. A detailed macromolecular analysis of steady-state cultures was performed in order to compile a molecular inventory of carbon-limited BCG growing at low and high rates. This analysis demonstrated that the mycobacterial cell is remodeled in response to changes in the growth rate in carbon-limited conditions (3).
In a previous study we established that the lipid, RNA, and protein contents of the mycobacterial cell are different in slow-growing M. bovis BCG and fast-growing M. bovis BCG (3). The genome is of course the same in both organisms, so the different physiological states must be maintained by differences at the level of the transcriptome, proteome, and metabolome. The chemostat system provides an ideal environment for functional genomic studies to determine these differences, since cells can be reproducibly obtained in highly defined conditions. Data obtained in a microarray analysis of organisms grown in a chemostat have been shown to be more reproducible and accurate than batch culture DNA array data (6, 12, 13, 17). A combination of a chemostat culture and DNA microarrays was used previously to study the effects of oxygen limitation on the gene expression profile of M. tuberculosis (2). In that study the workers demonstrated that chemostat cultures can be used to study the responses of mycobacteria to specific environmental conditions. Here, in order to investigate the molecular mechanisms underlying the observed shift in the macromolecular composition of M. bovis BCG in a carbon-limited chemostat, we performed a transcriptome analysis of fast-growing and slow-growing BCG cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. bovis BCG strain ATCC 35748 was cultured in a 2-liter bioreactor (Adaptive Biosystem Voyager) under aerobic conditions at pH 6.6 as previously described (3). Chemostat cultures were grown in Roisin's minimal medium at a constant dilution rate of 0.03 h−1 (equivalent to a doubling time [td] of 23 h) or 0.01 h−1 (td, 69 h). Culture samples were withdrawn from the chemostat to monitor the cellular dry weight, viable counts, optical density, nutrient utilization, and CO2 and O2 concentrations (3). We assumed that steady-state conditions were reached when the level of carbon dioxide evolution, optical density at 600 nm, and dry weight remained constant for three consecutive volume changes. Once the steady state was reached, cells were harvested for analysis. Independent triplicate chemostat cultures were used for each growth rate.
RNA preparation.
In order to stabilize the mRNA population, 20 ml of a culture sample was rapidly withdrawn from the chemostat by using a tube submerged in the culture broth and was directly injected into a sterile bottle containing 80 ml of GTC solution (5 M guanidinium thiocyanate, 0.5% sodium N-lauryl sarcosine, 25 mM trisodium citrate, 0.1 M dithiothreitol) (20). RNA was prepared essentially as described by Stewart et al. (33). The extraction protocol was modified slightly so that it included additional chloroform extractions to remove lipid contamination.
Fluorescently labeled cDNA was produced by reverse transcription of total M. tuberculosis RNA (2 to 10 μg) with Superscript III (Invitrogen) in the presence of Cy3-dCTP or Cy5-dCTP (Amersham Pharmacia) using random hexamer oligonucleotides to prime cDNA synthesis. RNA samples prepared from chemostat cultures at a dilution rate of 0.03 h−1 (td, 23 h) were directly compared to RNA samples prepared from chemostat cultures at a dilution rate of 0.01 h−1 (td, 69 h) by competitive microarray analysis with both dye arrangements.
Microarrays and hybridizations.
The DNA microarrays, provided by the Bacterial Microarray Group at St. Georges (http://bugs.sgul.ac.uk/index.php), were constructed from PCR-amplified open reading frame-specific DNA representing all of the predicted open reading frames of the M. tuberculosis H37Rv genome robotically spotted onto a poly-l-lysine-coated glass microscope slide (33). Prehybridization, hybridization, and washing were performed as described by Stewart et al. (33). All microarray experiments were technically replicated four times with cells from three independent chemostat experiments, resulting in 12 sets of microarray data.
Data processing and statistical analysis.
Microarrays were scanned using a GenePix 4000B (Axon Instruments) at a level just below saturation of the most intensely fluorescent spot on each array. Fluorescence intensity data from each array were quantified and quality control was fully automated using the BlueFuse for microarrays (version 3.3) software (Bluegnome). Hybridization intensity values were normalized using lowess local regression. This was followed by across-array normalization that gave the log2 ratios on each array the same median absolute deviation using limma, which is part of the Bioconductor package of R, version 2.4.0 (10). Technical replicates were averaged and filtered using a Perl script so that only genes for which there was a value for each dye orientation were used; this resulted in 3,476 genes whose significance was tested. Expression values were compared using an eBayes modified t test (32) and were also subjected to the Benjamini-Hochberg multiple-testing correction using the multtest package in R (26) to obtain a false-positive rate of 0.05, which corresponded to an uncorrected t test P value of approximately 0.0048. Expression data were clustered with the TIGR MeV software (29) technique, using the Pearson correlation coefficient as a distance measure (8).
In order to identify functionally related groups of genes that were significantly regulated, three thresholds were used (1.2-fold, 1.6-fold, and 2-fold). The following two lists were generated for each threshold: genes whose expression ratio (with no significance threshold) was greater than the 1.2-, 1.6-, or 2-fold threshold (upregulated genes) and genes whose expression ratio was less than 0.83, 0.625, or 0.5 (downregulated genes). Genes were divided into 101 functional categories based on the M. tuberculosis H37Rv simplified gene ontology (http://www.sanger.ac.uk/projects/M_tuberculosis/gene_list/). Any functional groups with less than eight members were not included in further analyses. Each set of functionally related genes was then analyzed as a group, and the chi-square test was performed to investigate whether the difference in the distribution of up- and downregulated genes in any set of genes differed significantly from the ratio expected by chance.
qRT-PCR analysis.
Oligonucleotides and probes were designed using the Primer 3 software (28) and are listed in Table S2 in the supplemental material. Quantitative reverse transcription PCRs (qRT-PCRs) were performed as described by Phillips et al., using 6-carboxyfluorescein reporter dye- and 6-carboxytetramethylrhodamine quencher dye-labeled probes in conjunction with specific primer sets (25). The quantity of DNA or cDNA in each reaction mixture was determined by using a standard curve generated by amplification of known amounts of a standard DNA prepared from M. bovis BCG. Pearson's and Spearman's correlation coefficients were used for comparison of qRT-PCR and microarray data.
RESULTS AND DISCUSSION
Chemostat culture.
BCG was inoculated into a chemostat, and after an initial phase in batch culture, continuous culture conditions were established using Roisin's minimal medium. Roisin's minimal medium is a chemically defined glycerol-limited medium containing only one source of carbon plus Tween 80. Steady-state conditions were established for BCG cells grown at a dilution rate of 0.01 h −1, corresponding to a td of 69 h (low growth rate), and at a dilution rate of 0.03 h−1, corresponding to a td of 23 h (high growth rate). The physiological characteristics of the cells have been described previously (3). Cells were harvested from three independent chemostat cultures, and RNA was extracted for transcriptome analysis.
DNA microarray analysis.
The gene expression of slow-growing M. bovis BCG cells and the gene expression of fast-growing M. bovis BCG cells were compared in order to identify the genes most important for slow growth and carbon limitation. We found that 338 genes, representing 8.1% of the genome, were significantly differentially expressed in the slow-growing cells adapting to carbon-limited conditions. Genes whose expression was significantly altered in response to slow growth in carbon-limited conditions were divided into functional categories using the gene ontology developed as part of the genome sequencing project (http://www.sanger.ac.uk/Projects/M_tuberculosis/Gene_list/).
We next tried to identify functional categories of genes whose expression was significantly changed by slow growth. A chi-square test was used to calculate the probability that the distribution of up- and downregulated genes within each functional group was consistent with the null hypothesis that the group represented a random sample of the total population of genes in the data set. For genes with known functions it was observed that at the low growth rate expression of genes belonging to the PE and PPE families, insertion (IS) elements, and genes associated with the biosynthesis of the cell envelope was significantly induced, whereas genes involved in the synthesis and modification of macromolecules were downregulated (Table 1). Data set S1 in the supplemental material provides supporting information for the transcriptomic data for all genes.
TABLE 1.
Summary of the most significant functional groups of genes with significantly altered expression in M. bovis BCG in a carbon-limited chemostat culture at a dilution rate of 0.01 h−1 (td, 69 h) when this expression was compared with the expression at a dilution rate of 0.03 h−1 (td, 23 h)
| Functional classificationa | Total no. of genes tested | No. of genes upregulated | No. of genes downregulated | P valueb |
|---|---|---|---|---|
| Whole genome | 3,476 | 179 | 159 | |
| Energy metabolism | 268 | 24 | 28 | 6.629 × 10−8 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 106 | 10 | 2 | 0.013 |
| Lipid biosynthesis | 64 | 1 | 8 | 0.001 |
| Synthesis and modification of macromolecules | 207 | 12 | 25 | 3.88 × 10−7 |
| Cell envelope | 323 | 22 | 8 | 0.0186 |
| Chaperone/heat shock | 16 | 2 | 2 | 0.0434 |
| Detoxification | 20 | 1 | 3 | 0.0294 |
| IS elements, repeated sequences, and phage | 71 | 11 | 0 | 9.882 × 10−6 |
| PE and PPE families | 114 | 10 | 3 | 0.0351 |
Functional categories as described at http://www.sanger.ac.uk/projects/M_tuberculosis/Gene_list/.
P values were determined by a chi-square test.
Independent validation of microarray analysis using qRT-PCR.
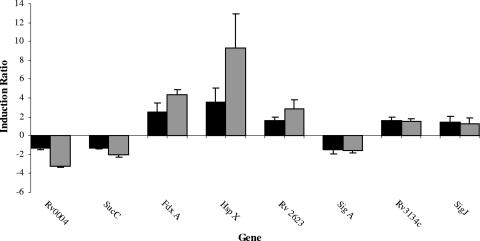
The microarray analysis results were independently validated for a subset of genes by qRT-PCR. The qRT-PCR results corroborated the microarray data for the eight genes tested (Spearman correlation coefficient, 0.87; Pearson correlation coefficient, 0.86). These data indicated that the fold changes determined by qRT-PCR were often greater than the fold changes for the same genes determined by microarrays (Fig. 1). Ratio underestimation is a commonly recognized feature of microarray technology (40). The excellent agreement between the microarray results and the qRT-PCR results substantiated the statistical approach that we utilized in this study.
FIG. 1.
Comparison of the gene expression ratios obtained by microarray analysis (solid bars) and qRT-PCR (gray bars). The bars indicate the average ratios for three independent biological replicates, and the error bars indicate the standard deviations.
Functional groups of genes that respond to modulation of the growth rate. (i) Synthesis and modification of macromolecules.
The downregulation of genes encoding ribosomal proteins observed during slow growth of M. bovis BCG is consistent with our previous finding that ribosome production in M. bovis BCG is subject to growth rate-dependent control (3) and has also been observed in other models of persistence (4). Despite the general decrease in ribosomal protein synthesis, four ribosomal protein genes, rpsR2, rpsN2, rpmG, and rpmB2, were significantly upregulated (2.58-, 6.02-, 4.81-, and 7.69-fold, respectively) at the low growth rate. These genes are arranged in an operon and are separated from the majority of the ribosomal genes. There is evidence suggesting that the RpmB and RpmG proteins of Escherichia coli have related roles in ribosome synthesis and function (19). Ribosome synthesis is defective and not very well coordinated when these proteins are absent from E. coli (19). The fact that these ribosomal proteins are regulated independently from the other members of their family in slow-growing M. bovis BCG suggests that they may also have a growth rate-related function in M. tuberculosis. Voskuil et al. (37) speculated that RpsN, RpmB, and RpmG may increase ribosomal fidelity, but further work is required to establish the functions of these proteins.
(ii) Energy metabolism.
Three tricarboxylic acid (TCA) cycle genes (sucB, sucC, and icd-2) were downregulated 0.68- to 0.78-fold, but we found no evidence that there was transcriptional activation of the glyoxylate shunt at the low growth rate; indeed, the glcB gene encoding malate synthase was downregulated (0.67-fold), whereas expression of the icl-1 and icl-2 genes encoding isocitrate lyase was unchanged. The glcB gene was also downregulated in M. tuberculosis cells starved in phosphate-buffered saline (PBS) (4).
Despite the fact that the oxygen levels in the chemostat vessel were continuously maintained at 70 to 100%, aerobic respiratory metabolism was downregulated, and three genes (frdA, fdhD and narJ) encoding proteins involved in anaerobic respiration were upregulated 1.35- to 1.46-fold at the low growth rate. This finding is consistent with the diminished need for respiration during slow growth. Anaerobic respiration has been described previously for persistent M. tuberculosis (38), and upregulation of other anaerobic genes has been demonstrated previously in a mouse infection model (31) and has also been reported under oxygen-sufficient conditions (11). Expression of the frdA gene was induced in bacteria residing in activated macrophages and also in the pericavity and distant lungs of humans with tuberculosis. The upregulation of 12 of the 48 genes controlled by the DosR two-component response regulator is also puzzling. Although expression of dosR (Rv3133c) was not significantly induced in the chemostat model (1.39-fold; P = 0.14), the Rv3134c gene, which is presumed to be a member of the same operon as dosR, was upregulated 1.8-fold (P = 0.028). The DosR system has been shown to be responsive to hypoxia in pathogenic and nonpathogenic mycobacteria (21, 24). However, it has been demonstrated that expression of dosR is also induced by nitrogen oxide and a number of other stresses in M. tuberculosis (15, 36), and upregulation of a subset of genes in the DosR regulon was also observed as M. tuberculosis entered the stationary phase of growth (37). Two kinases (DosS and DosT) that phosphorylate DosR in response to reduced oxygen tension and nitric oxide have been identified. Neither of the genes was induced during slow growth in the chemostat model. One explanation of these results is that expression of a subset of the DosR regulon is modulated by other environmental cues, such as a low growth rate or nutrient limitation, and then relayed through a different regulatory mechanism. Alternatively, it is possible that the remaining members of the DosR regulon were upregulated in the chemostat model but were not detected in this study due to the level of sensitivity of the microarrays. The qRT-PCR results presented here support this hypothesis. Irrespective of which scenario proves to be correct, our data provided further evidence that in addition to hypoxia, low growth rates and nutrient limitation modulate the expression of genes in the DosR regulon.
(iii) Lipid biosynthesis.
Array analysis revealed that there was significant downregulation of genes involved in modification of fatty and mycolic acids.
(iv) Cell envelope.
There are immense changes in the cell envelope of M. tuberculosis upon infection of both macrophages and human lungs, and modification of the cell wall is likely to be an important survival strategy for M. tuberculosis. Our study showed that a low growth rate and carbon limitation induced expression of 22 membrane proteins, suggesting that there was some remodeling of the cell envelope.
(v) Chaperones and heat shock.
We observed no trend in the transcription of chaperones and heat shock genes. The gene encoding the DosR regulon member alpha-crystallin protein, hspX, was upregulated 3.6-fold (P = 0.004). Evidence has suggested that HspX is an important protein in the pathogenesis of M. tuberculosis. The hspX gene has been found to be upregulated during the stationary phase (37), in several models of dormancy (23, 37), and during the growth of M. tuberculosis in macrophages (30), mice (31), and lung specimens from patients with TB (34). Recent work has demonstrated that HspX has a role in slowing the growth of M. tuberculosis both in vitro and in vivo (14). An hspX gene replacement mutant of M. tuberculosis exhibited increased growth in macrophages and also in a mouse model, whereas overexpression of the hspX gene resulted in a reduction in the growth rate of M. tuberculosis in vitro (14, 39). The demonstration that expression of hspX was induced during slow growth in a carbon-limited chemostat provides further evidence that this chaperone plays a role in reducing the growth rate of M. tuberculosis.
(vi) IS elements, repeated sequences, and phage.
The expression of IS elements was significantly elevated at a low growth rate, and 11 genes were upregulated. Expression of four of these genes was also induced during the stationary phase of growth and in Wayne's model of nonreplicating persistence (37).
(vii) PE and PPE families.
We observed significant induction of expression of the antigenic PE and PPE family of genes at a low growth rate under carbon-limiting conditions. One of these genes, Rv0834c, has been shown to be differentially expressed in the host cell (35). Localized at the cell surface, PE and PPE proteins participate in cell surface interactions and may be involved in immune evasion.
Comparisons of the chemostat model with other in vitro models.
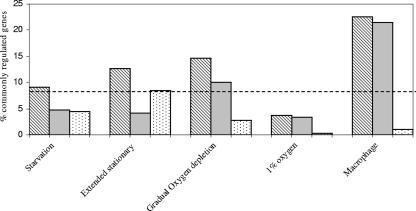
The transcriptional response to the low growth rate in the chemostat was compared to the expression profiles obtained for several in vitro models, including (i) a PBS starvation model (4), (ii) an extended stationary-phase model (11), and (iii) Wayne's model of persistence (23). These models were selected as they all use nutrient and/or oxygen limitation to induce low growth rates in M. tuberculosis. Wayne's model was developed to investigate the effects of hypoxic conditions on mycobacteria and involves cultivating mycobacteria in sealed slowly stirred tubes exposed to a limited volume of air in the headspace. The PBS model has been used to investigate the response of M. tuberculosis to nutrient starvation by resuspending mycobacterial cultures in PBS. During the extended stationary-phase model of TB, M. tuberculosis was cultivated in a bioreactor for 100 days. The data were also compared to the data obtained with another chemostat model investigating the effects of 1% oxygen on M. tuberculosis at a constant growth rate (2). In addition, in order to establish whether the requirements for survival in the chemostat model had any similarities to the requirements for growth in vivo, gene expression data for M. tuberculosis growing in activated macrophages (after 24 h of infection) were also compared with the data obtained in this study (30). The data were compared by downloading lists of genes whose expression was significantly altered in the models according to the criteria of the original researchers that were provided in supplemental material. The extraction of comparable data from the publications was complicated by the different formats used for presenting the data and also by differences in the criteria used for including genes in the lists, and therefore the analysis shown in Fig. 2 is only a preliminary analysis. The problems associated with comparing DNA expression data obtained in different studies have been discussed by Kendall et al. (16).
FIG. 2.
Comparison of the gene expression profiles obtained with the chemostat model of persistence and in other microarray studies. The striped bars indicate the percentages of genes significantly regulated in response to slow growth in the chemostat that were also significantly regulated in the microarray study. Approximately 9% of the genes were regulated in the chemostat model, so this value represents a baseline for the comparison (if there was no relationship between the paired data sets, then we would expect approximately 9% of genes in any randomly generated list to be also regulated in the chemostat) and is indicated by a dashed line. The gray bars indicate the percentages of genes that were concordantly regulated, whereas the dotted bars indicate the percentages of genes that were discordantly regulated. The data were obtained from studies performed by Betts et al. (4) (starvation), Hampshire et al. (11) (extended stationary), Muttucumaru et al. (23) (gradual oxygen depletion), Bacon et al. (2) (1% O2), and Schnappinger et al. (30) (macrophages).
Some similarity was observed between the low-growth rate response studied here and the results of the transcriptome study of Wayne's model of persistence reported by Muttucumaru et al. (23). The common response in this case was likely to be a response to slow growth in both models rather than to oxygen limitation, since there was very little correlation between the transcriptional profile of BCG in the chemostat model of TB and the response of M. tuberculosis to oxygen limitation in a chemostat (2).
Comparison of the chemostat model with adaptation to macrophage growth.
The transcriptional response to the low growth rate in the chemostat was compared to expression profiles for M. tuberculosis obtained from infected macrophages. Surprisingly, 76 genes whose expression was altered in M. tuberculosis after 24 h of macrophage infection were similarly regulated in BCG growing in the chemostat model (30). These results provided evidence that the chemostat model provides at least some of the conditions experienced by M. tuberculosis while it is growing inside the host.
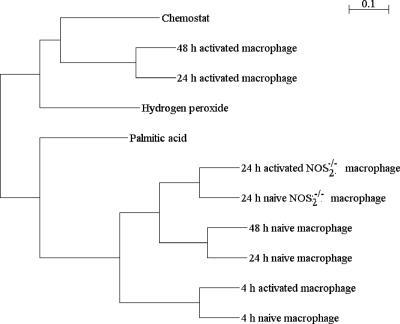
To determine the level of similarity, a cluster analysis was performed using the Pearson's algorithm to examine the correlation between transcriptomic profiles for the chemostat-grown BCG and the data sets described by Schnappinger et al. (30), including data for macrophage-adapted M. tuberculosis cells (naïve, activated, and nitric oxide synthase 2 [NOS2] knockout macrophages) and data for two in vitro conditions (hydrogen peroxide treatment and growth in palmitic acid medium). The clustering algorithm separated the normalized data into two distinct clusters (Fig. 3). The first cluster included data from the chemostat model described here and for activated macrophages after 24 and 48 h of infection and after hydrogen peroxide treatment, while all the other data formed a second cluster. Within the first cluster the chemostat and activated macrophage data sets were the most closely related. The clustering of the chemostat model data and the data obtained after hydrogen peroxide treatment is probably a reflection of the reduction in the growth rate induced by these treatments. The second cluster contained all the naïve macrophage data, the data for activated macrophages after 4 h of infection, and the NOS2 knockout macrophage data.
FIG. 3.
Hierarchical clustering of the transcriptional responses of M. bovis BCG growing in the chemostat model of TB and macrophage-adapted M. tuberculosis (naïve, activated, and NOS2 knockout macrophages) and also after hydrogen treatment and growth in palmitic acid (30). Expression data were clustered using the TMeV data analysis software with a Pearson correlation analysis (8, 29).
To further evaluate the correlation between the chemostat and macrophage data and to determine the level of correlated gene expression change, chi-square test significance values were calculated for the chemostat data and all of the data sets described by Schnappinger et al. (30). This analysis was performed by identifying genes as up- or downregulated or unchanged using three thresholds (2-fold, 1.6-fold, and 1.2-fold). The chi-square test was then used to determine whether the distribution of upregulated, downregulated, and unchanged genes in the paired data sets (e.g., macrophages and chemostat) was compatible with the null hypothesis that the levels of gene expression were independently distributed. High levels of correlation (low P values) were obtained for the chemostat data and macrophage data (30) for all fold changes tested (Table 2). With the 1.2-fold threshold, the macrophage data were most highly correlated with the chemostat data. However, when the twofold cutoff was used, the highest correlation with the macrophage data was the correlation with the hydrogen peroxide-treated cells. Together, these findings indicate that the common regulatory changes for the macrophages and slow growth in the chemostat involve a large set of genes but only small differences in gene expression (that are not observed when the twofold threshold is used), whereas the correlated changes for the macrophages and the hydrogen peroxide treatment involve a smaller set of genes but larger differences (more than twofold) in gene expression (Table 2).
TABLE 2.
Chi-square test significance valuesa
| Comparisonb | Fold change | P value |
|---|---|---|
| Chemostat data vs 48-h activated | 1.2 | 8.25 × 10−138 |
| macrophage data | 1.6 | 1.49 × 10−44 |
| 2.0 | 8.55 × 10−23 | |
| 48-h activated macrophage data vs | 1.2 | 1.91 × 10−13 |
| hydrogen peroxide data | 1.6 | 1.72 × 10−49 |
| 2.0 | 2.05. × 10−80 | |
| 48-h activated macrophage data vs | 1.2 | 2.23 × 10−8 |
| palmitic acid data | 1.6 | 1.41 × 10−16 |
| 2.0 | 4.78 × 10−15 |
The chi-square test was used to test the null hypothesis that the genes were independently distributed in the up- and downregulated categories.
Data for 48-h activated macrophages, hydrogen peroxide, and palmitic acid were obtained from reference 30.
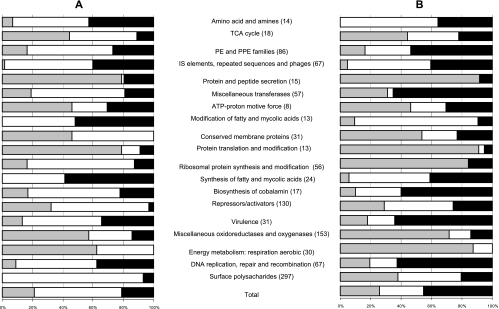
Simple extraction of gene lists based on fold changes and significance values is a useful way to reveal gross changes in gene expression. However, this strategy may miss important information that can be obtained from more subtle examination of genome-wide changes in the expression profile. Several methods have been developed to identify sets of functionally related genes that are significantly regulated in a particular data set, such as gene set enrichment analysis (22), GO::TermFinder (5), and related methods (1). The basic principle of these methods is that Gene Ontology terms are used to label genes, which are then ranked according to some measure of gene expression (gene expression level or significance), and the resulting ranking order is examined to identify terms which are not uniformly distributed throughout the list. Unfortunately, Gene Ontology is not currently available for M. tuberculosis, and so we developed a related approach that does not require Gene Ontology terms. As part of the genome sequencing project (http://www.sanger.ac.uk/Projects/M_tuberculosis/Gene_list/), all M. tuberculosis genes have previously been divided into 101 functional groups based on a gene ontology determined for E. coli (27). To identify the common functional components that characterize the response of M. tuberculosis and M. bovis BCG to the macrophage environment and slow growth in the chemostat, each data set (macrophages and chemostat) was first ranked to identify the mostly significantly regulated functional groups (using the 1.2-fold threshold that generated the highest level of correlation, as shown in Table 2). To do this, a chi-square test was used to calculate the probability that the distribution of up- and downregulated genes within each functional group was consistent with the null hypothesis that the group represented a random sample of the total population of genes in the data set. The functional categories were then ranked using the calculated significance values for rejection of the null hypothesis. The similarity of the resulting ranking orders (data not shown) indicated that the most significantly regulated groups were obtained for both the macrophages and the chemostat. To investigate whether regulatory changes were in the same direction or in opposite directions, the proportions of genes that were upregulated, downregulated, and unchanged were plotted (Fig. 4) for the most significantly regulated functional categories in each data set. The resulting plot provides a novel system-level comparison of the transcriptional response of the tubercle bacillus to slow growth in the chemostat and to growth inside a macrophage. The similar plots obtained indicated that the responses of the tubercle bacillus to slow growth in the chemostat and to adaptation to the macrophage environment (using the 1.2-fold cutoff) involve concordant changes in similar sets of functionally related genes. Concordance was observed particularly among genes involved in the tricarboxylic acid cycle (downregulated), aerobic respiration (downregulated), ATP proton motive force (downregulated), protein translation and modification (downregulated), protein and peptide secretion (downregulated), DNA replication and repair (upregulated), and IS elements (upregulated). Some important differences were also found. For example, the surface polysaccharides were downregulated in the macrophages but mostly unchanged in the chemostat, and the group of repressors and activators was not significantly changed in the chemostat but was upregulated in the macrophages. The results clearly demonstrate that a significant component of the adaptation of the tubercle bacillus to survival inside a macrophage is the global gene expression changes that are associated with adaptation to a low growth rate, which can be studied in a chemostat.
FIG. 4.
Proportions of upregulated, downregulated, and unchanged genes during growth in the chemostat model (A) and after 24 h in activated macrophages (B) in functional groups that were significantly regulated (P < 0.05, as determined by a chi-square test) in one or both of the data sets. Numbers in parentheses are the numbers of genes in the functional groups. The macrophage data were obtained from the study of Schnappinger et al. (30). The percentages of downregulated genes are indicated by gray bars, the solid bars indicate the percentages of upregulated genes, and the open bars indicate the percentages of genes that were unchanged in each functional group. The gene ontology was obtained from http://www.sanger.ac.uk/projects/M_tuberculosis/Gene_list/. TCA, tricarboxylic acid.
Conclusions.
The transcriptional response of M. bovis BCG to changes in the growth rate in carbon-limited chemostats was analyzed. In this system the BCG cells actively grew at suboptimal rates and in nutrient-limited conditions. This system, therefore, differed from other in vitro models of mycobacterial growth in that the bacteria were neither starving nor feasting, and therefore, the emphasis was on the transcriptional consequences of strict carbon limitation in actively growing but “hungry” cells. This model also has significant advantages over the batch models in that bacteria are in a defined and constant environment, allowing the growth rate to be investigated independent of any other environmental parameter.
There was some overlap between the transcriptional responses of M. bovis BCG in the chemostat model and the responses reported for M. tuberculosis in previously described models of persistence, and therefore, low growth rates and carbon limitation are likely to be common features of all these models (4). A high level of correlated gene expression was observed for BCG growing in the chemostat model and M. tuberculosis growing in macrophages (30), demonstrating that a significant proportion of the gene regulatory changes observed for the adaptation of mycobacteria to survival in the macrophages can be accounted for by adaptation of the bacillus to slow growth in carbon-limited conditions, as measured in the chemostat.
Supplementary Material
Acknowledgments
This work was supported by European Union grant ICA4-CT-2002-10063 and by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) (grant BB/D007208/1).
We thank the Wellcome Trust-funded multicollaborative microbial pathogen microarray group at St. George's Hospital Medical School in London, United Kingdom, for providing M. tuberculosis microarrays. We also thank G. R. Stewart for reading the manuscript and for his useful suggestions.
Footnotes
Published ahead of print on 23 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Al Shahrour, F., R. Diaz-Uriarte, and J. Dopazo. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20:578-580. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, J., B. W. James, L. Wernisch, A. Williams, K. A. Morley, G. J. Hatch, J. A. Mangan, J. Hinds, N. G. Stoker, P. D. Butcher, and P. D. Marsh. 2004. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis 84:205-217. [DOI] [PubMed] [Google Scholar]
- 3.Beste, D. J. V., J. Peters, T. Hooper, C. Avignone-Rossa, M. E. Bushell, and J. McFadden. 2005. Compiling a molecular inventory for Mycobacterium bovis BCG at two growth rates: evidence for growth rate-mediated regulation of ribosome biosynthesis and lipid metabolism. J. Bacteriol. 187:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, E. I., S. A. Weng, J. Gollub, H. Jin, D. Botstein, J. M. Cherry, and G. Sherlock. 2004. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daran-Lapujade, P., M. L. A. Jansen, J. M. Daran, W. van Gulik, J. H. de Winde, and J. T. Pronk. 2003. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae, a chemostat culture study. J. Biol. Chem. 10:9125-9138. [DOI] [PubMed] [Google Scholar]
- 7.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 8.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentleman, R., V. Carey, D. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampshire, T., S. Soneji, J. Bacon, B. W. James, J. Hinds, K. Laing, R. A. Stabler, P. D. Marsh, and P. D. Butcher. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis 84:228-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes, A., N. Zhang, J. Wu, P. R. Butler, N. C. Hauser, J. D. Hoheisel, F. L. Lim, A. D. Sharrocks, and S. G. Oliver. 2002. Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26:281-290. [DOI] [PubMed] [Google Scholar]
- 13.Hoskisson, P. A., and G. Hobbs. 2005. Continuous culture—making a comeback? Microbiology 151:3153-3159. [DOI] [PubMed] [Google Scholar]
- 14.Hu, Y. M., F. Movahedzadeh, N. G. Stoker, and A. R. M. Coates. 2006. Deletion of the Mycobacterium tuberculosis alpha-crystallin-like hspX gene causes increased bacterial growth in vivo. Infect. Immun. 74:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall, S. L., F. Movahedzadeh, S. C. G. Rison, L. Wernisch, T. Parish, K. Duncan, J. C. Betts, and N. G. Stoker. 2004. The Mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis 84:247-255. [DOI] [PubMed] [Google Scholar]
- 16.Kendall, S. L., S. C. G. Rison, F. Movahedzadeh, R. Frita, and N. G. Stoker. 2004. What do microarrays really tell us about M. tuberculosis? Trends Microbiol. 12:537-544. [DOI] [PubMed] [Google Scholar]
- 17.Kolkman, A., M. M. A. Olsthoorn, C. E. M. Heeremans, A. J. R. Heck, and M. Slijper. 2005. Comparative proteome analysis of Saccharomyces cerevisiae grown in chemostat cultures limited for glucose or ethanol. Mol. Cell. Proteomics 4:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Lillebaek, T., A. Dirksen, I. Baess, B. Strunge, V. Thomsen, and A. B. Andersen. 2002. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J. Infect. Dis. 185:401-404. [DOI] [PubMed] [Google Scholar]
- 19.Maguire, B. A., and D. G. Wild. 1997. The effects of mutations in the rpmB,G operon of Escherichia coli on ribosome assembly and ribosomal protein synthesis. Biochim. Biophys. Acta 1353:137-147. [DOI] [PubMed] [Google Scholar]
- 20.Mangan, J. A., I. M. Monahan, and P. D. Butcher. 2002. Gene expression during host-pathogen interactions: approaches to bacterial mRNA extraction and labelling for microarray analysis, p. 137-151. In B. W. Wren and N. Dorrell (ed.), Functional microbial genomics. Academic Press, London, United Kingdom.
- 21.Mayuri, G. Bagchi, T. K. Das, and J. S. Tyagi. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol. Lett. 211:231-237. [DOI] [PubMed] [Google Scholar]
- 22.Mootha, V. K., C. M. Lindgren, K. F. Eriksson, A. Subramanian, S. Sihag, J. Lehar, P. Puigserver, E. Carlsson, M. Ridderstrale, E. Laurila, N. Houstis, M. J. Daly, N. Patterson, J. P. Mesirov, T. R. Golub, P. Tamayo, B. Spiegelman, E. S. Lander, J. N. Hirschhorn, D. Altshuler, and L. C. Groop. 2003. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267-273. [DOI] [PubMed] [Google Scholar]
- 23.Muttucumaru, D. G. N., G. Roberts, J. Hinds, R. A. Stabler, and T. Parish. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 84:239-246. [DOI] [PubMed] [Google Scholar]
- 24.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips, A., T. Bullock, and N. Plant. 2003. Sodium valproate induces apoptosis in the rat hepatoma cell line, FaO. Toxicology 192:219-227. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 27.Riley, M. 1993. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57:862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and biologist programmers, p. 365-386. In Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 29.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 30.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi, L., Y. J. Jung, S. Tyagi, M. L. Gennaro, and R. J. North. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA 100:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Syst. Appl. Genet. Mol. Biol. 3:Article 3. [DOI] [PubMed]
- 33.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 34.Timm, J., F. A. Post, L. G. Bekker, G. B. Walther, H. C. Wainwright, R. Manganelli, W. T. Chan, L. Tsenova, B. Gold, I. Smith, G. Kaplan, and J. D. McKinney. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA 100:14321-14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triccas, J. A., F. X. Berthet, V. Pelicic, and B. Gicquel. 1999. Use of fluorescence induction and sucrose counterselection to identify Mycobacterium tuberculosis genes expressed within host cells. Microbiology 145:2923-2930. [DOI] [PubMed] [Google Scholar]
- 36.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voskuil, M. I., K. C. Visconti, and G. K. Schoolnik. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84:218-227. [DOI] [PubMed] [Google Scholar]
- 38.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 39.Yuan, Y., D. D. Crane, and C. E. Barry. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial alpha-crystallin homolog. J. Bacteriol. 178:4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.