Abstract
It has long been suspected that microbial biofilms harbor cells in a variety of activity states, but there have been few direct experimental visualizations of this physiological heterogeneity. Spatial patterns of DNA replication and protein synthetic activity were imaged and quantified in staphylococcal biofilms using immunofluorescent detection of pulse-labeled DNA and also an inducible green fluorescent protein (GFP) construct. Stratified patterns of DNA synthetic and protein synthetic activity were observed in all three biofilm systems to which the techniques were applied. In a colony biofilm system, the dimensions of the zone of anabolism at the air interface ranged from 16 to 38 μm and corresponded with the depth of oxygen penetration measured with a microelectrode. A second zone of activity was observed along the nutrient interface of the biofilm. Much of the biofilm was anabolically inactive. Since dead cells constituted only 10% of the biofilm population, most of the inactive cells in the biofilm were still viable. Collectively, these results suggest that staphylococcal biofilms contain cells in at least four distinct states: growing aerobically, growing fermentatively, dead, and dormant. The variety of activity states represented in a biofilm may contribute to the special ecology and tolerance to antimicrobial agents of biofilms.
Bacteria or yeast that adhere to implanted medical devices or damaged tissue can become the focus of persistent infections (11). These microorganisms encase themselves in an extracellular polymeric matrix forming a slimy layer known as a biofilm. Biofilm infections develop gradually and may be slow to produce overt symptoms. Once established, however, these infections become chronic. The notorious tolerance of microorganisms to killing by antimicrobial agents when in biofilms contributes to the persistence of these infections (27, 28). Staphylococci are among the most common pathogens associated with device-related infections (3, 9).
One hypothesis to explain the antimicrobial tolerance manifested in biofilms is that biofilms harbor cells that are growing slowly or not at all (8, 32). Since most antibiotics target macromolecule synthesis processes, it is plausible that nongrowing cells could be less susceptible to killing. Metabolically inactive bacteria could also be less vulnerable to other types of chemical and physical challenges. A biofilm might contain not only rapidly growing cells, but also cells that are growing slowly, cells that have entered a stationary-phase-like state, and cells that are completely dormant. Some cells might sustain basal metabolic activity and limited gene expression even though they are not growing. A biofilm could contain some bacteria growing aerobically and others growing fermentatively. If this conjecture is correct, even a single biofilm cell cluster could comprise a population spanning a spectrum of physiological states. These different states may be due to local variation in the availability of nutrients and electron acceptors or mechanisms such as phenotypic variation.
There is some evidence for substrate limitation and slow growth in staphylococcal biofilms. The average specific growth rate of bacteria in a biofilm can be calculated by dividing the overall rate of production of bacteria by the standing population of cells in the biofilm. Using this approach, Hodgson et al. (14) found that Staphylococcus aureus grew with a specific growth rate of 0.06 h−1 in biofilms, whereas the bacterium grew with a specific growth rate of 0.7 h−1 in the planktonic state in the same medium. The community-averaged growth rate of Staphylococcus epidermidis in colony biofilms was 0.035 h−1, much lower than the maximum specific growth rate of 0.82 h−1 measured in batch culture (39). These results suggest that the average growth rate in a staphylococcal biofilm may be only a few percent of the growth rate exhibited in conventional suspended-culture conditions. DNA microarray comparisons of transcriptional patterns in biofilms and planktonic cells reveal a general down-regulation of transcription, translation, and aerobic energy production (36) and up-regulation of anaerobic metabolism (6, 23).
A limitation of all of the preceding studies is that they reveal none of the heterogeneity in growth state and activity that is probably present in the biofilm population. These studies provide only whole-population averages. Consider that a biofilm in which the average specific growth rate is half the growth rate of planktonic cells might actually consist of a population of cells in which half are growing rapidly and half not at all. The average does not represent accurately either of the subpopulations. In this article, we report the integrated application of techniques to map spatial patterns of DNA synthesis, protein synthesis, and local oxygen concentration that enable the visualization and quantification of highly heterogeneous patterns of respiratory and anabolic activity in S. epidermidis and S. aureus biofilms.
MATERIALS AND METHODS
Bacteria and media.
S. epidermidis strain RP62A (ATCC 35984) and S. aureus strain RN6390 containing pALC2073::gfpuvr (ALC2085) were grown on tryptic soy broth (TSB) at 37°C. The green fluorescent protein (GFP) gene contained in this S. aureus strain was under the control of a tetracycline-inducible promoter (5). Engineering of gfpuvr is described elsewhere (16). Full-strength TSB was used to grow shake flask cultures that provided the inoculum for biofilm experiments.
Colony biofilm preparation.
Colony biofilms were grown on polycarbonate membranes resting on agar plates (31). Planktonic cultures of S. epidermidis and S. aureus were grown overnight in TSB and diluted to an optical density of 0.050 at 600 nm (with a 1-cm path length). Chloramphenicol (10 μg/ml) was added to the medium used to grow the S. aureus inoculum to ensure retention of the plasmid. Chloramphenicol was not included in the agar medium used to grow colony biofilms. One 10-μl drop of diluted planktonic culture was used to inoculate membranes (25-mm, 0.22-μm pore size; Osmonics, Inc., Minnetonka, MN) resting on tryptic soy agar (TSA) plates. The membranes were sterilized by UV exposure (10 min per side) prior to inoculation. The plates were inverted and incubated at 37°C, and the membranes were transferred to a fresh TSA plate every 24 h and were grown for 44 h. For the S. epidermidis strain, the biofilms were transferred to agar medium containing 1 mM 5-bromo-2-deoxyuridine (BrdU) for an additional 4 h. For the S. aureus strain, the biofilms were transferred to agar medium containing 1 μg/ml tetracycline for an additional 4 h.
Colony biofilms were subjected to various nutrient and oxygen conditions during the introduction of BrdU. For anaerobic growth experiments, agar plates were transferred into an anaerobic incubation pouch with a sealing bar (BBL GasPak Pouch, Becton, Dickinson and Company, Sparks, MD) in an anaerobic glove box (Coy Laboratory Products, Inc., Ann Arbor, MI). In some experiments, TSA was supplemented with glucose at 2.5 g/liter. To supply additional oxygen, agar plates were placed in a bag that was inflated with pure oxygen and sealed.
Planktonic culture preparation.
Planktonic batch cultures of S. epidermidis were grown overnight at 37°C in baffled flasks in TSB. BrdU (1 mM) was added to fresh medium inoculated with a 50-fold dilution of overnight culture after 1.5 h of growth for exponential-phase experiments and after 7 h of growth for stationary-phase experiments. For a killed control experiment, an overnight culture was autoclaved for 20 min prior to the introduction of BrdU. Planktonic cultures were grown for a total of 11 h prior to filtration onto a polycarbonate membrane using a filtration unit.
Capillary biofilm system.
Biofilms were grown in glass capillary tubes (Friedrich and Dimmock, Millville, NJ) under continuous flow conditions (22, 33). The glass tubes had square cross sections which allowed direct microscopic observation of the biofilm through the tube walls. The capillary tubes had a nominal inside dimension of 0.9 mm and were approximately 10 cm long. Capillaries were inoculated with 2 ml of overnight cultures of S. epidermidis or S. aureus, and flow was stopped for 4 h. Chloramphenicol (10 μg/ml) was added to the medium used to grow the S. aureus inoculum. No antibiotic was included in the medium used to grow the capillary biofilm. After 4 h of static incubation, the flow of medium (1/10-strength TSB) was initiated at a flow rate of 120 ml/h by gravity feed. The medium carboy and the capillary were placed in separate 37°C incubators stacked on top of each other. Biofilms were allowed to develop for 20 h prior to the introduction of medium containing 1 mM BrdU for an additional 4 h or 1 μg/ml tetracycline for an additional 7 h. For microscopic observation, capillary biofilm was placed in a holder (Biosurface Technologies, Bozeman, MT) that could be mounted onto the microscope stage.
Drip-flow biofilm system.
Drip-flow biofilm reactors were used to cultivate biofilms (15, 34). Stainless steel coupons (7.8 cm by 1.2 cm) were inclined at a 10° angle within the reactor, which was incubated at 37°C. After inoculation with an overnight culture of S. epidermidis or S. aureus, the reactor was allowed to stand for 24 h. Chloramphenicol (10 μg/ml) was added to the medium used to grow the S. aureus inoculum. No antibiotic was included in the medium used to grow the S. aureus drip-flow biofilm. After this time, the flow was initiated and the medium (1/10-strength TSB) dripped onto the coupons from a height of approximately 1 cm. For the S. epidermidis strain, the reactor was fed for 20 h at a flow rate of 10 ml/h before switching to medium containing 1 mM BrdU for an additional 4 h. For the S. aureus strain, the reactor was fed for 44 h at a flow rate of 10 ml/h before switching to medium containing 1 μg/ml tetracycline for an additional 4 h.
Cryoembedding and cryosectioning.
Colony biofilm, drip-flow biofilm, and planktonic culture samples were cryoembedded with Tissue-Tek O.C.T. compound (VWR Scientific Products, West Chester, PA) as described previously (31, 32). Membrane-supported biofilms were placed on a 7.8- by 1.2-cm stainless steel coupon, and the colonies were covered with the embedding medium. The coupon was placed on dry ice, and the medium was allowed to freeze. Colonies along with the membranes were removed from the coupon, and the edges of the membranes were trimmed. The colonies with the embedded side down were placed on the dry ice, and the embedding medium was used to cover the exposed membranes, which was then allowed to freeze. Embedded colonies were sectioned into 5-μm-thick cross sections using a Leica CM 1850 cryostat. The colony cross sections were placed on Superfrost Plus microscope slides (Fisher Scientific).
DNA synthetic activity.
The BrdU technique described in this paper was a modification of a published protocol (12). S. epidermidis colony cross sections were fixed for 5 min using a solution containing 80% ethanol in water. This fixation process was to stop metabolic activity and fix the sections onto the slides.
A solution of 50 mg/liter lysozyme was introduced for 5 min to permeabilize the cell envelope. Subsequently, 4 N hydrochloric acid was introduced for 10 min to denature DNA, as this protocol only recognizes single-stranded DNA. A blocking solution of 2% skim milk and 0.3% Tween 80 in Tris-buffered saline was introduced for 5 min to limit nonspecific binding of antibody.
A 20-fold dilution factor of anti-BrdU, mouse immunoglobulin G1, monoclonal PRB-1, and Alexa Fluor 488 conjugate (anti-BrdU, Alexa Fluor 488 conjugate) was introduced for 1 h, which illuminated regions of active replication in green fluorescence. Slides were covered to protect from photobleaching the antibody. Sections were probed with the antibody for 1 h. Sections were counterstained with 5 mg/liter rhodamine B for 5 min (Eastman Organic Chemicals, Rochester, NY) to reveal in red fluorescence the extent of the biomass independent of its activity. Sections were stained by placing 20-μl drops in succession along the cross section. After each step, slides were rinsed twice with phosphate-buffered saline (PBS) with excess solution blotted from the slide. Slides were fixed and stained at room temperature.
After introduction of BrdU for 4 h, glass capillary biofilms were processed with the same series of reagents detailed above and directly observed by confocal scanning laser microscopy (CSLM). Experiments were also conducted in which a capillary biofilm was labeled with BrdU for 4 h, cryoembedded, cryosectioned, and stained prior to observation by CSLM.
Drip-flow biofilms were labeled with BrdU for 4 h, cryoembedded, cryosectioned, stained, and then examined by CSLM.
Protein synthetic activity.
S. aureus colony biofilms were grown for 44 h, transferred to a TSA plate containing 1 μg/ml tetracycline for 4 h, cryoembedded, and cryosectioned into 5-μm slices using a Leica CM 1850 cryostat.
Glass capillary biofilms were observed with time-lapse CSLM to obtain a time series of GFP expression during induction with tetracyline.
Drip-flow biofilms were induced with 1 μg/ml tetracycline for 4 h, cryoembedded, cryosectioned, and then examined by CSLM.
Some experiments were performed in which both BrdU and tetracycline were introduced simultaneously to image DNA and protein synthesis in the same S. aureus colony. Colony biofilms were grown for 44 h and were then transferred to a TSA plate containing 10 mM BrdU and 1 μg/ml tetracycline. Images of frozen sections were first obtained in the green channel for GFP expression. The slides were then heat treated followed by the fixation and staining protocol to image DNA synthetic activity. A second set of images was taken to image DNA replication, which was also obtained in the green channel. Images were realigned and combined to view the combined activities. Since protein synthesis and DNA replication were both initially imaged in the green channel, DNA replication images were false-colored in red.
Oxygen concentration profiles and penetration.
Oxygen concentration profiles in colony biofilms were measured with a dissolved oxygen microelectrode as described elsewhere (24, 31). Dissolved oxygen microelectrodes are amperometric devices: oxygen diffuses through a silicone rubber membrane, arrives at a cathodically polarized working electrode, and is reduced to water. The microelectrode uses an Ag/AgCl half-cell as the counterelectrode and gold as the working electrode. Applying −0.8 V typically satisfies the limiting current condition, and the measured current is proportional to the dissolved oxygen concentration in the vicinity of the microelectrode tip. The oxygen microelectrodes were calibrated in water equilibrated with nitrogen and in air at 37°C.
To measure oxygen profiles, the microelectrode was mounted on a micromanipulator (model M3301L; World Precision Instruments, New Haven, CT) equipped with a stepper motor (model 18503; Oriel, Stratford, CT) controlled by an Oriel model 20010 interface. The microelectrode was introduced from the top of the plate perpendicular to the biofilm. The micromanipulator was interfaced with a computer, and the microelectrode movement was facilitated by a controller (CTC-283-3; Micro Kinetics) with a positioning precision of 0.1 μm. Custom software was used to control and coordinate microelectrode movement, data acquisition, and real-time display of the concentration profile.
The local reaction rate of oxygen was calculated from measured oxygen concentration profiles by evaluating the second derivative with a central difference formula. The width of the respiratory zone was taken as the distance from the biofilm-air interface to the distal edge of the oxygen reaction rate peak as measured at the peak half-height.
Plasmid stability.
S. aureus biofilm was tested for the stability of the GFP-bearing plasmid. A biofilm-covered membrane was aseptically removed from the agar plate and was placed in a test tube containing 9 ml phosphate buffer solution. The test tube was vortexed for 5 min to remove cells from the polycarbonate membrane. The resulting suspension was serially diluted in PBS before spread plating. Dispersed cells were spread plated on a TSA plate and a TSA plate supplemented with 10 μg/ml chloramphenicol. Agar plates were inverted and incubated at 37°C. CFU in both plates were determined, and the experiment was repeated twice. A sterile toothpick was used to take a single dab from one CFU from the TSA spread plate and was dabbed onto both the TSA plate and the TSA plate containing 10 μg/ml chloramphenicol. Agar plates were inverted and incubated at 37°C. Up to 100 CFU (using a sterile toothpick each time) was used to conduct the colony screening experiment. CFU in both plates were compared.
Microscopy and image analysis.
CSLM was performed with a Leica TCS NT confocal scanning laser microscope (33). For imaging in transmission mode, excitation from a 488-nm laser was used. For imaging DNA synthesis and GFP expression, a 488-nm laser was used and the emission was collected at 500 to 530 nm (green channel). For imaging rhodamine B, a 568-laser was used and the emission was collected at 585 to 615 nm (red channel). A ×10 dry objective lens was used for these experiments. Microscope images were analyzed using the linescan function in MetaMorph image analysis software (Universal Imaging Corporation, Downingtown, PA). For colony and drip-flow biofilm experiments, the relative fluorescent intensities were measured for at least three independently grown biofilms and for six to eight independent sections across each biofilm. For each image taken, fluorescent intensities were measured at three random locations, resulting in a total of between 54 and 72 data points per condition. Intensity profiles were measured perpendicular to the membrane supporting the colony biofilm. The dimension of active zones of DNA synthesis or protein synthesis was taken as the width of a peak at the peak half-height. Statistical comparisons were made using a two-sided t test assuming unequal variances. The biofilm thickness was measured as the distance from the membrane to the biofilm-air interface. For the S. aureus capillary biofilm experiment, a time series was initiated in which an image was collected every 2 min. The time series ran for 4 to 7 h.
Respirometry.
The overall rate of oxygen consumption in colony biofilms was measured using a Micro-Oxymax closed circuit respirometer (Columbus Instruments, Columbus, OH). Seven colony biofilms in nutrient agar bottles were attached to the respirometry equipment. A set of controls was performed in which the nutrient agar bottle did not contain a colony biofilm. Measurements of the rate of oxygen consumption were obtained at 1.5-h intervals for a total of 48 h. The rate of oxygen consumption arrived at a transient plateau between 15 h and 20 h, corresponding to the probable onset of mass transfer limitation of oxygen supply to the biofilm. The average rate of oxygen consumption between 15 h and 20 h was calculated from the replicate samples. Other colony biofilms were measured to determine their radii and also frozen and sectioned to determine thickness. The colony area was computed by treating the colony as circular. The thickness of colonies was determined by measuring the average thickness of the cross sections microscopically. The mean rate of oxygen consumption between 15 h and 20 h was divided by the average volume of colony biofilms to estimate the volumetric oxygen reaction rate.
Statistical analysis.
Statistical comparisons were made using a two-sided t test assuming unequal variances.
RESULTS
Mapping patterns of DNA synthesis in biofilms.
BrdU labeling has been used extensively in the study of replication and cell cycle phenomena in eukaryotes (13, 16, 17, 18) and to a lesser extent in the investigation of bacterial activity (4, 20, 21). BrdU is a thymidine analog that is incorporated into the DNA of replicating cells. Cells in which DNA synthesis occurred during the BrdU pulse can be subsequently illuminated using a fluorescently tagged antibody that specifically recognizes brominated DNA.
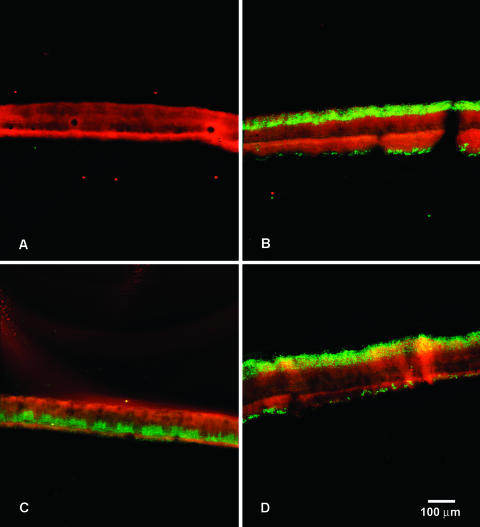
We developed BrdU labeling of staphylococcal biofilms using the colony biofilm system (1, 31, 39). These are dense aggregates of bacteria that build up on a filter membrane resting on a nutrient agar plate. Carbon and nitrogen sources and other required substrates are delivered to the bottom of the biofilm from the agar. Oxygen is delivered to the top of the biofilm via air. After 48 h of development, the mean thickness (and standard deviation) of S. epidermidis colony biofilms was 153 ± 18 μm. Mature colony biofilms were transferred to a BrdU-amended agar plate for 4 h and then frozen, sectioned, and probed with the anti-BrdU antibody. Highly stratified patterns of DNA synthetic activity, representative images of which are shown in Fig. 1, were observed. Colony biofilms not labeled with BrdU and probed with the antibody exhibited no green fluorescence (Fig. 1A). When grown in air, two bands of DNA synthetic activity were evident: one along the air interface of the biofilm and a thinner region along the membrane interface (Fig. 1B). Sometimes there was also a trace of activity in a centrally located stratum. Colony biofilms labeled with BrdU but not probed with the antibody likewise showed no fluorescence (image not shown).
FIG. 1.
Patterns of DNA synthesis in S. epidermidis colony biofilms. Panel A shows an unlabeled (no BrdU) colony biofilm probed with antibody. Panel B shows a colony biofilm labeled with BrdU under aerobic conditions. Panel C shows a colony biofilm labeled with BrdU under anaerobic conditions. Panel D shows a colony biofilm labeled with BrdU in an environment of pure oxygen. Green areas are due to BrdU incorporation into DNA and indicate active replication. Red areas are due to rhodamine B counterstaining that reveals the extent of the biomass independent of its activity. In each image, the membrane interface of the colony is on the bottom and the air interface is on the top.
One thing these experiments demonstrate is that the BrdU was able to diffuse out of the agar, through the membrane, and permeate throughout the colony biofilm during the 4-h labeling pulse. This is evidenced by the incorporation of BrdU into cells at the opposite edge of the biofilm from which this substrate was delivered.
Several planktonic controls were performed by BrdU labeling bacteria in suspension and then filtering the cells onto a membrane. The membrane was processed for antibody probing in the same manner that colony biofilms were. A batch culture of S. epidermidis pulse-labeled with BrdU while in exponential phase exhibited bright green fluorescence, indicating extensive DNA synthetic activity (data not shown). A culture labeled with BrdU in stationary phase exhibited relatively weak green fluorescence, indicating a much lower level of DNA synthetic activity (not shown). Bacteria killed by heat treatment and then labeled with BrdU evidenced no DNA synthetic activity by the immunofluorescence assay (not shown).
Spatial patterns of DNA synthesis are highly stratified in biofilms.
Stratified patterns of DNA synthetic activity were observed in all three of the biofilm systems to which the BrdU labeling technique was applied. In the colony biofilm system, the dimensions of the zones of DNA synthetic activity were quantified by image analysis (Table 1). When grown in air, the zone of DNA synthetic activity at the air interface averaged 31 μm in width and the zone of DNA synthetic activity at the membrane interface averaged 16 μm.
TABLE 1.
Summary of measured dimensions of zones of active DNA synthesis in S. epidermidis colony biofilms and of active protein synthesis and DNA synthesis in S. aureus colony biofilms
| Condition | Dimensions of zone of synthesis (μm)a
|
|||||
|---|---|---|---|---|---|---|
|
S. epidermidis DNA
|
S. aureus
|
|||||
| Gas | Membrane | Protein
|
DNA
|
|||
| Gas | Membrane | Gas | Membrane | |||
| Air | 31 ± 14 | 16 ± 11 | 38 ± 9 | 14 ± 5 | 25 ± 8 | 23 ± 13 |
| Oxygen | 45 ± 13 | 13 ± 7 | 59 ± 9 | 18 ± 6 | NM | NM |
| Anaerobic | BD | 19 ± 11 | BD | 17 ± 5 | NM | NM |
| Air with glucose | NM | NM | 36 ± 8 | 33 ± 10 | NM | NM |
The mean and standard deviation are reported. BD, below detection limit; NM, not measured.
Patterns of DNA synthetic activity in S. epidermidis colony biofilms changed in response to changes in the nutrient and oxygen conditions. When BrdU labeling was performed in an anaerobic environment, the band of DNA synthetic activity along the air interface was eliminated (Fig. 1C). When BrdU labeling was performed in a gaseous environment of pure oxygen, the region of DNA synthetic activity at the gas interface increased in thickness (Fig. 1D). The expansion in the zone of DNA synthetic activity at the gas interface from 31 μm in air to 45 μm in pure oxygen, a factor of 1.5, was statistically significant (P < 0.001). The dimension of the zone of DNA synthetic activity along the membrane interface did not change significantly as the gaseous environment was changed from anaerobic, to air, to pure oxygen (P = 0.37 and P = 0.10 for aerobic to anaerobic and aerobic to pure oxygen comparisons, respectively). When the agar was supplemented with glucose during the BrdU labeling, variable but generally enlarged regions of DNA synthetic activity were observed. These specimens did not lend themselves to image analysis because the activity patterns were so irregular.
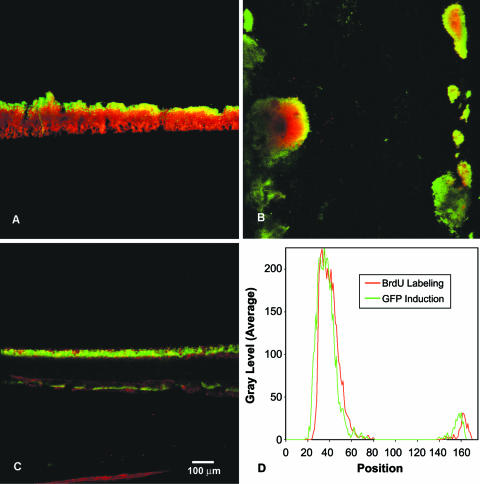
Biofilms grown in a continuous flow system, the drip-flow reactor, demonstrated a stratified pattern of DNA synthetic activity after BrdU pulse-labeling and immunofluorescent detection (Fig. 2A). A single band of bright green fluorescence averaging 39 μm wide and corresponding to active replication was observed along the biofilm-fluid interface.
FIG. 2.
Patterns of DNA synthesis in S. epidermidis drip-flow (A) and capillary (B) biofilms and overlay of DNA synthetic activity and protein synthetic activity in the same S. aureus colony biofilm (C and D). In panels A and B, green areas are due to BrdU labeling and red areas are due to rhodamine B counterstain. In panel A, the substratum was on the bottom and the air interface was on the top. Flow was from top to bottom in panel B. In panel C, green areas are due to GFP expression and red areas are due to BrdU labeling. In panel C, the substratum was on the bottom, with the air interface on the top. Panel D shows the average fluorescence intensity along a linear transect across the section for the two signals. The position indicated on the x axis is arbitrary. The intensities indicated on the y axis of panel D are arbitrary as they result from automatic scaling by the digital image acquisition system.
Biofilms grown under continuous flow conditions in glass capillary tubes were pulse-labeled with BrdU, probed with antibody, and examined in situ by CSLM. Rings of green fluorescence, corresponding to DNA synthetic activity, were observed at the periphery of cell clusters (Fig. 2B). In places where the biofilm was very thin, all of the biomass was bright green, indicating intense DNA synthetic activity. In larger cell clusters, no DNA synthetic activity could be detected in the cluster interior.
We questioned whether these patterns could be an artifact of incomplete permeation of the antibody into the biofilm. Extension of the antibody incubation period to 10 h produced patterns of activity similar to those seen with a 1-h antibody incubation. A second control was performed. A BrdU-labeled capillary biofilm was perfused with a tissue-embedding medium and frozen on dry ice. The glass was chipped away with a triangular file, and the specimen was sectioned. Frozen sections were probed with antibody. Though this procedure perturbs biofilm structure, it yielded stratified patterns qualitatively consistent with those shown in Fig. 2B. These controls show that the zone of DNA synthetic activity observed in capillary reactor biofilms was not limited by penetration of the antibody into cell clusters.
Mapping patterns of protein synthesis in biofilms.
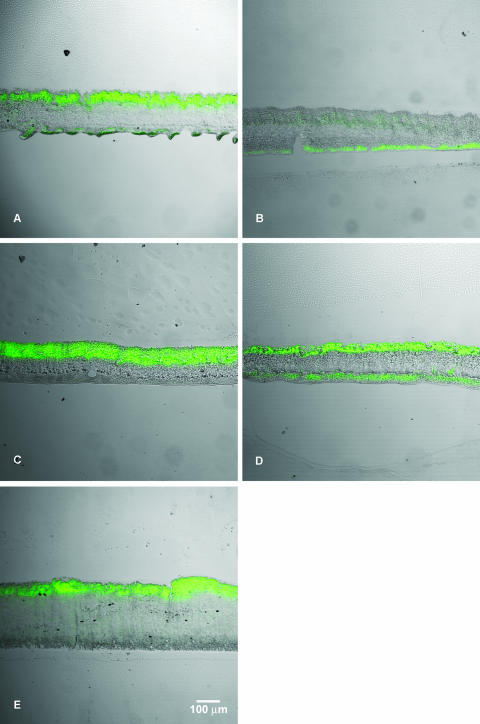
We visualized the cellular capacity for de novo protein synthesis using an S. aureus strain containing a tetracycline-inducible GFP (5). In the presence of subinhibitory concentrations of tetracycline, bacterial cells that can synthesize protein make GFP and can be imaged by fluorescence microscopy. Colony biofilms formed by this strain, then induced with tetracycline for 4 h, generated banded patterns of green fluorescence when cross sections were examined (Fig. 3). The mean thickness (and standard deviation) of 48-h-old S. aureus colony biofilms was 173 ± 22 μm.
FIG. 3.
Patterns of protein synthetic activity (green) imaged in S. aureus colony biofilms induced for GFP expression in air (A), in an anaerobic environment (B), in pure oxygen (C), or in air on medium supplemented with glucose (D) and in a S. aureus drip-flow reactor-grown biofilm (E). The green fluorescent image is overlaid with a transmitted light image of the section to allow visualization of the extent of the biomass independent of activity. In panel E, the substratum is on the bottom and the air interface is on the top.
This inducible GFP construct is plasmid borne, so it is important to establish the stability of the plasmid. Dark regions of a biofilm could be regions in which there is no protein synthesis occurring, or they could just be regions in which most of the bacteria have lost the plasmid. To address this issue, colony biofilms were dispersed and the cell suspension was plated onto TSA and also onto TSA containing 10 μg/ml chloramphenicol. The number of colonies that formed on the antibiotic-containing plates was 94% of the number that grew on TSA plates. One hundred colonies from the TSA plates were picked and applied as spots onto chloramphenicol-containing plates. All 100 colonies grew in the presence of chloramphenicol, indicating that these bacteria contained the plasmid. These results show that the plasmid is retained in most or all of the cells in biofilms.
Bateman et al. (5) previously demonstrated the induction of GFP in planktonic cultures of S. aureus ALC2085. We performed an experiment to demonstrate that even in a stationary-phase culture, this strain was able to express GFP in response to tetracycline. An overnight culture was divided into two flasks, both incubated at 37°C. One flask received tetracycline to give a final concentration of 1 μg ml−1, and the other flask was unamended. The percentage of cells that expressed green fluorescence was scored by analysis in a Becton-Dickinson flow cytometer. The percentage of cells scored as bright green rose from approximately 2% at time zero to approximately 70% after 4 h in the tetracycline-amended culture. The uninduced control showed no change over the same time period.
The inducing agent, tetracycline, was delivered to the colony biofilm from the membrane interface and was able to induce expression of the fluorescent protein along the air interface of a colony biofilm (Fig. 3A). This shows that tetracycline permeated throughout the biofilm.
Spatial patterns of protein synthesis are highly stratified in biofilms.
Stratified patterns of protein synthetic activity were observed in colony biofilms and in biofilms formed in the drip-flow and capillary reactors when the inducible GFP technique was applied. The dimensions of the zones of protein synthetic activity in colony biofilms were quantified by image analysis (Table 1). When grown in air, the zone of protein synthetic activity at the air interface averaged 38 μm in width and the zone of protein synthetic activity at the membrane interface averaged 14 μm.
Patterns of protein synthetic activity in S. aureus colony biofilms measured by GFP induction changed in response to changes in the nutrient and oxygen conditions (Table 1). When tetracycline induction was performed in an anaerobic environment, the band of protein synthetic activity along the air interface was eliminated while the band of GFP expression along the membrane interface was preserved (Fig. 3B). When pure oxygen was introduced into the gas phase during tetracycline induction, the region of protein synthetic activity at the air interface increased in thickness, whereas the zone of GFP expression along the membrane interface was unchanged (Fig. 3C). The green fluorescence along the membrane interface in panel 3C is faint and cannot be discerned in the transmission overlay. The expansion in the zone of protein synthetic activity at the gas interface from 38 μm in air to 59 μm in pure oxygen, a factor of 1.6, was statistically significant (P < 0.001). When tetracycline induction was performed on agar supplemented with glucose, the region of protein synthetic activity at the membrane interface enlarged to 33 μm, whereas there was no significant change in the dimension of the zone of GFP expression at the air interface (Fig. 3D). The expansion in the zone of protein synthetic activity at the membrane interface from 14 μm in medium lacking glucose to 33 μm in medium with added glucose was statistically significant (P < 0.001).
S. aureus colony biofilms were simultaneously labeled with BrdU and induced for GFP expression by adding tetracycline. A sequential imaging approach was used because the protocol for preparing specimens for antibody probing destroys GFP fluorescence. Frozen sections were imaged for GFP fluorescence immediately after sectioning and prior to processing for immunofluorescence. A transmitted light image of the section was also collected. The slide was removed from the microscope and processed for antibody probing. This processing includes treatment with hydrochloric acid, which eliminates all GFP fluorescence. After probing with antibody, the fluorescence pattern corresponding to DNA synthetic activity was imaged. This image was false-colored red to distinguish it from the GFP signal (Fig. 2C). The two fluorescent images were overlaid using image analysis software cueing off of structural features visible in transmitted light images to align them. Stratified patterns of DNA synthetic activity and protein synthetic activity were observed, and these patterns coincided closely with each other (Fig. 2D).
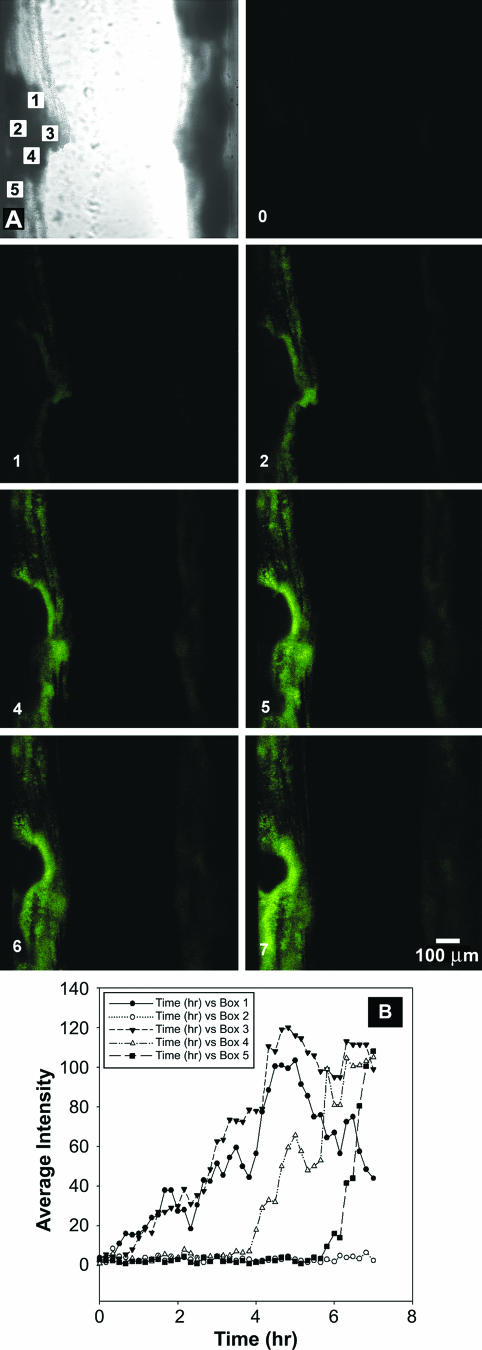
We imaged the transient expression of GFP in S. aureus bioflms grown in capillary reactors (Fig. 4). After the biofilm had been grown for 24 h, tetracycline was added to the medium. Fluorescence appeared first at the surfaces of cell clusters adjoining the fluid flow (Fig. 4). GFP fluorescence progressed inward into the biofilm over time, but did not ever develop in the center of a large cell cluster in the course of the 7-h experiment (Fig. 4). A movie of this experiment can be viewed at http://www.erc.montana.edu/Res-Lib99-SW/Movies/Database/MD_DisplayScript.asp. Image analysis of the green fluorescent intensity at several different locations in the biofilm demonstrates the large variation from spot to spot in the local rate of protein synthesis (Fig. 4B). The movie seems to indicate a slow downstream flow of some of the biomass.
FIG. 4.
Time series of GFP expression in S. aureus biofilm during tetracycline induction in a capillary reactor biofilm. The numeric label in the lower left corner of a panel is the time, in hours, after the first appearance of tetracycline in the flow cell. Green areas are due to GFP expression. A movie of this sequence can be viewed at http://www.erc.montana.edu/Res-Lib99-SW/Movies/Database/MD_DisplayScript.asp. Panel A shows a transmission image of the same region. Flow was from top to bottom. Panel B quantifies the variable dynamics of GFP expression during induction in S. aureus biofilm. Each numbered curve corresponds to a different spot in the biofilm as indicated by the boxes in panel A. The pixel intensity in the area indicated by the box was averaged at each time point.
Biofilms grown in the drip-flow reactor were induced with tetracycline; these also exhibited a stratified pattern of protein synthetic activity (Fig. 3E). A single band of bright green fluorescence, averaging 41 μm in width, was observed at the air interface of the biofilm.
If tetracycline affected cells in aerobic and anaerobic states differently, this could influence the observed patterns of protein synthesis. We measured the MIC of tetracycline against the S. aureus strain containing the inducible GFP under both aerobic and anaerobic conditions. The bacterium was more sensitive to tetracycline under anaerobic conditions (MIC, 0.4 μg/ml) than it was under aerobic conditions (MIC, 2 μg/ml). It is possible then that anaerobic expression of GFP was inhibited by the applied tetracycline (1 μg/ml), whereas expression would not be expected to be affected in aerobic zones. The observation of GFP expression in a colony biofilm induced under anaerobic conditions (Fig. 3B) and in a band along the anoxic nutrient interface of colony biofilms (e.g., Fig. 3D) argues against this possibility. Tetracycline may have inhibited overall growth in anaerobic regions, but it still permitted sufficient protein synthesis to enable GFP to be visualized.
Oxygen is locally depleted in biofilms.
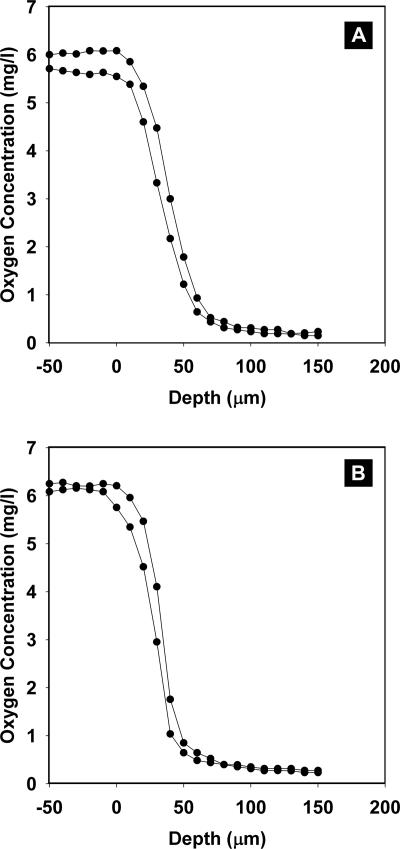
Local oxygen concentrations within staphylococcal colony biofilms were measured with microelectrodes (Fig. 5). Oxygen penetrated approximately 50 μm into biofilms formed by either S. epidermidis (Fig. 5A) or S. aureus (Fig. 5B). These measurements show that the lower two-thirds of these colony biofilms was anoxic. The dimension of the zone of respiration can be determined from these profiles by computing the local second derivative of the oxygen concentration profile. This quantity represents the curvature of the oxygen concentration data and is proportional to the local reaction rate of oxygen. Though derived from the raw microelectrode data by this mathematical analysis, the result is properly regarded as an experimental observation; the calculation requires no adjustable parameters. The dimensions of the zones of respiratory activity measured in this way were 31 ± 7 μm in S. epidermidis colony biofilms and 38 ± 9 μm in S. aureus colony biofilms.
FIG. 5.
Oxygen concentration profiles in S. epidermidis (A) and S. aureus (B) colony biofilms. Zero on the x axis corresponds to the biofilm-air interface. Negative positions are in air, and positive distances are in the biofilm. Representative measurements from two different colonies are plotted in each panel.
A comment on the eyeball reading of a penetration depth from oxygen concentration profiles, as done above from Fig. 5, may be helpful here. This is a subjective determination and should probably not be regarded as a quantitative measurement. The depth of oxygen concentration that one comes up with (we say “approximately 50 μm”) all depends on exactly what low value of oxygen concentration is selected for the penetration endpoint. We prefer the dimension of the zone of respiratory activity derived from concentration profiles because this measurement involves no subjective set point.
No significant quantity of oxygen is delivered to the biofilm from the plates. This is evident from the oxygen concentration profiles, which show a monotone decreasing concentration with increasing depth. If oxygen were diffusing into the biofilm from the agar, there would be an increase in oxygen concentration in close proximity to the agar. This is not seen. The reason for the lack of oxygen delivery from the plates is probably the long diffusion path length from air-exposed agar to the underside of the colony biofilm.
The penetration of oxygen into a biofilm is governed by a reaction-diffusion interaction. As oxygen diffuses into the biofilm, it is consumed by respiring bacteria. The relative rates of oxygen diffusion and oxygen consumption determine the extent of its penetration. The depth of oxygen penetration is accessible theoretically through the following equation: a = (2DeSo/ko)1/2, where a is the penetration depth of the reacting solute, De is the effective diffusion coefficient of the solute in the biofilm, So is the solute concentration at the biofilm interface, and ko is the volumetric reaction rate of the solute inside the biofilm (29).
We have made independent estimates of each of these parameters to enable an a priori calculation of oxygen penetration depth. The effective diffusive permeability of oxygen in biofilm at 37°C was taken as 57% of the diffusion coefficient in pure water at this temperature, or 1.53 × 10−5 cm2 s−1 (29). The concentration of dissolved oxygen at the biofilm-air interface was taken as 6.0 mg liter−1, which is the equilibrium concentration at the barometric pressure in Bozeman, MT. The volumetric consumption rates of oxygen within the biofilm were determined by respirometry of colony biofilms to be 27 mg liter−1 s−1 for S. epidermidis and 39 mg liter−1 s−1 for S. aureus. Given these estimates, the predicted penetration depths of oxygen are 26 μm in S. epidermidis colony biofilms and 22 μm in S. aureus colony biofilms.
Most biofilm cells are viable.
Bacteria dispersed from a 48-h-old colony biofilm were enumerated for colony formation by plating and for total cell numbers by direct microscopic counting of filtered aliquots. The mean viable cell count of triplicate specimens was 7 × 108 CFU per membrane. The mean total cell count in the same specimens was 3 × 108 cells per membrane. Clearly it is not possible for viable cell numbers to exceed total cell numbers by a factor of 2. This discrepancy may be due to coalescence of cells on filter membranes during the last stages of filtration resulting in two cells being counted as one. This result does suggest, however, that the viability of bacteria in colony biofilms is high. Bacteria dispersed from colony biofilms and stained with the commercial viability kit BacLight were scored microscopically as green (live) or red (dead). A total of 10.1% of the total population (n = 4,428) was scored red, indicating that the large majority of bacteria were viable.
DISCUSSION
Anabolic and respiratory activities in staphylococcal biofilms are stratified. Regions of anabolic activity are localized at the interface between the biofilm and a nutrient or oxygen source. These regions are a few tens of micrometers in dimension. In the experiments reported here, zones of anabolic activity were less than about 60 μm wide. These results demonstrate conclusively the presence of metabolically active cells in the biofilm.
The data imply the existence of a substantial population of inactive cells in the biofilm. In S. epidermidis colony biofilms grown in air, for example, DNA synthetic activity was detected in a 31-μm zone at the air interface of the colony and a 16-μm zone at the membrane interface (Table 1). The average thickness of the biofilm was 153 μm, so one can estimate that an internal region of 106 μm, or 69% of the total biomass thickness, was dominated by inactive cells. Similarly, in S. aureus colony biofilms, protein synthetic activity was detected in a 38-μm zone at the air interface of the colony and a 14-μm zone at the membrane interface (Table 1). The average thickness of the biofilm in this case was 173 μm, leaving by difference an inactive centrally located stratum of 121 μm, or 70% of the total biomass thickness. Most of the bacteria in such inactive regions are still viable as only 10% of the cells in a colony biofilm were dead. Even if all of the dead cells were concentrated in the inactive internal 70% of the biofilm, the relative viability in this region would be 86% viable. Viability in the inactive middle of the biofilm is also suggested by the fact that additions of glucose or pure oxygen to the system expand the zone of anabolic activity into regions that were previously inactive. This shows that at least some of the cells in an inactive region can be stimulated to become active when nutrients or oxygen are made available. We conclude that staphylococcal biofilms are composed of physiologically heterogeneous populations that include active and also inactive cells. This result jibes with the interpretation advanced by Yarwood et al. (37), who observed complex spatial patterns of GFP expression, and putative dead cells, in S. aureus biofilms.
The double-banded patterns of anabolic activity in colony biofilms can be interpreted in the following way. At the membrane interface of the biofilm, no oxygen is available and the bacteria grow by fermentation. Staphylococci ferment glucose, fructose, or serine, with lactate and acetate being predominant metabolic products in each case (25). Once fermentable carbon sources have been exhausted, no further growth is possible except along the air interface of the biofilm, where oxygen is available. At this location, bacteria grow aerobically on nonfermentable carbon sources. In the interior stratum of the biofilm, where both oxygen and fermentable carbon sources are absent, bacteria are metabolically inactive. Similar twin bands of fermentative and aerobic activity have been inferred previously in the study of Klebsiella pneumoniae colony biofilms (2, 32, 38). In biofilm systems in which carbon sources and oxygen are delivered to the same interface, a single band of anabolic activity is observed. It is possible, however, that this activity is actually a contiguous layer of aerobic growth at the biofilm surface and fermentative growth just below the aerobic layer that are seamlessly adjoined. The data reported here do not afford a test of this hypothesis because the BrdU labeling and GFP induction techniques do not discriminate between aerobic metabolism and anaerobic anabolism.
The use of GFP promoter fusions to genes that are up-regulated in certain environmental conditions, such as anaerobiosis, would be one approach to demonstrating the metabolic state of cells more specifically than is possible through the measures of macromolecular synthesis that we have employed (30).
While the predominant zones of activity were along the air and membrane interfaces of colony biofilms, DNA synthetic activity was occasionally observed in thin strata in the colony interior. The authors do not have an explanation for this activity. This observation suggests that the spatial distribution of metabolic activity within a biofilm can indeed be complex and may involve phenomena beyond diffusion limitation of substrates.
Measurements of the dimensions of the zones of DNA synthetic activity, protein synthetic activity, and oxygen respiration correspond. For example, in S. epidermidis colony biofilms grown in air, the zones of DNA synthetic activity and active respiration along the air interface of the biofilm were both 31 μm in width. In S. aureus colony biofilms in which the BrdU labeling and GFP induction techniques were applied to the same colony, the patterns of these two anabolic activities were similar (Fig. 2C and D). Similarly, at the membrane interface, zones of anabolic activity were consistently approximately 15 to 25 μm in width.
Collectively these results suggest that staphylococcal biofilms contain cells in at least four distinct states: growing aerobically, growing fermentatively, dead, and dormant.
There are relatively few experimental visualizations of general anabolic activity in biofilms. Besides the investigations of K. pneumoniae biofilms mentioned above, most studies have focused on Pseudomonas aeruginosa biofilms. These investigations have demonstrated oxygen concentration gradients (31, 33, 34), localized protein synthetic activity as mapped by induction of alkaline phosphatase (15, 34, 35) or GFP (7, 31, 33), and growth rate as revealed by a growth rate-dependent promoter fusion to an unstable GFP (26). Collectively the research on P. aeruginosa biofilms indicates stratified activity located adjacent to the source of oxygen. A recent study of Shewanella oneidensis biofilm activity revealed that bacterial growth was predominantly localized near the biofilm-nutrient interface, but the expression of an anaerobically-induced gene was concentrated in interior regions of the biofilm in which growth was limited (30). This serves as a reminder that the absence of bulk growth in a particular region of a biofilm does not imply a complete absence of metabolic activity or gene expression.
The patterns of respiratory and anabolic activity observed in staphylococcal biofilms are consistent with a reaction-diffusion interaction governing the ingress of nutrients and oxygen into the biofilm. For oxygen in particular, sufficient information is available to make the analysis quantitative by solving the differential equation that describes simultaneous Fickian diffusion and oxygen reaction (19, 29). Oxygen penetration depths calculated in this way (22 to 26 μm) are in reasonable agreement with the experimentally measured dimensions of zones of respiratory activity (31 to 38 μm), DNA synthetic activity (25 to 31 μm), and protein synthetic activity (38 μm) measured at the air interface.
Reaction-diffusion theory predicts that oxygen penetration depth should increase in proportion to the square root of the interfacial oxygen concentration. Experiments that substitute pure oxygen for air, representing an oxygen concentration ratio of 100/21, are expected to result in an increase in the depth of oxygen penetration by a factor of (4.76)1/2 or 2.2. We found factors of 1.5 and 1.6 for the expansion of zones of DNA synthetic activity and protein synthetic activity, respectively, comparing biofilm growth in oxygen to growth in air. In an analogous experimental comparison of active regions in P. aeruginosa biofilms grown in pure oxygen and ambient air environments, a factor of 1.6 for the ratio of the width of the active zones under these two conditions was reported (34). One reason why experimentally determined factors are smaller than the theoretical ratio of 2.2 is that the calculated ratio assumes that the rate of oxygen utilization does not depend on the oxygen concentration. If in fact the oxygen consumption rate increases with increased oxygen concentration, this would tend to yield bands of bacterial activity thinner than calculated.
An immediate practical implication of this work is to the understanding of antimicrobial tolerance exhibited by microorganisms in biofilms. An inactive subpopulation harbored within a biofilm is a plausible contributing factor to the recalcitrance of biofilm infections. If inactivity is an important protective mechanism, then surely there are avenues to interdicting this protection. What nutritive and other cues are needed to awaken inactive bacteria and render them susceptible to antimicrobial agents? What specific molecular defenses are deployed when cells enter an inactive state, and how might these defenses be subverted? What residual metabolic activity and gene expression are retained in nongrowing cells? Which antimicrobial agents are, relatively speaking, effective against inactive cells (10)? Improved approaches for controlling biofilm infections may turn on the answers to such questions.
Acknowledgments
This work was supported by an award from the W. M. Keck Foundation.
We thank A. Cheung for providing the S. aureus strain containing the inducible GFP.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. J. Antimicrob. Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderl, J. N., J. Zahller, F. Roe, and P. S. Stewart. 2003. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. J. Antimicrob. Chemother. 47:1251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreoli-Pinto, T. J., and K. U. Graziano. 1999. Important aspects of the colonization of central venous catheter. Boll. Chim. Farm. 138:19-23. [PubMed] [Google Scholar]
- 4.Artusson, V., R. D. Finlay, and J. K. Jansson. 2005. Combined bromodeoxyuridine immunocapture and terminal-restriction fragment length polymorphism analysis highlights differences in the active soil bacterial metagenome due to Glomus mosseae inoculation or plant species. Environ. Microbiol. 7:1952-1966. [DOI] [PubMed] [Google Scholar]
- 5.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 48:2659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, M. R. W., D.G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-783. [DOI] [PubMed] [Google Scholar]
- 9.Cacoub, P., P. Leprince, P. Nataf, P. Hausfater, R. Dorent, B. Wechsler, V. Bors, A. Pavie, J. C. Peitte, and I. Gandjbakhch. 1998. Pacemaker infective endocarditis. Am. J. Cardiol. 82:480-484. [DOI] [PubMed] [Google Scholar]
- 10.Coates, A. R., and H. Yu. 2006. New strategies for antibacterial drug design: targeting non-multiplying latent bacteria. Drugs R&D 7:133-151. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 12.Dolbeare, F., and J. R. Selden. 1994. Immunochemical quantitation of bromodeoxyuridine: application to cell-cycle kinetics. Methods Cell Biol. 41:297-316. [PubMed] [Google Scholar]
- 13.Elias, M. C. Q. B., M. Faria, R. A. Mortara, M. C. M. Motta, W. de Souza, M. Thiry, and S. Schenkman. 2002. Chromosome localization changes in the Trypanosoma cruzi nucleus. Eukaryot. Cell 1:944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgson, A. E., S. M. Nelson, M. R. W. Brown, and P. Gilbert. 1995. A simple in vitro model for growth control of bacterial biofilms. J. Appl. Bacteriol. 79:87-93. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C.-T., K. D. Xu, G. A. McFeters, and P. S. Stewart. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl. Environ. Microbiol. 64:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahl, B. C., M. Goulian, W. Van Wamel, M. Herrmann, S. M. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect. Immun. 68:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichtenwalner, R. J., M. E. Forbes, W. E. Sonntag, and D. R. Riddle. 2006. Adult-onset deficiency in growth hormone and insulin-like growth factor-I decreases survival of dentate granule neurons: insights into the regulation of adult hippocampal neurogenesis. J. Neurosci. Res. 83:199-210. [DOI] [PubMed] [Google Scholar]
- 18.Oike, H., I. Matsumoto, and K. Abe. 2006. Group IIA phospholipase A2 is coexpressed with SNAP-25 in mature taste receptor cells of rat circumvallate papillae. J. Comp. Neurol. 494:876-886. [DOI] [PubMed] [Google Scholar]
- 19.Perez, J., C. Picioreanu, and M. van Loosdrecht. 2005. Modeling biofilm and floc diffusion processes based on analytical solution of reaction-diffusion equations. Water Res. 39:1311-1323. [DOI] [PubMed] [Google Scholar]
- 20.Pernthaler, A., J. Pernthaler, M. Schattenhofer, and R. Amann. 2002. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernthaler, A., and J. Pernthaler. 2005. Diurnal variation of cell proliferation in three bacterial taxa from coastal North Sea waters. Appl. Environ. Microbiol. 71:4638-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rani, S. A., B. Pitts, and P. S. Stewart. 2005. Rapid diffusion of fluorescent tracers into Staphylococcus epidermidis biofilms visualized by time lapse microscopy. J. Antimicrob. Chemother. 49:728-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Resch, A., R. Rosenstein, C. Nerz, and G. Götz. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revsbech, N. P. 1989. Diffusion characteristics of microbial communities determined by use of oxygen microsensors. J. Microbiol. Methods 9:111-122. [Google Scholar]
- 25.Sivakanesan, R., and E. A. Dawes. 2000. Anaerobic glucose and serine metabolism in Staphylococcus epidermidis. J. Gen. Microbiol. 118:143-157. [DOI] [PubMed] [Google Scholar]
- 26.Sternberg, C., B. B. Christensen, T. Johansen, A. T. Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart, P. S., G. A. McFeters, and C. T. Huang. 2000. Biofilm control by antimicrobial agents, p. 373-405. In J. D. Bryers (ed.), Biofilms. John Wiley & Sons, New York, NY.
- 28.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 29.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teal, T. K., D. P. Lies, B. J. Wold, and D. K. Newman. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walters, M. C., F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. J. Antimicrob. Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wentland, E. J., P. S. Stewart, C. T. Huang, and G. A. McFeters. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12:316-321. [DOI] [PubMed] [Google Scholar]
- 33.Werner, E., F. Roe, A. Bugnicourt, M. J. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]
- 36.Yao, Y., D. E. Sturdevant, and M. Otto. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191:289-298. [DOI] [PubMed] [Google Scholar]
- 37.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahller, J., and P. S. Stewart. 2002. Transmission electron microscopic study of antibiotic action on Klebsiella pneumoniae biofilm. J. Antimicrob. Chemother. 46:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, Z., and P. S. Stewart. 2002. Penetration of rifampin through Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 46:900-903. [DOI] [PMC free article] [PubMed] [Google Scholar]