Abstract
The histidine protein kinase DcuS of Escherichia coli senses C4-dicarboxylates and citrate by a periplasmic domain. The closely related sensor kinase CitA binds citrate, but no C4-dicarboxylates, by a homologous periplasmic domain. CitA is known to bind the three carboxylate and the hydroxyl groups of citrate by sites C1, C2, C3, and H. DcuS requires the same sites for C4-dicarboxylate sensing, but only C2 and C3 are highly conserved. It is shown here that sensing of citrate by DcuS required the same sites. Binding of citrate to DcuS, therefore, was similar to binding of C4-dicarboxylates but different from that of citrate binding in CitA. DcuS could be converted to a C4-dicarboxylate-specific sensor (DcuSDC) by mutating residues of sites C1 and C3 or of some DcuS-subtype specific residues. Mutations around site C1 aimed at increasing the size and accessibility of the site converted DcuS to a citrate-specific sensor (DcuSCit). DcuSDC and DcuSCit had complementary effector specificities and responded either to C4-dicarboxylates or to citrate and mesaconate. The results imply that DcuS binds citrate (similar to the C4-dicarboxylates) via the C4-dicarboxylate part of the molecule. Sites C2 and C3 are essential for binding of two carboxylic groups of citrate or of C4-dicarboxylates; sites C1 and H are required for other essential purposes.
Escherichia coli is able to use C4-dicarboxylates as substrates for anaerobic growth by fumarate respiration, which requires the synthesis of fumarate reductase (frdABCD genes) and the fumarate/succinate antiporter DcuB (dcuB gene) (for reviews, see references 6, 15, and 28). Expression of the frdABCD and dcuB genes is stimulated by the DcuSR two-component system (12, 14, 15, 29). DcuS responds to C4-dicarboxylates and related compounds through a periplasmic sensing domain (19). The two carboxylic groups of the C4-dicarboxylates are crucial for stimulus perception, whereas other parts of the C4-dicarboxylates such as ligands at position C2 or C3 are of minor significance. The apparent KD values of C4-dicarboxylates for stimulating the expression of DcuS-regulated genes are in the range of 0.45 to 3 mM.
DcuS is a member of the CitA/DcuS family of sensory histidine kinases and shares significant sequence similarities with CitA (4, 5, 15, 17, 18). CitA is the highly specific and high-affinity citrate sensor kinase of the CitAB two-component system that controls expression of the citrate fermentation genes in Klebsiella pneumoniae and E. coli. The CitA and DcuS sensors have similar membrane topologies, with a periplasmic sensory domain and a cytoplasmic kinase domain (15, 18, 29). CitA and DcuS are typical members of the periplasmic sensing histidine kinases (20). The structure of the periplasmic domain of DcuS has been solved by nuclear magnetic resonance (NMR) spectroscopy, and that of CitA from K. pneumoniae has been solved by crystallography and X-ray analysis (23, 25). The overall structures for the two domains are similar and resemble the PAS (Per-Arnt-Sim) domain of the photoactive yellow protein of Halorhodospira halophila (3). The periplasmic domains of DcuS and CitA are composed of a core of four (DcuS) or five (CitA) β-strands that form the basis of the binding pocket, which is surrounded by α-helices.
CitA binds H-citrate2− with high affinity and specificity in the μM range (17, 18). Binding occurs via the three carboxylic/carboxylate groups and the hydroxyl group of H-citrate2− (10, 25). The carboxylic/carboxylate groups are liganded by sites C1, C2, and C3 of the sensor, which includes the essential residues K152C, R109C, and H112C: hereafter, all residues are labeled with a subscript C if they are from CitA and with a subscript D if they are from DcuS (Fig. 1). The essential residue R150C is part of the hydroxyl-binding site H. The periplasmic domain of DcuS contains the corresponding residues F149D, R107D, and H110D for sites C1 to C3 and R147D for site H, all of which are essential for C4-dicarboxylate sensing by DcuS (19). Therefore, C4-dicarboxylate binding by DcuS requires all four sites (C1 to C3 and H sites) known from CitA, although the third carboxylate and the hydroxyl group of citrate are not present in C4-dicarboxylates. Carboxyl sites C2 and C3 are well conserved in DcuS, whereas C1 shows the largest difference from the corresponding sequence of CitA, suggesting that sites C2 and C3 represent the actual carboxylate binding sites of DcuS (19).
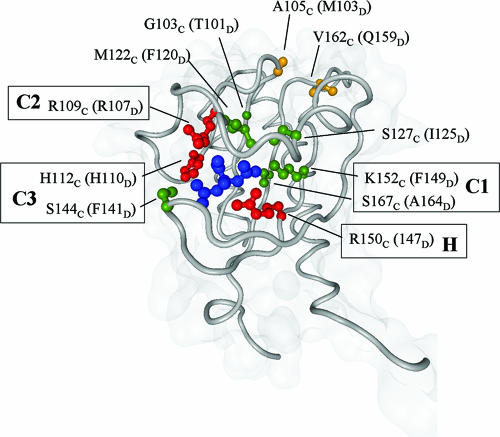
FIG. 1.
Positions of conserved and subtype-specific residues relative to the citrate ligand in CitA. Residues are labeled as they occur in CitA (subscript C) and in parentheses as they are found in DcuS (subscript D). Universally conserved residues are shown in red, subtype-specific residues in the binding pocket in green, and residues outside the binding pocket in orange.
DcuS is able to use citrate as a stimulus as well (12, 29). The apparent KD (7 mM) is 2.3-fold higher than that for succinate induction. The KD of CitA for citrate, by contrast, is in the μM range, and the sensor is highly specific for C6-tricarboxylates (17, 18). The present study examined whether citrate sensing by DcuS involves the same residues and sites (C1 to C3 and the H site) that are required for C4-dicarboxylate sensing. To learn more about C4-dicarboxylate/citrate differentiation by DcuS, available sequence and structural information was used in addition to convert DcuS to a sensor that is specific either for C4-dicarboxylates or for citrate.
MATERIALS AND METHODS
Growth of bacteria and expression of dcuB′-′lacZ.
The strains and plasmids used in this study are listed in Table 1. For genetic experiments, E. coli was grown in LB broth (26). For expression studies, E. coli IMW260 containing a dcuB′-′lacZ reporter gene fusion and a dcuS insertional inactivation was grown under anoxic conditions in enriched M9 mineral medium with glycerol (50 mM), dimethyl sulfoxide (DMSO; 20 mM) as an electron acceptor (19, 29), and various effectors. Anoxic conditions were adjusted by growth in stoppered glass tubes under an atmosphere of N2. The effectors were added at 20 mM, and nitropropionate was added at 5 mM. Cell densities were measured as absorbance at an optical density of 578 nm (OD578). Samples were withdrawn from exponentially growing cultures (OD578 of 0.5 to 0.8), and β-galactosidase activity (21) was determined in four replicates in two independent growth experiments with three cultures each. P1 transduction and lysogenization with λRZ5 derivatives were carried out as described previously (29).
TABLE 1.
Strains of Escherichia coli and plasmids used in this study
| E. coli strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flB530 deoC1 ptsF25 rbsR | 27 |
| SC1088citA | lacZ4075 mini Tet del citA::Kanr | 13 |
| IMW237 | MC4100, λ[Φ(dcuB′-′lacZ)Hyb Ampr] | 30 |
| IMW260 | MC4100 but λ[Φ(dcuB′-′lacZ)Hyb Ampr] dcuS::Camr | 30 |
| IMW238 | MC4100 but λ[Φ(dcuB′-′lacZ)Hyb Ampr] dcuR::Kanr | 30 |
| IMW280 | MC4100 but λ[Φ(dcuB′-′lacZ)Hyb Ampr] citA::Kanr | IMW237 × P1 (Sc1088citA) |
| IMW239 | MC4100 but λ[Φ(dcuB′-′lacZ)Hyb Ampr] citB::Specr | 30 |
| Plasmids | ||
| pET28a | Novagen | |
| pMW145 | pET28a, comprising the periplasmic domain of DcuS from amino acid Ser45 to Arg180 | 23 |
| pMW423 | pMW145 including periplasmic domain DcuS-PD(T101G F120M I125S)45-180 | This study |
| pMW181 | pET28a with dcuS | 19 |
| pMW345 | pMW181 with dcuS mutant encoding DcuS(F149K) | This study |
| pMW352 | pMW181 with dcuS mutant encoding DcuS(F149K A164S) | This study |
| pMW344 | pMW181 with dcuS mutant encoding DcuS(T101G) | This study |
| pMW368 | pMW181 with dcuS mutant encoding DcuS(F120M) | This study |
| pMW373 | pMW181 with dcuS mutant encoding DcuS(I125S) | This study |
| pMW390 | pMW181 with dcuS mutant encoding DcuS(F120M I125S) | This study |
| pMW351 | pMW181 with dcuS mutant encoding DcuS(T101G F120M) | This study |
| pMW369 | pMW181 with dcuS mutant encoding DcuS(T101G I125S) | This study |
| pMW353 | pMW181 with dcuS mutant encoding DcuS(T101G F120M I125S) | This study |
| pMW354 | pMW181 with dcuS mutant encoding DcuS(F141S) | This study |
| pMW355 | pMW181 with dcuS mutant encoding DcuS(Q159V) | This study |
| pMW356 | pMW181 with dcuS mutant encoding DcuS(Q159V M103A) | This study |
| pMW370 | pMW181 with dcuS mutant encoding DcuS(T101G F120M I125S F149K) | This study |
| pMW374 | pMW181 with dcuS mutant encoding DcuS(T101G F120M I125S A164S) | This study |
| pMW371 | pMW181 with dcuS mutant encoding DcuS(T101G F120M I125S Q159V) | This study |
| pMW372 | pMW181 with dcuS mutant encoding DcuS(T101G F120M I125S F149K A164S) | This study |
| pMW398 | pMW181 with dcuS mutant encoding DcuS(R107A) | This study |
| pMW409 | pMW181 with dcuS mutant encoding DcuS(F149K A164S F141S) | This study |
| pMW410 | pMW181 with dcuS mutant encoding DcuS(T101G F120M I125S F141S) | This study |
| pMW236 | pMW181 with dcuS mutant encoding DcuS(H110A) | This study |
| pMW237 | pMW181 with dcuS mutant encoding DcuS(R147A) | This study |
| pMW277 | pMW181 with dcuS mutant encoding DcuS(F149A) | This study |
| pMW298 | pMW181 with dcuS mutant encoding DcuS(A164S) | |
| pMW296 | pMW181 with dcuS mutant encoding DcuS(M103A) |
Molecular genetic methods.
Standard molecular genetic methods were used according to Sambrook et al. (26) or as recommended by the suppliers. Plasmids were isolated using the QIAprep Spin Miniprep kit (QIAGEN Hilden). Site-directed mutagenesis was carried out with the QuickChange kit (Stratagene) as follows: E. coli strains were transformed with plasmids by electroporation (8). The dcuS coding region, including the complete promoter, was PCR amplified by the hot-start method and cloned into pET28a (19). The resulting plasmid, pMW181, served as the template for site-directed mutagenesis by PCR with synthetic primers containing the desired mutation. The mutations were verified by DNA sequencing. Oligonucleotide primers used for mutagenesis are listed in Table 2. For overproduction of the periplasmic domain containing the triple mutation T101G F120M I125S, the domain was amplified from pMW353 by PCR using the primers pDcuS-Nde2 and pDcuS-HindIII and cloned into pET28a.
TABLE 2.
Site-directed mutagenesis of dcuS in the region coding for the periplasmic domain of DcuS in plasmid pMW181
| Mutation(s) | Sequence (5′→3′)a |
|---|---|
| DcuS(A164S) | CAAATTGGCGTGGTGTCTATCGGCCTTGAGTTAAGCC |
| DcuS(F149K) | CGCAGGCTTTACGCGTAAAAACCCCCATCTACGATGAAAATC |
| DcuS(T101G) | GATCTGCTGTTTATTGTCGTTGGCGATATGCAAAGTCTTC |
| DcuS(F120M) | CGTATTGGTCAGCCAATGAAAGGTGATGAC |
| DcuS(I125S) | CCATTTAAAGGTGATGAC/TCGCTTAAAGCGCTGAATGGCG |
| DcuS(F120M I125S) | CCAATGAAAGGTGATGACTCGCTTAAAGCGCTGAATGGCG |
| DcuS(F141S) | CTATCAATCGCGGTTCTCTGGCGCAGGCTTTAC |
| DcuS(Q159V) | CTACGATGAAAATCATAAAGTAATTGGCGTGGTGGCGATC |
| DcuS(M103A) | TGTCGTTACCGATGCGCAAAGTCTTCGCTACTCG |
| DcuS(I125S) | CCATTTAAAGGTGATGACTCGCTTAAAGCGCTGAATGGCG |
| DcuS(R107A) | CCGATATGCAAAGTCTTGCCTACTCGCATCCTGAAG |
| DcuS(H110A) | CTTCGCTACTCGGCACCTGAAGCCCAGC |
| DcuS(R147A) | GGCGCAGGCTTTAGCAGTATTTACCCCC |
| DcuS(F149A) | GCAGGCTTTACGCGTAGCTACCCCCATCTACG |
| pdcuS-Nde2 | ATTTACTTCTCGCATATGAGTGATATG |
| pdcuS-Hind | GACCAGATAAAGCTTCAGCGACTG |
Oligonucleotides for mutagenesis are complementary (reverse) to the sequences shown; nucleotides resulting in amino acid exchanges in DcuS are underlined.
NMR study of the isolated periplasmic domain of DcuS (DcuS-PD).
The periplasmic domains of the wild-type DcuS and the DcuS(T101G F120M I125S)-PD triple mutant were overproduced, respectively, from plasmids pMW145 and pMW423 in the presence of [15N]NH4Cl (23). DcuS(T101G F120M I125S)-PD was the same construct as the previously described DcuS-PD (19, 23), but contained the T101G F120M I125S mutations. The periplasmic domains, which contained an N-terminal His tag, were isolated and obtained in buffer containing 500 mM imidazole, 200 mM NaCl, and 50 mM sodium/potassium phosphate at pH 7.0. The sample was adapted to buffer containing 5 mM imidazole, 200 mM NaCl, and 50 mM sodium/potassium phosphate at pH 7.0 by dialysis, and the His tag was removed with thrombin protease. The protein was concentrated by centrifugation at maximally 7.500 × g in a Vivaspin concentration tube (10,000-molecular-weight cutoff). Samples (300 μl) for NMR studies contained 25 to 30 mg protein/ml, 4.4 mM imidazole, 176 mM NaCl, 44 mM sodium/potassium phosphate at pH 7.0, 9.99% D2O, 0.01% NaN3, 50 μM Pefabloc, and 50 mM glycine.
Bioinformatics.
Homologs of the periplasmic domains of K. pneumnoniae CitA (1P0Z) and E. coli DcuS (1OJG) were identified from the nonredundant protein sequence database using PSI-BLAST (1). The database was filtered at 90% sequence identity for the first iteration and at 70% for the remaining six iterations. The domains identified at E values less than 10−3 were extracted and clustered in CLANS (9) with default parameters, except for the attraction parameters, which were set to a value of 5 and an exponent of 1 (compare Fig. 5). Clustering at an E-value cutoff of 10−3 yielded a central group containing all CitA and DcuS orthologs. This group was reclustered at progressively more restrictive E values, until CitA and DcuS could be cleanly separated at an E value of 10−27. Sequences found in the two groups at this cutoff were aligned in ProbCons (7) and subjected to alignment subtyping with SDPpred (16) at default parameters. Three residues were universally conserved. In the following, all residues are labeled with a subscript C if they are from CitA and with a subscript D if they are from DcuS: R109C/R107D, H112C/H110D, and R150C/R147D (red in Fig. 1). The program identified five positions as subtype specific: three in the ligand-binding pocket (G103C/T101D, M122C/F120D, and K152C/F149D [green in Fig. 1]) and two outside (V162C/Q159D [orange in Fig. 1] and Y170C/L167D). Based on the inspection of sequence alignments and structures, we identified three additional residues likely to be subtype specific in the ligand-binding pocket (S127C/I125D, S144C/F141D, and S167C/A164D [green in Fig. 1]) and one outside (A105C/M103D [orange in Fig. 1]), which were not identified by SDPred due to the restrictive cutoff settings. We also did not consider that Y170C/L167D would be subtype specific and attribute its identification by SDPred to the high self-score of tyrosine in replacement matrices.
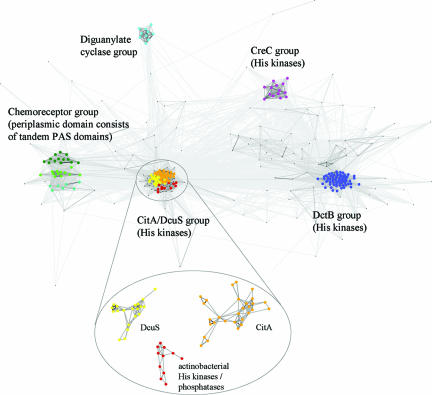
FIG. 5.
Cluster analysis of DcuS and CitA homologs. Sequences were identified on the basis of their similarities to the DcuS and CitA periplasmic domains, as described in Materials and Methods. The clusters are labeled according to the main putative activity of their members, but most clusters do not consist exclusively of one type. Most strikingly, the chemoreceptor cluster has two subclusters of chemoreceptors and one (cyan) made up mainly of histidine kinases and some diguanylate cyclases. The cluster containing DcuS and CitA was reclustered at a more stringent cutoff in order to separate the two groups.
RESULTS
Subtype-specific residues for differentiation of DcuS and CitA.
Citrate binding in CitA is accomplished by sites C1, C2, and C3 for the carboxylic groups and site H for the hydroxyl group. Sites C2 with ligands R109C and T101C for the central carboxylate and C3 with ligands H112C, S144C, and L145C for one of the distal carboxylates (25) are located closest to the exit of the pocket (Fig. 1 and Fig. 2). The buried part of the pocket provides the third carboxylate-binding site C1 with ligands K152C and S167C and the hydroxyl-binding site H (R150C). Residues R109C, H112C, K152C, and R150C of sites C1 to C3 play a central role in citrate binding by CitA (10, 18). In DcuS, the homologous residues R107D, H110D, F149D, and R147D are essential for C4-dicarboxylate sensing (19). This suggested that the sites for C4-dicarboxylate (or fumarate) binding by DcuS are similar to the citrate-binding site of CitA.
FIG. 2.
Alignment of the sequences from the C4-dicarboxylate and tricarboxylate binding sites in DcuS and CitA proteins. The sequences are limited to residues 90 to 170 of DcuS from E. coli. These represent the major part of the periplasmic domain (amino acids 42 to 181) and the corresponding sequences from the homologous proteins. Residues in the part of the pocket close to the exit in DcuS and CitA are shown in green, those from the buried part of the pocket in red, and those located in between in magenta. Conserved and essential residues of DcuS and CitA and the ligands of citrate in CitA are highlighted. The subtype-specific residues are indicated by arrows. EC, E. coli; BS, Bacillus subtilis; BH, Bacillus halodurans; KP, K. pneumoniae.
Sequence alignment of the periplasmic domains of DcuS and CitA showed that of the residues essential for ligand binding, R109C/R107D, H112C/H110D, and R150C/R147D, are universally conserved, whereas K152C/F149D is subtype specific for DcuS versus CitA. We identified seven additional positions as subtype specific for DcuS, as described in Materials and Methods: T101D, M103D, F120D, I125D, F141D, Q159D, and A164D (Fig. 1 and 2). These and the essential residues R107D, H110D, R147D, and F149D (19) were mutated singly and in various combinations in DcuS in order to characterize citrate binding and to differentiate C4-dicarboxylate binding from citrate binding.
Sensing of citrate and C4-dicarboxylates requires the same sites of DcuS.
Response to citrate was tested in vivo by the use of the DcuS-dependent reporter gene fusion dcuB′-′lacZ. Mutation of each of the residues essential for fumarate binding in sites C1 to C3 and H (R107D, H110D, R147D, and F149D) to alanine completely inactivated induction of the dcuB′-′lacZ reporter gene fusion by citrate (Table 3). Therefore, sensing of citrate and C4-dicarboxylate by DcuS requires each of the C1, C2, C3, and H sites and the amino acid residues identified earlier as essential for C4-dicarboxylate sensing (19).
TABLE 3.
Requirement of essential residues in sites C1, C2, C3, and H and of DcuS subtype-specific residues (T101, M103, F120, I125, F141, Q159, A164) for citrate and fumarate sensing
| Straina | Type of DcuS |
dcuB′-′lacZ fusion β-galactosidase activity (Miller units)b
|
|
|---|---|---|---|
| + Fumarate | + Citrate | ||
| IMW260c(pdcuS+) | DcuSWt | 189 | 89 |
| Essential residues in sites C1, C2, C3, and Hd | |||
| IMW260(pMW234) | DcuS(R107A) | 7 | 5 |
| MW260(pMW236) | DcuS(H110A) | 6 | 4 |
| IMW260(pMW237) | DcuS(R147A) | 12 | 3 |
| IMW260(pMW277) | DcuS(F149A) | 9 | 3 |
| IMW260(pMW345) | DcuS(F149K) | 110 | 5 |
| DcuS subtype-specific residuese | |||
| IMW260(pMW344) | DcuS(T101G) | 210 | 13 |
| IMW260(pMW296) | DcuS(M103A) | 177 | 50 |
| IMW260(pMW292) | DcuS(F120A) | 4 | 17 |
| IMW260(pMW368) | DcuS(F120M) | 11 | 6 |
| IMW260(pMW373) | DcuS(I125S) | 212 | 17 |
| IMW260(pMW354) | DcuS(F141S) | 159 | 9 |
| IMW260(pMW355) | DcuS(Q159V) | 212 | 23 |
| IMW260(pMW298) | DcuS(A164S) | 200 | 63 |
Growth in supplemented M9 mineral salts medium under anoxic conditions with glycerol (50 mM) plus DMSO (20 mM) and citrate (20 mM) or fumarate (20 mM) as inductors.
β-Galactosidase activity measured at OD578 of the cultures of 0.5 to 0.8 for four replicates from two independent growth experiments with three cultures each (deviation, ≤20%).
dcuS deletion strain (IMW260).
R107, H110, R147, and F149.
T101, M103, F120, I125, F141, Q159, and A164.
In the same way, the significance of the DcuS subtype-specific residues (T101D, M103D, F120D, I125D, F141D, Q159D, and A164D) was tested by mutating the residues to either alanine or the corresponding residues of CitA (Table 3). Remarkably, mutations in most of the residues (T101, I125, F141, and Q159) retained sensitivity to fumarate, whereas the response to citrate was lost completely or for the most part, even when the residues were converted to CitA-specific residues. Mutation of F120 to either alanine or the CitA homologous methionine residue resulted in DcuS proteins which were inactive towards fumarate and citrate as well. Only mutating residues M103 and A164 had no major effect on either of the stimuli. The DcuS subtype-specific residues therefore can be used to differentiate interaction of DcuS with fumarate and citrate, and citrate has more stringent requirements for the composition of the binding site in DcuS.
Optimizing carboxylate-binding ligands in sites C1 and C3: formation of C4-dicarboxylate-specific DcuS (DcuSDC).
For closer characterization of carboxylate binding, residues in and around sites C1 to C3 and H were adapted by mutation to the ligands found in the citrate sensor CitA at the corresponding sites. In site C1, which is the least-conserved carboxylate-binding site in DcuS, mutations F149K A164S (class I mutation) were introduced to create a site with ligands homologous to K152C and S167C of CitA (Fig. 1 and 2). The ligands at site C2 of DcuS (R107D V99D) were in agreement with the corresponding residues in CitA of E. coli (R109C and V101C) and very similar to K. pneumoniae CitA (R109C and T101C) and were not changed. To create an “optimized” C3 site in DcuS, mutation F141S (class II mutation) was introduced.
The activities of class I and II mutants were tested in vivo after complementation of a chromosomal dcuS mutant with mutated dcuS alleles on plasmids. Plasmid-borne wild-type dcuS stimulates the expression of dcuB—lacZ by fumarate or citrate to high levels (Table 4), in agreement with earlier reports (19). Strains carrying the class I mutants DcuS(F149K A164S) and DcuS(F149K) retained fumarate sensitivity, and the response to fumarate (fumarate induced/noninduced) was similar to or even higher than that for the wild type. However, induction by citrate was lost, and the fumarate/citrate induction ratio increased from 2.1 in the wild type to 42 in the mutants. Similarly, strains carrying the class II mutant DcuS(F141S) retained a high capacity for induction by fumarate, but not by citrate. Therefore, mutations optimizing the ligands in the C1 and C3 sites resulted in DcuS specific for C4-dicarboxylates (“DcuSDC”). In mutant DcuS(F149K A164S F141S), mutations I and II of sites C1 and C3 were combined (Table 4). Surprisingly, the resulting mutant was completely unresponsive to stimulation by either fumarate or citrate.
TABLE 4.
Effect of class I, class II, class III, and class IV mutations in DcuS on the fumarate and citrate sensitivities of DcuS-dependent expression of dcuB′-′lacZ
| Mutanta | Protein (mutation) | Class (site in DcuS) |
dcuB′-′lacZ fusion β-galactosidase activity (Miller units)b
|
Fumarate/citrate ratioc | ||
|---|---|---|---|---|---|---|
| No addition | + Fumarate | + Citrate | ||||
| IMW260d(pdcuS+) (wild type) | DcuS | Wild type | 17 | 189 | 89 | 2.1 |
| IMW260(pMW352) | DcuS(F149K A164S) | Class I (ligands in C1) | 4 | 42 | 1 | 42.0 |
| IMW260(pMW345) | DcuS(F149K) | Class I (ligands in C1) | 3 | 110 | 5 | 22.0 |
| IMW260(pMW354) | DcuS(F141S) | Class II (ligands in C3) | 7 | 159 | 9 | 17.7 |
| IMW260(pMW409) | DcuS(F149K A164S F141S) | Class I + II | 2 | 4 | 4 | 1 |
| IMW260(pMW353) | DcuS(T101G F120M I125S) | Class III (size of C1/C2) | 5 | 1 | 58 | 0.02 |
| IMW260(pMW372) | DcuS(T101G F120M I125S F149K A164S) | Class I + III | 2 | 2 | 27 | 0.07 |
| IMW260(pMW410) | DcuS(T101G F120M I125S F141S) | Class II + III | 3 | 3 | 4 | 0.75 |
| IMW260(pMW355) | DcuS(Q159V) | Class IV (access binding site) | 8 | 212 | 23 | 9.2 |
| IMW260(pMW356) | DcuS(Q159V M103A) | Class IV (access binding site) | 5 | 194 | 17 | 11.4 |
Anaerobic growth of the transformed bacteria in glycerol-plus-DMSO medium in the presence of fumarate or citrate. Other conditions are as in Fig. 4.
β-Galactosidase activities for four replicates each in two independent growth experiments with three cultures each (deviation, ≤15%).
Ratio of fumarate- and citrate-induced β-galactosidase activities.
dcuS deletion strain (IMW260).
Optimizing the size of the carboxylate-binding sites C1 and C2: formation of citrate-specific DcuSCit.
A third class of mutations was introduced by optimizing the size and steric properties of the binding pocket to facilitate citrate binding (class III mutation). The class III triple mutant (T101G F120M I125S) was designed to increase the size of the carboxylate pocket C1. Residues T101D, F120D, and I125D of DcuS form a network of interactions at the bottom of this pocket and are bulkier and more hydrophobic than the corresponding residues in CitA (G103C, M122C, and S127C). Thus, the triple mutant DcuS(T101G F120M I125S) was expected to provide a better fit for citrate in the binding pockets C1 and, albeit to a lesser extent, C2. In the dcuB′-′lacZ reporter gene assay, the mutant (Table 4) remained sensitive to citrate but lost fumarate sensitivity completely. The high citrate selectivity was reflected by the fumarate/citrate ratio of dcuB′-′lacZ induction, which was about 0.02 for the mutant compared to 2.1 for plasmid-borne wild-type DcuS.
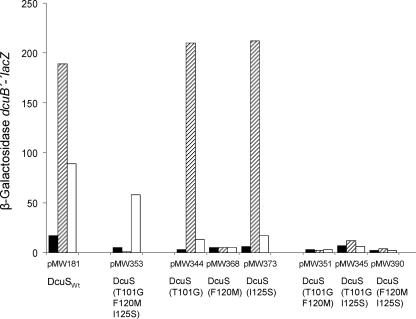
To identify the mutations responsible for citrate specificity in the triple mutant, each of the precursor single and double mutants was tested for C4-dicarboxylate and citrate stimulation of dcuB′-′lacZ expression (Fig. 3). Remarkably, two of the single mutants, DcuS(T101G) and DcuS(I125S), were C4-dicarboxylate specific, whereas the third mutant, DcuS(F120M), was inactive toward either effector type. The double mutants were inactive toward each of the two effectors. Thus, citrate specificity depended on the combined effect of the three mutations, rather than proceeding by a stepwise increase in selectivity. This finding supports the notion of a synergistic interaction between these three residues.
FIG. 3.
Effect of the class III DcuS mutation, DcuS(T101G F120M I125S), on fumarate- and citrate-induced expression of dcuB′-′lacZ. Cells of the E. coli IMW260 (ΔdcuS) strain carrying low-copy plasmids coding for dcuS alleles DcuSWt (wild type with plasmid pMW181) or the class III mutant DcuS(T101G F120M I125S) (plasmid pMW353) were used. In addition, the effects of the corresponding single and double mutants as the precursors of the triple mutant are shown. The bacteria were grown on glycerol plus DMSO under anoxic conditions in enriched M9 medium with the addition of fumarate or citrate. β-Galactosidase activity was assayed after growth without inductor (black bar), in the presence of fumarate (hatched bar), or with citrate (open bar) to the mid-exponential phase of growth (A578 = 0.5 to 0.8). Activities are the mean of at least six independent growth experiments and four replicates each.
The class III triple mutant, in which the carboxylate-binding pockets C1/C2 were enlarged, was combined with the DcuS mutants containing optimized carboxylate ligands in C1 (class I mutant) or C3 (class II mutant) (Table 4). Combination of the (citrate specific) triple mutant with the (fumarate specific) class I mutant DcuS(F149K A164S) resulted in a quintuple mutant that was again citrate specific; however, dcuB′-′lacZ expression and the specificity for citrate were lower than in the triple mutant. Combination of the triple mutant with each of the single mutations (F149K and A164S) of the class I mutant resulted in an inactive protein that was sensitive to neither fumarate nor citrate (not shown). Similarly, the triple mutant was combined with the class II mutant (F141S), in which the ligands for carboxylate binding in site C3 were optimized (Table 4). The resulting quadruple mutant was inactive for both fumarate and citrate. Thus, combination of the citrate-specific triple mutant with the fumarate-specific mutant in sites C1 or C3 did not result in the accumulation of positive properties.
Generation of fumarate specificity by mutation of residues outside the binding pocket.
A surprising outcome of alignment subtyping was the identification of two subtype-specific residues outside the ligand-binding pocket (Fig. 1 and Fig. 2). In DcuS, one of these (Q159D) forms a side-chain-backbone contact with the base of the “lid” that closes off the substrate-binding pocket. In CitA, this interaction by the corresponding residue (V162) is absent. Q159D, therefore, seemed an attractive mutation target singly and in combination with a neighboring residue (M103D), which to a lesser degree also shows subtype specificity. In this class IV mutation, M103A Q159V, residues M103D and Q159D were replaced with the corresponding residues of CitA. Under the control of DcuS(M103A Q159V), expression of the dcuB′-′lacZ reporter gene fusion became specific for fumarate and sensitivity to citrate was lost (Table 4). Fumarate specificity was already shown for the single mutant DcuS(Q159V).
Effector specificity of DcuS, DcuSDC, and DcuSCit for di- and tricarboxylates.
Sensitivity to C6-tricarboxylates (citrate, isocitrate, and tricarballylate) and C4-dicarboxylates was tested for citrate- and fumarate-specific forms of DcuS by comparing induction of the dcuB′-′lacZ reporter gene fusion (Fig. 4). In the strain with the DcuS wild type, citrate yielded the highest response among the tricarboxylates, followed by isocitrate. Tricarballylate showed no induction compared to background levels. Glutarate, in which two carboxylic groups are separated by three (CH2) groups, principally differs from citrate in that it lacks the central carboxylic group. It did not cause induction, in contrast to citrate and isocitrate. This suggested that the central carboxylate plays an important role in effector binding. Among the C4-dicarboxylates, induction by fumarate, aspartate, and d-tartrate was comparable to that by the structurally related 3-nitropropionate and significantly higher than that for citrate. Only mesaconate (2-methylfumarate) was a poor inducer, as previously reported (19).
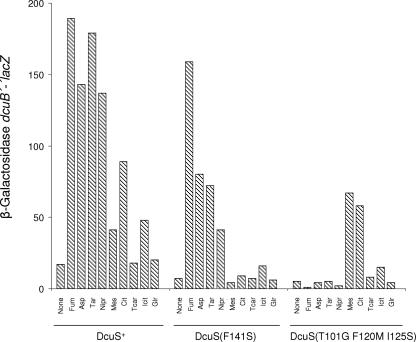
FIG. 4.
Effect of C4-dicarboxylates, C6-tricarboxylates, and related compounds on the expression of dcuB′-′lacZ in strains carrying wild-type DcuS, C4-dicarboxylate-specific DcuSDC(F141S), or citrate-specific DcuSCit(T101G F120M I125S). E. coli IMW260 (ΔdcuS) was transformed with low-copy plasmids containing alleles of dcuS encoding either DcuS, DcuSDC(F141S), or DcuSCit(T101G F120M). The bacteria were grown in the presence of the effectors (20 mM [5 mM for nitropropionate]), and β-galactosidase activity was measured at an OD578 of 0.5 to 0.8. The activities are the mean of at least six independent growth experiments and four repeats each. Fum, fumarate; Asp, aspartate; Tar, d-tartrate; Nipr, nitropropionate; Mes, mesaconate; Cit, citrate; Icit, isocitrate; Tcar; tricarballylate; Glr, glutarate.
In the C4-dicarboxylate-specific mutant DcuS(F141S), fumarate showed the strongest induction; induction by other C4-dicarboxylates was decreased significantly and was missing for mesaconate (2-methylfumarate) and the tricarboxylates (Fig. 4). The citrate-specific triple mutant DcuS(T101G F120M I125S) did not respond to the typical C4-dicarboxylates (fumarate, aspartate, and d-tartrate) or to 3-nitropropionate. While the mutant responded to citrate with high activities, the sensitivity to isocitrate was largely decreased (Fig. 4). However, induction by mesaconate was much higher than in the wild type. Thus, DcuSDC(F141S) and DcuSCit(T101G F120M I125S) represent C4-dicarboxylate- and citrate-specific sensors with complementary substrate specificity. Mesaconate, which is a C4-dicarboxylate with a bulky hydrophobic side group, behaves very similar to the tricarboxylates. It was the strongest inducer of citrate-specific DcuS(T101G F120M I125S).
The apparent KD values for the induction of dcuB′-′lacZ by maleate, succinate, and citrate with wild-type DcuS were about 2, 3, and 7 mM, respectively (19). The apparent KD values of mutants DcuSDC(T101G), DcuSDC(F149K), and DcuSCit(T101G F120M I125S) for the C4-dicarboxylates or citrate were similar and differed from the wild type maximally by factors of 3 (not shown).
Structural properties of the periplasmic domain of the triple mutant.
The periplasmic domain DcuS-PD(T101G F120M I125S)45-180 of the triple mutant was overproduced in the presence of [15N]NH4Cl. The purified domain of the mutated protein was used for measuring 15N-1H heteronuclear single-quantum correlation spectra (19, 23). The spectra showed a small chemical shift dispersion characteristic for unfolded protein. Since DcuS(T101G F120M I125S) is active in vivo, unfolding probably occurs in the separate domain, suggesting an increased lability of the mutated protein. Even the wild-type periplasmic domains of DcuS and even more of CitA (M. Sevvana, V. Vijayan, M. Zweckstetter, D. R. Madden, G. M. Sheldrick, M. Bott, C. Griesinger, and S. Becker, submitted for publication) are rather labile and show extended regions of slow chemical exchange. Therefore, it is not surprising that mutants further destabilize the periplasmic domain when it is in isolation while the conformational restriction through the interaction with the membrane in the full construct conveys sufficient stability.
Citrate-dependent regulation by DcuS: relationship to the citrate sensor system CitAB?
E. coli contains the citrate-specific CitAB two-component system, which plays a role in regulating anaerobic citrate fermentation (5, 17). CitAB induces in the presence of citrate the expression of the citCDEF gene cluster for citrate lyase (CitDEF) and citrate lyase ligase (CitC) (4, 5). Whether CitAB contributes to citrate regulation of dcuB′-′lacZ was tested. Expression and the response to citrate were studied in a series of mutants, each lacking a sensor kinase or regulator of one of the two systems (DcuS, DcuR, CitA, or CitB) (Table 5). Stimulation of dcuB expression by citrate was completely lost upon deletion of dcuS or dcuR, similar to that by fumarate, demonstrating the essential role of DcuS and DcuR for citrate induction of dcuB′-′lacZ. Genetic inactivation of citB, encoding the response regulator CitB, had only a small effect on dcuB expression during growth on citrate. In contrast, inactivation of the citrate histidine kinase CitA significantly decreased the expression of dcuB′-′lacZ in the presence of citrate. The effect of CitA may be direct by interaction of CitA with DcuS in the presence of citrate, or indirect, e.g., by affecting dcuSR expression. In the CitAB wild-type background, however, citrate regulation of dcuB′-′lacZ expression depends completely on DcuSR and therefore reflects changes in DcuS properties. Therefore, in the site-directed mutagenic studies of DcuS in the above paragraphs, expression of dcuB′-′lacZ reflects properties of DcuS.
TABLE 5.
Transcriptional regulation of the dcuB′-′lacZ reporter gene fusions by fumarate or citrate: involvement of the DcuSR and CitAB two-component systemsa
| Growth |
dcuB′-′lacZ fusion β-galactosidase activity (Miller units)
|
||||
|---|---|---|---|---|---|
| Wild type | Mutant
|
||||
| dcuS | dcuR | citA | citB | ||
| Glucose | 8 | 2 | 2 | 2 | NDb |
| Glucose + fumarate | 45 | 1 | 2 | 34 | 43 |
| Glucose + citrate | 30 | 2 | 2 | 9 | 26 |
Bacterial strains with a dcuB′-′lacZ reporter gene fusion in either a wild-type (IMW237), dcuS (IMW260), dcuR (IMW238), citA (IMW280), or citB (IMW239) background were grown anaerobically on glucose (20 mM) and fumarate (20 mM) or citrate (20 mM) as the inductors to an A578 of 0.5 to 0.8. β-Galactosidase activities were determined from three independent growth experiments. Activities are the average of at least six independent measurements (deviation, ≤12%).
ND, not determined.
For these reasons, DcuSR represents a second regulatory system for responding to citrate in E. coli. Despite the high apparent Kd value compared to the CitAB system, regulation by citrate via DcuSR might be physiologically relevant, since the Kd values are in the same range as those for C4-dicarboxylates. Citrate as an additional effector of DcuSR is plausible from a physiological view since citrate fermentation includes the reactions of fumarate respiration, which makes induction of the fumarate respiratory system by citrate sensible.
DISCUSSION
Citrate binding in DcuS via the C4-dicarboxylate-binding site.
DcuS functions as a citrate sensor with an apparent Kd that is only by a factor of 2 to 3 higher than that for C4-dicarboxylates. Binding of citrate requires the same sites that are known from fumarate sensing, namely sites C1 to C3 and H. However, the DcuS subtype-specific residues T101, I125, F141, and Q159 allow differentiation of C4-dicarboxylate from citrate binding, although the basis for the differences is not clear. From citrate, only the two neighboring (i.e., the central and one of the distal) carboxylate groups are essential, whereas the presence of the two distal groups is not sufficient. C2 and C3 are supposed to represent the sites for binding of the two essential carboxylate groups of citrate (and the two carboxylates of the C4-dicarboxylates), whereas sites C1 and H may have different functions (19). Thus, citrate and C4-dicarboxylates are recognized in a similar way involving the same principal sites in DcuS and the effector molecule. This type of citrate binding would be in contrast to that in CitA, with three sites for carboxylate and one site for hydroxyl binding, which are all specifically involved in citrate binding.
The third (distal) carboxylate of citrate apparently is not required for binding by DcuS but tolerated in the binding pocket, as demonstrated by the similarity of citrate and mesaconate binding and by the fact that citrate specificity is increased by changing sites C1/C2. Thus, the third carboxylate group might be close to the C1 site, which is conserved to a lesser extent and has to provide a pocket of sufficient size. The mutation and effector studies altogether suggest that citrate is bound in DcuS by the C4-dicarboxylate portion of the molecule. With the triple mutant DcuS(T101G F120M I125S), a citrate-specific form of DcuS was generated. Citrate binding by this form of DcuS, however, is still clearly different from that of CitA and is reminiscent of that of DcuS since it responds to mesaconate (2-methylfumarate) in addition to citrate. The mutant presumably has an optimized binding pocket at C1 which allows an improved placement of the bulky side groups in citrate (—CH2-COO−) or mesaconate (methyl group).
Different ways to generate C4-diarboxylate- and citrate-specific forms of DcuS.
C4-dicarboxylate-specific DcuSDC can be generated in several ways: first and most paradoxically by adapting the ligands in the carboxylate-binding sites to those of CitA and, second, by optimizing access to the binding pocket. In this way, DcuS is converted on the sequence level to a CitA-adapted protein, but, surprisingly, the changes resulted in a loss of citrate sensitivity and gain of C4-dicarboxylate specificity. In contrast, a form of DcuS with citrate specificity, DcuS(T101G F120M I125S), could only be generated by widening the size of the binding pocket around sites C1 and C2. This supports the view that citrate is bound by DcuS, similar to a C4-dicarboxylate, and binding of the residual “acetate” portion of citrate is improved by enlarging the binding pocket. These changes improve as well binding of the bulky methyl group in mesaconate. Structural studies are complicated by the structural lability of the periplasmic domain after separation from the holoprotein, resulting in unfolding or aggregation of the isolated periplasmic domains of all mutant forms of DcuS as shown here and earlier (19).
Phylogenetic relationships between DcuS and CitA.
The sequences of the periplasmic domains of DcuS and CitA proteins were compared by PSI-BLAST (1) and clustered using CLANS (9). Clustering at an E-value cutoff of 10−3 yielded a central group comprising the DcuS and CitA orthologs (Fig. 5). The nearest paralogous groups were formed by DctB and CreC histidine kinases, diguanylate cyclases, and chemoreceptors with periplasmic domains consisting of tandem PAS domains. Both the cyclase and chemoreceptor groups consist of putative, uncharacterized open reading frames, found primarily in environmental, phylogenetically diverse organisms such as Shewanella, Magnetospirillum, Desulfotalea, and Geobacter. DctB represents succinate (or C4-dicarboxylate)-responsive histidine kinases from aerobic gram-negative bacteria, such as Rhizobium or Sinorhizobium (11, 22, 24). In E. coli, the CreC histidine kinase (catabolite sensory kinase, or PhoM) responds to an unknown catabolite during growth of the bacteria on minimal media and it is thought to function as a central carbon regulator (2). Inspection of multiple alignments for the individual groups showed that proteins in the phylogenetically deepest branch of the DcuS/CitA group, formed by putative His kinases from actinobacteria, lack polar or charged residues at site C1 and thus resemble DcuS. The nearest paralogous group to DcuS and CitA, formed by putative chemoreceptors, also resembles DcuS in this respect. These observations imply that the polar residues at the C1 site of CitA represent a derived phenotype and suggest that the specific, high-affinity CitA binding site may have evolved from a binding site with less specificity and lower binding affinity, as is still present in DcuS.
Acknowledgments
We thank Johannes Söding for advice in alignment subtyping and Holger Scheib for modeling the DcuS mutants.
The work in the laboratory of G.U. was supported by grants from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avison, M. B., R. E. Horton, T. R. Walsh, and P. M. Bennett. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J. Biol. Chem. 276:26955-26961. [DOI] [PubMed] [Google Scholar]
- 3.Borgstahl, G. E., D. R. Williams, and E. D. Getzoff. 1995. 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 34:6278-6287. [DOI] [PubMed] [Google Scholar]
- 4.Bott, M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167:78-88. [PubMed] [Google Scholar]
- 5.Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18:533-546. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., C. Condon, B. D. Lemire, and J. H. Weiner. 1985. Molecular biology, biochemistry and bioenergetics of fumarate reductase, a complex membrane-bound iron-sulfur flavoenzyme of Escherichia coli. Biochim. Biophys. Acta 811:381-403. [DOI] [PubMed] [Google Scholar]
- 7.Do, C. B., M. S. Mahabhashyam, M. Brudno, and S. Batzoglou. 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 15:330-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farinha, M. A., and A. M. Kropinski. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 58:221-225. [DOI] [PubMed] [Google Scholar]
- 9.Frickey, T., and A. Lupas. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702-3707. [DOI] [PubMed] [Google Scholar]
- 10.Gerharz, T., S. Reinelt, S. Kaspar, L. Scapozza, and M. Bott. 2003. Identification of basic amino acid residues important for citrate binding by the periplasmic receptor domain of the sensor kinase CitA. Biochemistry 42:5917-5924. [DOI] [PubMed] [Google Scholar]
- 11.Giblin, L., J. Archdeacon, and F. O'Gara. 1996. Modular structure of the Rhizobium meliloti DctB protein. FEMS Microbiol. Lett. 139:19-25. [DOI] [PubMed] [Google Scholar]
- 12.Golby, P., S. Davies, J. R. Kelly, J. R. Guest, and S. C. Andrews. 1999. Identification and characterization of a two-component sensor kinase and response regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingmer, H., C. A. Miller, and S. N. Cohen. 1998. Destabilized inheritance of pSC101 and other Escherichia coli plasmids by DpiA, a novel two-component system regulator. Mol. Microbiol. 29:49-59. [DOI] [PubMed] [Google Scholar]
- 14.Janausch, I. G., I. Garcia-Moreno, and G. Unden. 2002. Function of DcuS from Escherichia coli as a fumarate-stimulated histidine protein kinase in vitro. J. Biol. Chem. 277:39809-39814. [DOI] [PubMed] [Google Scholar]
- 15.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kröger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 16.Kalinina, O. V., P. S. Novichkov, A. A. Mironov, M. S. Gelfand, and A. B. Rakhmaninova. 2004. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 32:W424-W428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaspar, S., and M. Bott. 2002. The sensor kinase CitA (DpiB) of Escherichia coli functions as a high-affinity citrate receptor. Arch. Microbiol. 177:313-321. [DOI] [PubMed] [Google Scholar]
- 18.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, L. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33:858-872. [DOI] [PubMed] [Google Scholar]
- 19.Kneuper, H., I. G. Janausch, V. Vijayan, M. Zweckstetter, V. Bock, C. Griesinger, and G. Unden. 2005. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J. Biol. Chem. 280:20596-20603. [DOI] [PubMed] [Google Scholar]
- 20.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Nan, B., Y. Zhou, Y. H. Liang, J. Wen, Q. Ma, S. Zhang, Y. Wang, and X. D. Su. 2006. Purification and preliminary X-ray crystallographic analysis of the ligand-binding domain of Sinorhizobium meliloti DctB. Biochim. Biophys. Acta 1764:839-841. [DOI] [PubMed] [Google Scholar]
- 23.Pappalardo, L., I. G. Janausch, V. Vijayan, E. Zientz, J. Junker, W. Peti, M. Zweckstetter, G. Unden, and C. Griesinger. 2003. The NMR structure of the sensory domain of the membraneous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 178:39185-39188. [DOI] [PubMed] [Google Scholar]
- 24.Reid, C. J., and P. S. Poole. 1998. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J. Bacteriol. 180:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinelt, S., E. Hofmann, T. Gerharz, M. Bott, and D. R. Madden. 2003. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 278:39189-39196. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 27.Silhavy, T., J. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Unden, G., and A. Kleefeld. July 2004. C4-dicarboxylate degradation in aerobic and anaerobic growth. Chapter 3.4.5. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org. [DOI] [PubMed]
- 29.Zientz, E., J. Bongaerts, and G. Unden. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR) two-component regulatory system. J. Bacteriol. 180:5421-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]