Abstract
Streptomyces coelicolor contains paralogous versions of seven ribosomal proteins (S14, S18, L28, L31, L32, L33, and L36), which differ in their potential to bind structural zinc. The paralogues are termed C+ or C− on the basis of the presence or absence of putative cysteine ligands. Here, mutational studies suggest that the C− version of L31 can functionally replace its C+ paralogue only when expressed at an artificially elevated level. We show that the level of expression of four transcriptional units encoding C− proteins is elevated under conditions of zinc deprivation. Zur controls the expression of three transcriptional units (including rpmG2, rpmE2, rpmB2, rpsN2, rpmF2, and possibly rpsR2). Zur also controls the expression of the znuACB operon, which is predicted to encode a high-affinity zinc transport system. Surprisingly, the zinc-responsive control of the rpmG3-rpmJ2 operon is dictated by σR, a sigma factor that was previously shown to control the response to disulfide stress in S. coelicolor. The induction of σR activity during zinc limitation establishes an important link between thiol-disulfide metabolism and zinc homeostasis.
Bacterial 70S ribosomes are assembled on mRNA from 30S and 50S subunits during the process of translation initiation. The ribosome is approximately two-thirds RNA and one-third protein. The 30S subunit usually contains 21 proteins and 16S rRNA, whereas the 50S subunit comprises up to 36 proteins together with 23S and 5S rRNA. A major role of ribosomal proteins (R proteins) is to stabilize the rRNA (31), although several proteins play additional roles in ribosome function (8). The production of ribosomes is a major drain on cellular energy and material resources, and so the synthesis of ribosomal components is tightly controlled in most, if not all, bacteria. rRNA synthesis is the major control point, and various mechanisms, for example, ppGpp-dependent stringent control, ensure that synthesis is commensurate with growth requirements and nutrient availability (45). The control of R-protein gene transcription is also subject to stringent control (9, 11), although feedback posttranscriptional control of translation initiation is an additional important control mechanism (12).
Although most bacteria contain multiple copies of rRNA operons, the ∼55 R proteins are usually encoded by single, highly conserved genes. However, it was recently discovered that this is not the case for all R proteins. Seven R proteins are found in phylogenetically distinct forms both among different bacteria and within single genomes. These proteins are L31 (rpmE), L32 (rpmF), L33 (rpmG), L36 (rpmJ), L28 (rpmB), S18 (rpsR), and S14 (rpsN) (30). Strikingly, the two forms of each protein differ by the presence or absence of predicted (and in the cases of L31, L36, and S14, proven [20, 32, 50]) Zn ribbon motifs that usually consist of two pairs of conserved cysteines. The presence or absence of cysteine zinc ligands in these R proteins has led to their designation as C+ or C−, respectively, and these proteins are known collectively as the C+/− group (30). It was proposed previously that an ancient duplication of the ancestral C+ form of each gene was followed by the evolution of C− forms and the subsequent alternative loss of C+ or C− forms in different lineages (30). However, some genomes maintain both C+ and C− forms of certain R-protein genes, in which case the protein products are considered paralogues. For example, the Escherichia coli genome encodes mostly C− R proteins but encodes both C+ and C− forms of L31, whereas Bacillus subtilis uses either C+ or C− R proteins in most cases but contains both forms of S14, L31, and L33 (30). In rapidly growing cells, ribosomes that contain up to seven C+ R proteins clearly place a major demand on zinc resources. It seems likely, therefore, that zinc limitation was a driving force behind the evolution of the C− R proteins (42). Indeed, bioinformatic analyses of the promoter regions of paralogous C− R-protein genes suggested that they might be controlled by zinc-responsive regulators in several bacteria (42). Recently, studies with B. subtilis (2, 32) confirmed that zinc deficiency stimulates the expression of the C− gene rpmE2 and the incorporation of  into ribosomes.
into ribosomes.
Although zinc is an essential metal in bacteria, high levels can be toxic. In E. coli, the levels of free zinc are therefore tightly controlled by the action of the zinc-responsive regulators Zur and ZntR, which control uptake and export systems, respectively (7, 15, 44). These regulators have extremely high affinities for zinc, implying that there is essentially no free zinc in the cell (34). The question of how zinc metalloproteins acquire their zinc despite the absence of free zinc in the cytoplasm thus remains a crucial question.
Our studies of the antibiotic-producing actinomycete Streptomyces coelicolor have revealed that rpmE1, which encodes 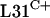 , is transcribed from two promoters. The rpmE1-proximal promoter rpmE1p1 is active during normal growth, whereas the rpmE1-distal promoter rpmE1p2 is controlled by the disulfide stress response sigma factor σR (41). The genome sequence of S. coelicolor (4) revealed two genes predicted to encode C− forms of L31 (rpmE2 and rpmE3), which prompted our investigation into the functional roles of the three L31-encoding genes. In this paper, we show that rpmE2 and rpmE3 are alternative, nonessential genes and provide evidence that rpmE1 is essential. We show that S. coelicolor contains paralogous versions of all seven C+/− R proteins and that the C− genes are controlled by zinc availability. The major regulator is the zinc-responsive regulator Zur, although we also show that the zinc-dependent control of two C− R-protein genes is dependent on σR. Our data reveal regulatory interplay between the responses to zinc deprivation and disulfide stress.
, is transcribed from two promoters. The rpmE1-proximal promoter rpmE1p1 is active during normal growth, whereas the rpmE1-distal promoter rpmE1p2 is controlled by the disulfide stress response sigma factor σR (41). The genome sequence of S. coelicolor (4) revealed two genes predicted to encode C− forms of L31 (rpmE2 and rpmE3), which prompted our investigation into the functional roles of the three L31-encoding genes. In this paper, we show that rpmE2 and rpmE3 are alternative, nonessential genes and provide evidence that rpmE1 is essential. We show that S. coelicolor contains paralogous versions of all seven C+/− R proteins and that the C− genes are controlled by zinc availability. The major regulator is the zinc-responsive regulator Zur, although we also show that the zinc-dependent control of two C− R-protein genes is dependent on σR. Our data reveal regulatory interplay between the responses to zinc deprivation and disulfide stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. coelicolor A3(2) strains were cultivated on minimal medium, R5, supplemented minimal medium (SMMS) and mannitol-soya agars and modified liquid minimal medium (NMMP) as described previously (24). For liquid growth, strains were grown to mid- to late exponential phase in NMMP by using glucose as the carbon source. Zinc deficiency was achieved by using NMMP without added zinc, along with ultrapure reagents and acid-treated plasticware. Presumably, trace amounts of zinc remained under these conditions because there was no obvious effect on the growth rate up to at least mid- to late exponential phase. ZnSO4 was added as indicated in the figure legends at concentrations of up to 3.5 μM, the level normally added in the preparation of NMMP medium. A list of strains is provided in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3)pLysS | Overexpression host | 48 |
| ET12567(pUZ8002) | Nonmethylating donor for E. coli-Streptomyces conjugations | 38 |
| S. coelicolor strains | ||
| M145 | A3(2) SCP1− SCP2− | 24 |
| M600 | A3(2) SCP1− SCP2− | 10 |
| J1915 | M145 ΔglkA119 SCP1− SCP2− | 23 |
| J2139 | M600 ΔsigR | 40 |
| J1915(pSX110-X1) | J1915 ΔrpmE1; single-crossover recombinant | This study |
| S103 | J1915 ΔrpmE2 | This study |
| S116 | M600 ΔrpmE3::apr | This study |
| S117 | J1915 ΔrpmE2 ΔrpmE3::apr | This study |
| S118(pSX125) | J1915 ΔrpmE1(pSX125) | This study |
| S118(pSX122) | J1915 ΔrpmE1(pSX122) | This study |
| S118(pSX128) | J1915 ΔrpmE1(pSX128) | This study |
| S121 | M145 Δzur::apr | This study |
| Plasmids | ||
| pBluescript II SK(+) | Multicopy cloning vector (bla lacZa) | Stratagene |
| pHJL401 | SCP2-derived bifunctional plasmid with low copy number (bla tsr lacZa) | 25 |
| pIJ6650 | Conjugative, counterselectable suicide plasmid (apr glkA lacZa oriT) | 37 |
| pMT3000 | pUC18-based cloning vector (bla lacZa) | 39 |
| pSET152 | Conjugative, integrative vector (apr lacZa oriT ΦC31 attP); ΦC31 based | 5 |
| pET20b | E. coli T7-based expression vector (bla) | Novagen |
| pSX110 | pIJ6650 into which 2.5-kb ΔrpmE1 allele has been cloned | This study |
| pSX121 | pIJ6650 into which 3-kb ΔrpmE2 allele has been cloned | This study |
| pSX122 | pSET152::rpmE1 | This study |
| pSX125 | pHJL401::rpmE1 | This study |
| pSX128 | pSET152::Φ(rpmE1′-rpmE2) | This study |
Construction of plasmids.
rpmE1, along with 158 bp of upstream DNA including the two native promoters (41), was amplified by PCR by using the primers rpmE5 and rpmE7 (Table 2) and then cloned into EcoRI-BamHI-digested pSET152 and pHJL401 to give pSX122 and pSX125, respectively. To construct pSX128, rpmE2 was amplified by PCR and then cloned into EcoRV-digested pBluescript II SK(+) to give pSX126. rpmE1, along with its upstream transcriptional-translational control region (159 bp), was isolated by PCR as an EcoRI fragment and subcloned into EcoRI-digested pSX126. In this construct, rpmE1 is adjacent to, and in the same orientation as, rpmE2. Inverse PCR mutagenesis with the primers rpmE2-P2-1 and rpmE2-P2-2 (Table 2) was used to fuse the promoters and first four residues of rpmE1 to rpmE2 at the fifth codon (see Fig. 2). The rpmE1-rpmE2 transcriptional-translational fusion, Φ(rpmE1′-rpmE2), was subcloned as an EcoRI-BamHI fragment into pSET152 to give pSX128.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| rpmE5 | 5′ GGGGAATTCCGAGGCGAAGGGCGTCGTCAAC |
| rpmE7 | 5′ CGGGGATCCTGCTACTGCGTGGGAAAGG |
| rpmE2-P2-1 | 5′ CAAGGTGTCTCCTAGTGTTCC |
| rpmE2-P2-2 | 5′ AAGCGCGACATCCACCCCGAGTACCGCCCGGTC |
| Zur-F | 5′ CCGAATTCATATGACCACCGCTGGACCGCCCGTG |
| Zur-R | 5′ CCGGATCCTCATCAACCGCCGGAGGCCCCCGC |
| znuAF1 | 5′ CGACGTCGAACTTGTCGGTGTTGC |
| znuAR1 | 5′ CGAGATGCTTCCAGAGAATGCTTCAG |
| F2S1a | 5′ ACCGGCACACAGCGGCGATGA |
| F2S1b | 5′ ACCGTCACGGGCACCAGATTC |
| G3S1b | 5′ CAGCCGGTCGGGGTCGTTGCGGC |
| G3S1c | 5′ GTACCGCGCGACGATCTCCCGG |
FIG. 2.
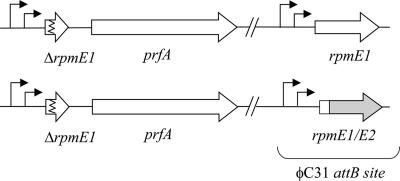
Expression of rpmE2 in association with the transcription and translation initiation signals of rpmE1. The Φ(rpmE1′-rpmE2) fusion [bottom; strain S118(pSX128)] or the rpmE1 control [top; strain S118(pSX122)] gene was introduced into the chromosome by using the integrative vector pSET152, which integrates at the ΦC31 attB site. Both constructs allowed the in-frame deletion of rpmE1 at the natural locus.
Construction of mutant strains.
An in-frame ΔrpmE1 allele was constructed by amplifying 1.3-kb upstream and downstream rpmE1-flanking regions by PCR and then combining the fragments into the cloning vector pMT3000. This procedure resulted in an in-frame rpmE1 allele containing three codons corresponding to the N terminus of the product, two codons corresponding to the C terminus, a central EcoRI site, and flanking BamHI-HindIII sites. The ∼2.5-kb ΔrpmE1 allele was subcloned into the counterselectable gene replacement vector pIJ6650 to give pSX110 and then introduced into S. coelicolor J1915 by conjugation from E. coli ET12567(pUZ8002) (24). A single-crossover recombinant was isolated and designated J1915(pSX110-X1). Double-crossover ΔrpmE1 mutants could not be isolated.
An in-frame ΔrpmE2 allele was constructed by amplifying ∼1.5-kb flanking regions and combining them into pMT3000 as described above, giving a fragment with 4 codons corresponding to the N terminus of the product, 21 codons corresponding to the C terminus, a central BamHI restriction site, and flanking BglII sites. The 3-kb BglII fragment was subcloned into pIJ6650 to give pSX121 and then introduced by conjugation into S. coelicolor J1915. Initial exconjugants were subjected to counterselection by using 2-deoxyglucose (2-DOG) to remove the integrated plasmid and were screened for the presence of the ΔrpmE2 allele by PCR; a ΔrpmE2 mutant was designated S103.
ΔrpmE3::apr and Δzur::apr alleles were constructed in the cosmids StG8A and StC121 (46), respectively, by using a PCR-mediated mutagenesis approach (18). Briefly, in each case a transferable apr-oriT resistance cassette was amplified from pIJ773 (18) by using primers designed to delete the DNA between the start and stop codons of each gene upon the recombination of the PCR product into the cosmid. The ΔrpmE3::apr mutant allele was recombined by a double-crossover event into the M600 genome to generate S116 and into the S103 genome to generate S117. The Δzur::apr allele was recombined by a double-crossover event into the genome of M145 to generate a strain designated S121.
RNA preparation and transcription analysis.
RNA was isolated from liquid-grown mycelium by using the Kirby mix method as described previously (24). Probes for S1 nuclease mapping were generated by PCR, generally by using a primer corresponding to an internal region of the open reading frame (ORF) and labeled at the 5′ end with [γ-32P]ATP and an unlabeled primer that hybridizes ∼400 bp upstream of the ORF. Labeled probes were purified by using a QIAGEN PCR purification kit and then hybridized with 30 μg of total RNA as described previously (24). Following S1 nuclease treatment, protected DNA fragments were separated by denaturing electrophoresis and visualized by using a phosphorimager (Storm; GE Healthcare).
Overproduction and purification of Zur.
The primers Zur-F and Zur-R (Table 2) were used to amplify the zur ORF by PCR, thereby generating an NdeI site corresponding to the N terminus of the gene product and a BamHI site corresponding to the C terminus for cloning into pET20b (Novagen). pET20b::zur was introduced into E. coli BL21(DE3)pLysS, and zur overexpression was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. Cell pellets in buffer A (20 mM Tris-HCl [pH 8], 50 mM NaCl, 5% glycerol, 1 mM dithiothreitol, 1 μM ZnCl2, complete mini EDTA-free proteinase inhibitor cocktail [Roche]) were disrupted by using a French press. Following the removal of cell debris by centrifugation, the soluble extract was subjected to ion exchange chromatography with a HiPrep 16/10 QFF column (GE Healthcare), heparin affinity chromatography with a HiPrep 16/10 heparin FF column (GE Healthcare), and gel filtration with a Sephadex 200 column (GE Healthcare), which resulted in ∼95% pure protein. Following the concentration of Zur to ∼1.4 mg/ml, purified protein in buffer B (20 mM Tris-HCl [pH 8], 400 mM NaCl, 5% glycerol, 5 mM dithiothreitol, 1 μM ZnCl2) was snap-frozen in small aliquots and stored at −80°C.
EMSAs and DNase I footprint assays.
Probes that included the promoter region of znuA, rpmF2, and the intergenic region of a cluster of seven genes (SCO3425 to SCO3431) that includes five C− R-protein genes, referred to herein as the 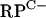 cluster, were generated by PCR by using the primer pairs znuAF1-znuAR1 (252 bp), F2S1a-F2S1b (203 bp), and G3S1b-G3S1c (472 bp), respectively (Table 2), and then 5′ end labeled with [γ-32P]ATP. Electrophoretic mobility shift assay (EMSA) mixtures contained the following in a final volume of 10 μl: DNA probe (<1 nM), binding buffer (10 mM Tris-HCl [pH 8.0], 5% [vol/vol] glycerol, 50 mM NaCl, 1 μM ZnCl2, 2 μg of bovine serum albumin), purified Zur, and 1 μg of herring sperm DNA (Promega). Following 15 min of incubation at room temperature, the binding reaction mixtures were separated on a 6% Tris-borate (no EDTA) polyacrylamide gel at 4°C. Bands were visualized by using a phosphorimager (Storm; GE Healthcare). Experiments were performed at least twice, and representative data sets are presented.
cluster, were generated by PCR by using the primer pairs znuAF1-znuAR1 (252 bp), F2S1a-F2S1b (203 bp), and G3S1b-G3S1c (472 bp), respectively (Table 2), and then 5′ end labeled with [γ-32P]ATP. Electrophoretic mobility shift assay (EMSA) mixtures contained the following in a final volume of 10 μl: DNA probe (<1 nM), binding buffer (10 mM Tris-HCl [pH 8.0], 5% [vol/vol] glycerol, 50 mM NaCl, 1 μM ZnCl2, 2 μg of bovine serum albumin), purified Zur, and 1 μg of herring sperm DNA (Promega). Following 15 min of incubation at room temperature, the binding reaction mixtures were separated on a 6% Tris-borate (no EDTA) polyacrylamide gel at 4°C. Bands were visualized by using a phosphorimager (Storm; GE Healthcare). Experiments were performed at least twice, and representative data sets are presented.
For DNase I footprinting, the 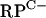 cluster, rpmF2, and znuA templates were prepared by PCR by using the oligonucleotides described above. To label one strand, the primers G3S1b (with a sequence corresponding to an internal region of rpmG2), F2S1a (with a sequence corresponding to a region upstream from rpmF2), and znuAR1 (with a sequence corresponding to an internal region of znuA) were end labeled with [γ-32P]ATP. Binding reactions were performed with a final volume of 25 μl by using 30 ng of template DNA and various concentrations of Zur. DNase I digestion was carried out according to the method of Takano et al. (49). To determine the extent of protection conferred by Zur, dideoxynucleotide sequencing reactions were performed by using a Sequenase 2.0 DNA sequencing kit (GE Healthcare) and the above-described 5′-end-labeled primers.
cluster, rpmF2, and znuA templates were prepared by PCR by using the oligonucleotides described above. To label one strand, the primers G3S1b (with a sequence corresponding to an internal region of rpmG2), F2S1a (with a sequence corresponding to a region upstream from rpmF2), and znuAR1 (with a sequence corresponding to an internal region of znuA) were end labeled with [γ-32P]ATP. Binding reactions were performed with a final volume of 25 μl by using 30 ng of template DNA and various concentrations of Zur. DNase I digestion was carried out according to the method of Takano et al. (49). To determine the extent of protection conferred by Zur, dideoxynucleotide sequencing reactions were performed by using a Sequenase 2.0 DNA sequencing kit (GE Healthcare) and the above-described 5′-end-labeled primers.
RESULTS
S. coelicolor encodes C+ and C− versions of seven ribosomal proteins.
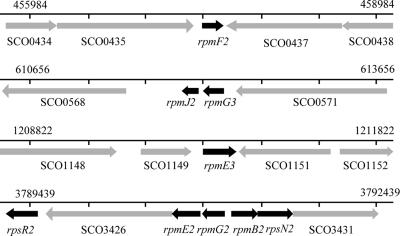
An analysis of the annotated S. coelicolor genome sequence revealed that both C+ and C− forms of all seven of the C+/− group R proteins are encoded (Table 3). In most cases, a single C+ and a single C− gene are present. However, pairs of closely related C− genes for L31 and L33 are present, along with single C+ genes for these proteins. We hypothesized that, under standard growth conditions, the C+ protein forms would act as the primary R proteins, a prediction that is supported by the genomic context of each gene. Of the seven C+ genes, six are closely linked to other genes with predicted essential roles in translation (data not shown). In contrast, many of the C− R-protein genes are linked to other C− R-protein genes but not to other genes involved in translation. The C− R-protein genes are organized among four loci (Fig. 1). Of particular interest is the 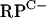 cluster. As well as the C− R-protein genes, the
cluster. As well as the C− R-protein genes, the 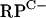 cluster includes SCO3426, encoding a protein related to YciC from B. subtilis (28% identity), which is thought to play a role in zinc homeostasis (13, 15). The remaining gene in the
cluster includes SCO3426, encoding a protein related to YciC from B. subtilis (28% identity), which is thought to play a role in zinc homeostasis (13, 15). The remaining gene in the 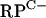 cluster, SCO3431, lies downstream of rpsN2 and encodes a putative membrane-spanning protein with no predicted function. The annotated start codon of SCO3431 overlaps with the stop codon of rpsN2, although note that an alternative start codon is located 60 bp downstream, which is consistent with the proposed start codon in Streptomyces avermitilis. It is therefore presently unclear whether rpsN2 and SCO3431 are cotranscribed. Elsewhere in the genome, two further C− genes (rpmG3 and rpmJ2) are organized as a likely operon (SCO0569-SCO0570), and rpmE3 may be organized in an operon downstream from a gene with no known function. Finally, rpmF2 appears to be transcribed as a monocistronic mRNA (see below). The sequences of the seven types of S. coelicolor putative C− R protein are aligned with those of their C+ counterparts from S. coelicolor as well as those of homologues from other bacteria in Fig. S1 in the supplemental material.
cluster, SCO3431, lies downstream of rpsN2 and encodes a putative membrane-spanning protein with no predicted function. The annotated start codon of SCO3431 overlaps with the stop codon of rpsN2, although note that an alternative start codon is located 60 bp downstream, which is consistent with the proposed start codon in Streptomyces avermitilis. It is therefore presently unclear whether rpsN2 and SCO3431 are cotranscribed. Elsewhere in the genome, two further C− genes (rpmG3 and rpmJ2) are organized as a likely operon (SCO0569-SCO0570), and rpmE3 may be organized in an operon downstream from a gene with no known function. Finally, rpmF2 appears to be transcribed as a monocistronic mRNA (see below). The sequences of the seven types of S. coelicolor putative C− R protein are aligned with those of their C+ counterparts from S. coelicolor as well as those of homologues from other bacteria in Fig. S1 in the supplemental material.
TABLE 3.
S. coelicolor C+ R proteins and putative C− R proteins
| R protein | C+ gene (gene no.) | C− gene (gene no.) |
|---|---|---|
| S14 | rpsN1 (SCO4715) | rpsN2 (SCO3430) |
| S18 | rpsR1 (SCO3908) | rpsR2 (SCO3425) |
| L28 | rpmB1 (SCO5564) | rpmB2 (SCO3429) |
| L31 | rpmE1 (SCO5359) | rpmE2 (SCO3427) |
| rpmE3 (SCO1150) | ||
| L32 | rpmF1 (SCO5571) | rpmF2 (SCO0436) |
| L33 | rpmG1 (SCO4635) | rpmG2 (SCO3428) |
| rpmG3 (SCO0570) | ||
| L36 | rpmJ1 (SCO4726) | rpmJ2 (SCO0569) |
FIG. 1.
Genome organization of the genes that encode C− R proteins in S. coelicolor. In each case, a ∼3-kb region of the annotated genome is shown. Arrows indicate the directions of transcription. Black arrows represent the C− R-protein genes.
rpmE1 appears to be essential.
Our initial functional studies of the C+/− group of R proteins focused on the three genes that encode L31 paralogues. The gene that encodes L31, rpmE, appears on several compiled lists of essential bacterial genes (16). However, it was conceivable that each rpmE gene in S. coelicolor might be individually redundant if the 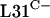 and
and 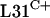 paralogues were functionally equivalent. To test this, an attempt was made to delete the
paralogues were functionally equivalent. To test this, an attempt was made to delete the 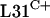 gene, rpmE1. An rpmE1 in-frame deletion allele was recombined via a single crossover into the S. coelicolor J1915 (ΔglkA) genome to generate a strain designated J1915(pSX110-X1). pSX110 contains the glucose kinase gene glkA, which enables the positive selection of double-crossover recombinants on medium containing the toxic glucose analogue 2-DOG (24). More than 20 double-crossover isolates resistant to 2-DOG and sensitive to apramycin were screened for the presence of the ΔrpmE1 allele by Southern analysis or PCR, but each had reverted to the wild type, suggesting that rpmE1 is essential. In support of this conclusion, the introduction of a functional copy of rpmE1 into J1915(pSX110-X1) on a low-copy-number plasmid (pSX125) permitted the subsequent replacement of the chromosomal wild-type gene with the ΔrpmE1 allele. The resulting strain was designated S118(pSX125), and the chromosomal structure was confirmed by Southern blot analysis (data not shown). These data imply that under the growth conditions used, rpmE2 and rpmE3 cannot compensate for the loss of rpmE1.
gene, rpmE1. An rpmE1 in-frame deletion allele was recombined via a single crossover into the S. coelicolor J1915 (ΔglkA) genome to generate a strain designated J1915(pSX110-X1). pSX110 contains the glucose kinase gene glkA, which enables the positive selection of double-crossover recombinants on medium containing the toxic glucose analogue 2-DOG (24). More than 20 double-crossover isolates resistant to 2-DOG and sensitive to apramycin were screened for the presence of the ΔrpmE1 allele by Southern analysis or PCR, but each had reverted to the wild type, suggesting that rpmE1 is essential. In support of this conclusion, the introduction of a functional copy of rpmE1 into J1915(pSX110-X1) on a low-copy-number plasmid (pSX125) permitted the subsequent replacement of the chromosomal wild-type gene with the ΔrpmE1 allele. The resulting strain was designated S118(pSX125), and the chromosomal structure was confirmed by Southern blot analysis (data not shown). These data imply that under the growth conditions used, rpmE2 and rpmE3 cannot compensate for the loss of rpmE1.
rpmE2 and rpmE3 are nonessential.
To investigate the functions of rpmE2 and rpmE3, single and double mutants were constructed. An in-frame ΔrpmE2 allele was introduced into the J1915 genome to generate a strain designated S103, an rpmE3::apr allele was recombined into the S. coelicolor M600 genome to generate S116, and a ΔrpmE2 ΔrpmE3::apr mutation in the J1915 background was created to generate S117. The three strains, S103, S116, and S117, were easily obtained and appeared to be identical to their parental counterparts in terms of growth, sporulation, and antibiotic production. In addition, no mutant appeared to be disadvantaged compared to the parental strains when growing in medium lacking added zinc. We therefore conclude that rpmE1 encodes the primary, essential L31 R protein in S. coelicolor and that rpmE2 and rpmE3 are nonessential. A similar conclusion was reached for B. subtilis (2).
Expression of rpmE2 partially rescues an rpmE1 null mutant.
The failure of rpmE2 or rpmE3 to suppress a ΔrpmE1 mutation might be because 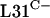 and
and 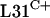 are not functionally equivalent or because rpmE2 and rpmE3 are not expressed at levels sufficient for ribosome biogenesis. To test the latter possibility, we constructed a transcriptional-translational fusion between rpmE1 and rpmE2. The rpmE2 reading frame, from codon 5 onward, was fused to rpmE1 after the fourth codon in the integrative plasmid pSX128. The transcriptional-translational fusion, Φ(rpmE1′-rpmE2), was introduced into S. coelicolor S118(pSX125), which allowed the subsequent loss of pSX125 (pHJL401::rpmE1). As a control, pSX122, which includes rpmE1 along with its promoter region, was integrated into the S118 genome (Fig. 2). S118(pSX122) was indistinguishable from the parental strain J1915, whereas S118(pSX128) grew and sporulated poorly and formed irregular colonies. In addition, S118(pSX128) overproduced the pigmented antibiotic actinorhodin on SMMS agar medium. Therefore, it appears that rpmE2 cannot fully compensate for the loss of rpmE1, even when expressed in association with the rpmE1 transcription and translation initiation signals. Nonetheless, these data suggest that
are not functionally equivalent or because rpmE2 and rpmE3 are not expressed at levels sufficient for ribosome biogenesis. To test the latter possibility, we constructed a transcriptional-translational fusion between rpmE1 and rpmE2. The rpmE2 reading frame, from codon 5 onward, was fused to rpmE1 after the fourth codon in the integrative plasmid pSX128. The transcriptional-translational fusion, Φ(rpmE1′-rpmE2), was introduced into S. coelicolor S118(pSX125), which allowed the subsequent loss of pSX125 (pHJL401::rpmE1). As a control, pSX122, which includes rpmE1 along with its promoter region, was integrated into the S118 genome (Fig. 2). S118(pSX122) was indistinguishable from the parental strain J1915, whereas S118(pSX128) grew and sporulated poorly and formed irregular colonies. In addition, S118(pSX128) overproduced the pigmented antibiotic actinorhodin on SMMS agar medium. Therefore, it appears that rpmE2 cannot fully compensate for the loss of rpmE1, even when expressed in association with the rpmE1 transcription and translation initiation signals. Nonetheless, these data suggest that 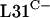 encoded by rpmE2 is able to partially provide the essential L31 function. The ability of rpmE3-encoded
encoded by rpmE2 is able to partially provide the essential L31 function. The ability of rpmE3-encoded 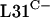 to functionally replace
to functionally replace 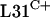 remains unknown.
remains unknown.
Zinc regulation of C− R-protein genes.
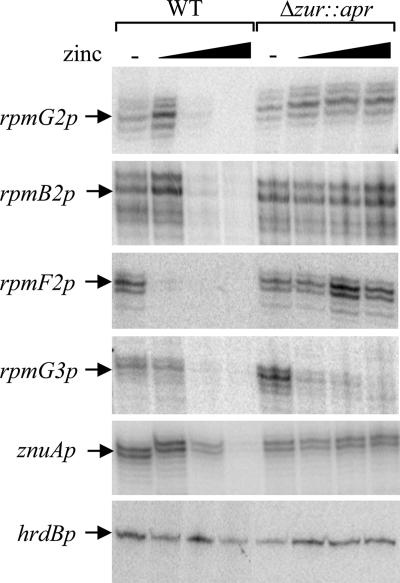
Initial S1 nuclease mapping experiments using RNA extracted from NMMP-grown cultures to investigate the divergent 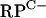 cluster promoters (rpmG2p and rpmB2p), as well as the promoter regions upstream of rpmG3, rpmE3, and rpmF2, failed to reveal transcription activity. To test the hypothesis that C− R proteins might replace C+ proteins during zinc depletion, RNA was extracted from S. coelicolor M145 grown in NMMP from which ZnSO4 had been initially omitted and then added back in at concentrations in the range of 0 to 3.5 μM. Transcripts were detected for four of the five promoter regions under conditions of zinc depletion but were absent at higher levels of zinc. For the leftward arm of the
cluster promoters (rpmG2p and rpmB2p), as well as the promoter regions upstream of rpmG3, rpmE3, and rpmF2, failed to reveal transcription activity. To test the hypothesis that C− R proteins might replace C+ proteins during zinc depletion, RNA was extracted from S. coelicolor M145 grown in NMMP from which ZnSO4 had been initially omitted and then added back in at concentrations in the range of 0 to 3.5 μM. Transcripts were detected for four of the five promoter regions under conditions of zinc depletion but were absent at higher levels of zinc. For the leftward arm of the 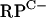 cluster, a single rpmG2 promoter was detected, with the +1 transcriptional start site coinciding with the first base of the rpmG2 start codon (Fig. 3; also, see Fig. 6). The coincidence of transcription and translation start sites is generally a rare phenomenon in bacteria but appears to be quite prevalent in Streptomyces (21). For the rightward arm of the
cluster, a single rpmG2 promoter was detected, with the +1 transcriptional start site coinciding with the first base of the rpmG2 start codon (Fig. 3; also, see Fig. 6). The coincidence of transcription and translation start sites is generally a rare phenomenon in bacteria but appears to be quite prevalent in Streptomyces (21). For the rightward arm of the 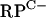 cluster, two closely spaced 5′ ends were detected ∼70 bp upstream of the rpmB2 start codon. The rpmG2 and rpmB2 transcript 5′ ends are ∼20 bp apart, indicating that the promoters overlap (Fig. 3; also, see Fig. 6). The rpmG3-rpmJ2 operon was induced under conditions of zinc depletion from a single promoter located ∼60 bp upstream of the start codon (Fig. 3; also, see Fig. 6). Finally, rpmF2 was induced under conditions of zinc depletion from a single promoter located 22 bp upstream of the start codon. The remaining C− R-protein gene, rpmE3, was not expressed under any of the growth conditions tested.
cluster, two closely spaced 5′ ends were detected ∼70 bp upstream of the rpmB2 start codon. The rpmG2 and rpmB2 transcript 5′ ends are ∼20 bp apart, indicating that the promoters overlap (Fig. 3; also, see Fig. 6). The rpmG3-rpmJ2 operon was induced under conditions of zinc depletion from a single promoter located ∼60 bp upstream of the start codon (Fig. 3; also, see Fig. 6). Finally, rpmF2 was induced under conditions of zinc depletion from a single promoter located 22 bp upstream of the start codon. The remaining C− R-protein gene, rpmE3, was not expressed under any of the growth conditions tested.
FIG. 3.
Zinc-dependent control of the rpmG2, rpmB2, rpmF2, rpmG3, and znuA promoters and possible involvement of zur in regulation. Wild-type (WT; M145) and Δzur::apr (S121) strains were grown in NMMP liquid medium containing various concentrations of added ZnSO4 (0 [−], 0.9, 1.7, and 3.5 μM) to mid- to late exponential phase prior to RNA extraction. Transcripts derived from each promoter were assayed by S1 nuclease mapping by using promoter-specific probes radiolabeled at one end. Constitutive hrdBp controls the expression of the principle sigma factor σHrdB and was used as a positive control. Note that the zinc-dependent regulation of rpmG3 is maintained in S121.
FIG. 6.
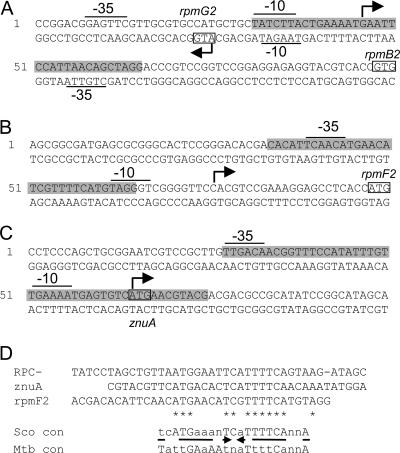
Transcriptional organization and Zur binding site locations in the rpmG2, rpmB2, rpmF2, and znuA promoter regions. Transcriptional start points are indicated by arrows, and the putative −10 and −35 promoter elements are underlined. Boxes indicate predicted translation start points. The extents of Zur protection on the labeled strands are indicated by gray highlighting. (A) 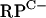 cluster intergenic region. (B) rpmF2 promoter region. (C) znuA promoter region. (D) Alignment of Zur binding sites in S. coelicolor, indicating a derived consensus sequence (Sco con), and comparison of this sequence to the consensus binding site sequence for M. tuberculosis Zur (Mtb con) (29). Asterisks and uppercase letters in the consensus sequences correspond to nucleotides that are identical among all the Zur binding sites analyzed; n represents any nucleotide.
cluster intergenic region. (B) rpmF2 promoter region. (C) znuA promoter region. (D) Alignment of Zur binding sites in S. coelicolor, indicating a derived consensus sequence (Sco con), and comparison of this sequence to the consensus binding site sequence for M. tuberculosis Zur (Mtb con) (29). Asterisks and uppercase letters in the consensus sequences correspond to nucleotides that are identical among all the Zur binding sites analyzed; n represents any nucleotide.
Zur controls the expression of the 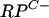 cluster and rpmF2.
cluster and rpmF2.
The increased transcription of C− R-protein genes when S. coelicolor was growing in zinc-depleted medium suggested that a zinc-responsive regulator controls their expression. A likely candidate was the product of SCO2508, which is homologous to Zur proteins in other bacteria (e.g., 29% identical to B. subtilis Zur) and which is henceforth designated Zur. In support of its potential role as a zinc regulator, the zur gene lies immediately downstream from three genes that encode predicted components of a ZnuABC high-affinity zinc uptake system (SCO2505 to SCO2507, or znuACB) (44). The zur reading frame was deleted in M145 to generate a strain designated S121 (Δzur::apr). S1 nuclease mapping revealed that the promoters rpmG2p, rpmB2p, and rpmF2p were deregulated in S121 compared to the parent, with significant activity detected even in the presence of 3.5 μM zinc (Fig. 3). However, rpmG3p retained its association with zinc control, suggesting that an alternative regulator may modulate its expression. The genomic location of zur suggested that it controls the expression of the znuACB operon, and this situation was confirmed by S1 nuclease mapping (Fig. 3). The single znuA promoter was induced under conditions of zinc limitation and deregulated in the zur mutant (Fig. 3). Like those of rpmG2p, the znuAp transcription and translation initiation sites coincide (Fig. 3; also, see Fig. 6).
Zur binds to the 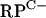 cluster, rpmF2, and znuA promoter regions.
cluster, rpmF2, and znuA promoter regions.
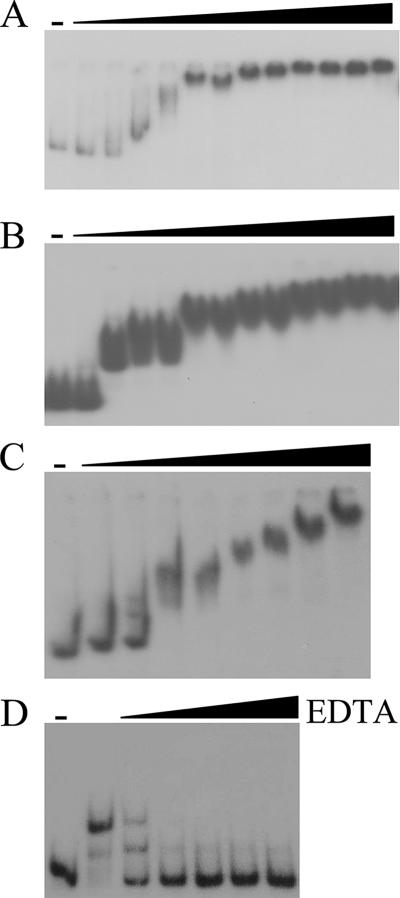
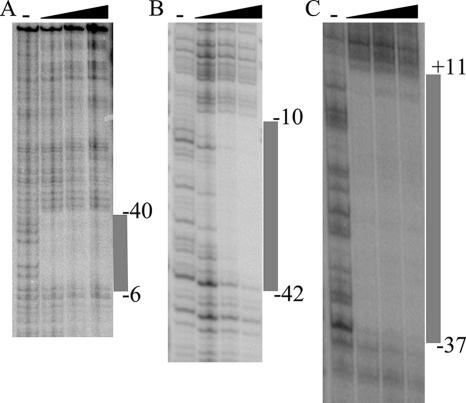
To confirm that Zur directly controls rpmG2p, rpmB2p, rpmF2p, and znuA, Zur was purified from an overproducing E. coli strain and used in EMSAs. DNA fragments containing the 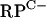 cluster, rpmF2, or znuA promoter region revealed specific binding (Fig. 4). In each case, increasing concentrations of Zur gave rise to slower-migrating protein-DNA complexes, suggesting that higher-order Zur-DNA complexes can be formed. In each case, the Zur-DNA complexes were resistant to nonspecific DNA but dissociated upon incubation with specific unlabeled DNA, indicating that binding was specific (data not shown). Like that of other members of the Fur family of regulators, the DNA binding activity of previously characterized Zur proteins is diminished when the bound regulatory metal is removed (13, 35, 43). In support of this pattern, the Zur-DNA complexes were dissociated when EDTA was included in the binding reaction mixtures (Fig. 4D). DNase I footprinting was used to determine the Zur binding site in each promoter region (Fig. 5 and 6). Zur protected ∼35 bp centered 22 bp upstream and 2 bp downstream from the rpmG2p and rpmB2p transcription initiation sites, respectively (Fig. 6A). Zur protected ∼32 bp centered 27 bp upstream from the rpmF2p transcription initiation site (Fig. 6B). Zur protected a more extensive stretch (48 bp) of the znuA promoter region, an area which overlapped with the transcription initiation site. In each case, multiple binding sites were not detected, which suggests that the level of Zur used fully occupied the binding sites. An alignment of the three Zur binding regions revealed significant homology to one another and to the recently described consensus binding site of Mycobacterium tuberculosis Zur (29). Like that in M. tuberculosis, the Zur binding site exhibits inverted dyad symmetry (Fig. 6D).
cluster, rpmF2, or znuA promoter region revealed specific binding (Fig. 4). In each case, increasing concentrations of Zur gave rise to slower-migrating protein-DNA complexes, suggesting that higher-order Zur-DNA complexes can be formed. In each case, the Zur-DNA complexes were resistant to nonspecific DNA but dissociated upon incubation with specific unlabeled DNA, indicating that binding was specific (data not shown). Like that of other members of the Fur family of regulators, the DNA binding activity of previously characterized Zur proteins is diminished when the bound regulatory metal is removed (13, 35, 43). In support of this pattern, the Zur-DNA complexes were dissociated when EDTA was included in the binding reaction mixtures (Fig. 4D). DNase I footprinting was used to determine the Zur binding site in each promoter region (Fig. 5 and 6). Zur protected ∼35 bp centered 22 bp upstream and 2 bp downstream from the rpmG2p and rpmB2p transcription initiation sites, respectively (Fig. 6A). Zur protected ∼32 bp centered 27 bp upstream from the rpmF2p transcription initiation site (Fig. 6B). Zur protected a more extensive stretch (48 bp) of the znuA promoter region, an area which overlapped with the transcription initiation site. In each case, multiple binding sites were not detected, which suggests that the level of Zur used fully occupied the binding sites. An alignment of the three Zur binding regions revealed significant homology to one another and to the recently described consensus binding site of Mycobacterium tuberculosis Zur (29). Like that in M. tuberculosis, the Zur binding site exhibits inverted dyad symmetry (Fig. 6D).
FIG. 4.
EMSAs using DNA fragments containing the 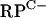 cluster intergenic region (A), the rpmF2p region (B and D), or the znuA promoter region (C). (A to C) Assay mixtures contained either no added Zur (−) or increasing concentrations of Zur, as follows: 0.003, 0.034, 0.085, 0.175, (0.255), 0.340, (0.51), (0.68), 0.85, (1.02), (1.36), 1.7, and 3.4 μM. The concentrations in parentheses were not used for the znuA probe, and the 3.4 μM concentration was used only for the znuA probe. (D) Assay mixtures contained either no added Zur (−) or 0.85 μM Zur. In assay mixtures containing Zur, EDTA was added at increasing concentrations, as follows: 0, 5 μM, 50 μM, 0.5 mM, 1.25 mM, and 2.5 mM.
cluster intergenic region (A), the rpmF2p region (B and D), or the znuA promoter region (C). (A to C) Assay mixtures contained either no added Zur (−) or increasing concentrations of Zur, as follows: 0.003, 0.034, 0.085, 0.175, (0.255), 0.340, (0.51), (0.68), 0.85, (1.02), (1.36), 1.7, and 3.4 μM. The concentrations in parentheses were not used for the znuA probe, and the 3.4 μM concentration was used only for the znuA probe. (D) Assay mixtures contained either no added Zur (−) or 0.85 μM Zur. In assay mixtures containing Zur, EDTA was added at increasing concentrations, as follows: 0, 5 μM, 50 μM, 0.5 mM, 1.25 mM, and 2.5 mM.
FIG. 5.
DNase I footprinting of Zur-operator complexes. Promoter fragments were 5′ end labeled on one strand and mixed with increasing concentrations of Zur (0.14, 0.27, and 0.7 μM) prior to DNase I treatment. (A) 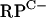 cluster intergenic region; (B) rpmF2 promoter region; (C) znuA promoter region. The extents of the interactions between Zur and the intergenic regions, as determined using dideoxynucleotide sequencing, are indicated by vertical gray bars (data not shown). The positions of the protected regions with respect to the transcription initiation points are indicated; for the
cluster intergenic region; (B) rpmF2 promoter region; (C) znuA promoter region. The extents of the interactions between Zur and the intergenic regions, as determined using dideoxynucleotide sequencing, are indicated by vertical gray bars (data not shown). The positions of the protected regions with respect to the transcription initiation points are indicated; for the 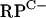 cluster, this position corresponds to the rpmG2 promoter.
cluster, this position corresponds to the rpmG2 promoter.
rpmG3 and rpmJ2 are controlled by σR.
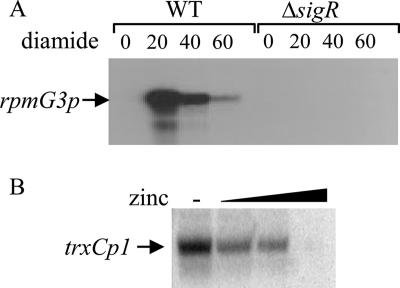
As mentioned above, the level of transcription of rpmG3-rpmJ2 increased in medium deprived of added zinc, and this regulation was retained in S121 (Δzur::apr), suggesting that an alternative zinc-responsive regulator controls transcription. An analysis of the promoter region of rpmG3 revealed an exact match of the σR consensus promoter sequence (GGAAT[18 bp]GTT) (41). σR activity is controlled by the cellular thiol-disulfide redox state and can be induced by the thiol oxidant diamide. S1 nuclease mapping confirmed that rpmG3p was induced by diamide in the wild-type strain M600 but not in the ΔsigR mutant J2139, confirming that σR controls this promoter (Fig. 7A). Furthermore, we were unable to detect rpmG3p activity in a ΔsigR background even under zinc-limited conditions. Experiments using S1 nuclease mapping probes that extended into rpmJ2 indicated that rpmG3 and rpmJ2 form an operon with a single promoter (data not shown). These data suggest that σR activity increases during zinc deprivation. To investigate this further, we analyzed the expression of another σR target, trxC, during zinc depletion. trxC encodes a thioredoxin and is transcribed from two promoters, trxCp1 and trxCp2, the former completely dependent on σR (37). As expected, S1 nuclease mapping revealed that trxCp1 is induced by zinc deficiency (Fig. 7B). Taken together, these data indicate that the rpmG3-rpmJ2 operon is regulated by both zinc deficiency and disulfide stress and that σR is a novel zinc-responsive regulator in S. coelicolor.
FIG. 7.
Transcriptional analysis of the σR-controlled promoters rpmG3p and trxCp1. (A) The rpmG3-rpmJ2 operon is induced by diamide in a σR-dependent manner. Diamide was added at time 0 to a final concentration of 0.5 mM to cultures of wild-type (WT; M600) and sigR mutant (J2139) strains. Numbers indicate minutes postaddition. (B) The σR-dependent promoter trxCp1 in M145 (wild type) was induced under conditions of zinc limitation. The strain was cultured in NMMP liquid medium containing various concentrations of added ZnSO4 (0 −, 0.9, 1.7, and 3.5 μM) to mid- to late exponential phase prior to RNA extraction. Transcripts were assayed by S1 nuclease mapping by using promoter-specific probes radiolabeled at one end.
DISCUSSION
There is considerable variation among bacteria in the potential for producing members of the C+/− R-protein family (30). Indeed, an analysis of annotated genomes (36) suggests that all three of the following extreme situations exist: organisms can encode only C+ forms, organisms can encode only C− forms, and organisms can encode both forms of each member of the C+/− family, the latter being the case for S. coelicolor. These extreme situations appear to be rare, with most bacteria at intermediate positions on this three-way scale, producing C− or C+ forms of some proteins and both paralogous versions of others. In cases in which C+/− paralogues are present in the same organism, data from studies with B. subtilis (2, 32) and the experiments reported here suggest that the C− forms are alternative proteins that are not produced under standard growth conditions. We demonstrated here that most, if not all, of the C− genes in S. coelicolor are not transcribed under standard zinc-replete growth conditions. However, the majority of C− R-protein genes were induced in response to zinc deprivation, which substantiates the proposal that these genes act to provide a selective advantage when zinc is limited, presumably by reducing the overall cellular demand for zinc (42) and possibly releasing zinc for use by other zinc metalloproteins. Studies with E. coli have estimated that each cell contains 2 × 105 to 4 × 105 zinc atoms (34) and up to 70,000 ribosomes at high growth rates (28). Although zinc levels and ribosome numbers in S. coelicolor have not been determined, it is clear that a large proportion of cellular zinc would be held by the ribosome. Therefore, the production of C− R proteins and the quantitative replacement of C+ proteins are likely to have a major influence on the availability of zinc to other zinc metalloproteins.
While the use of C− R proteins for ribosomal function presents an apparently clear advantage for organisms under conditions of zinc deprivation, the benefit of maintaining the C+ proteins is not obvious. One possibility is that the C− and C+ forms are not functionally equivalent. We showed here that even when fused to the transcriptional and translational initiation signals of rpmE1, the rpmE2 gene only partially suppressed the apparently lethal phenotype of a ΔrpmE1 mutation. However, it remains possible that the translational fusion did not confer sufficient 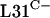 production for normal growth and development. Further, it is also important to emphasize that some organisms contain only the gene for
production for normal growth and development. Further, it is also important to emphasize that some organisms contain only the gene for 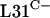 , implying that the C− forms are fully functional, at least in these bacteria. One possible benefit of maintaining the C+ forms is to allow the ribosome to act as a repository of cellular zinc. Thus, S. coelicolor may accumulate zinc in the ribosomes under zinc-replete conditions and be able to release that zinc during zinc deficiency by replacing the C+ proteins with C− proteins. This possibility is supported by studies with B. subtilis (2) in which it was shown that B. subtilis
, implying that the C− forms are fully functional, at least in these bacteria. One possible benefit of maintaining the C+ forms is to allow the ribosome to act as a repository of cellular zinc. Thus, S. coelicolor may accumulate zinc in the ribosomes under zinc-replete conditions and be able to release that zinc during zinc deficiency by replacing the C+ proteins with C− proteins. This possibility is supported by studies with B. subtilis (2) in which it was shown that B. subtilis 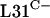 can expel
can expel 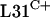 from ribosomes irrespective of the presence or absence of zinc. The ability to release zinc from ribosomes may be particularly important for a morphologically complex organism such as S. coelicolor. During sporulation, S. coelicolor produces specialized aerial hyphae that extend away from the substrate and eventually divide to form spores. In aerial hyphae, metal uptake from the environment would be limited, and the hyphae would instead rely on metal storage proteins or the intrahyphal transfer of metals from the substrate mycelium. The use of ribosomal zinc may be important in such a situation. In eukaryotes, metallothioneins are important cysteine-rich proteins that act as repositories for excess zinc, with the potential for donating zinc to zinc metalloproteins (52). However, analogous storage systems in bacteria appear to be rare. SmtA from Synechococcus strain PCC 7942 is a zinc metallothionein that is required for resistance to toxic levels of zinc and is induced by zinc (22). Other metallothionein-like proteins in some bacteria have been detected (6), although their biological function(s) remains uncharacterized. The role, if any, of the C+/− group of proteins in providing a system of zinc homeostasis requires further analysis.
from ribosomes irrespective of the presence or absence of zinc. The ability to release zinc from ribosomes may be particularly important for a morphologically complex organism such as S. coelicolor. During sporulation, S. coelicolor produces specialized aerial hyphae that extend away from the substrate and eventually divide to form spores. In aerial hyphae, metal uptake from the environment would be limited, and the hyphae would instead rely on metal storage proteins or the intrahyphal transfer of metals from the substrate mycelium. The use of ribosomal zinc may be important in such a situation. In eukaryotes, metallothioneins are important cysteine-rich proteins that act as repositories for excess zinc, with the potential for donating zinc to zinc metalloproteins (52). However, analogous storage systems in bacteria appear to be rare. SmtA from Synechococcus strain PCC 7942 is a zinc metallothionein that is required for resistance to toxic levels of zinc and is induced by zinc (22). Other metallothionein-like proteins in some bacteria have been detected (6), although their biological function(s) remains uncharacterized. The role, if any, of the C+/− group of proteins in providing a system of zinc homeostasis requires further analysis.
We demonstrated here that the promoters rpmG2p, rpmB2p, and rpmF2p are controlled by Zur in a zinc-dependent manner. Zur is one of four members of the Fur family of metalloregulators in S. coelicolor. The other characterized members are FurS and CatR, which are thought to be redox-responsive regulators that control the expression of antioxidant genes, and Nur, which controls nickel uptake (1, 19, 33). The regulation of several C− R proteins by Zur in S. coelicolor concurs with results from studies of B. subtilis and M. tuberculosis (2, 29). In addition, Zur homologues have been predicted to regulate C− R-protein genes in several other bacteria, including E. coli and Yersinia pestis (42), emphasizing that the control of paralogous C− proteins by Zur is widespread in bacteria.
Unexpectedly, we discovered that Zur was not involved in the zinc-dependent regulation of the rpmG3-rpmJ2 operon and that instead σR plays a key role. Although the mechanism behind the zinc responsiveness of σR is not yet clear, the known control mechanism of σR provides one possibility. σR is controlled posttranslationally by RsrA, a member of the ZAS (zinc binding anti-sigma) family of anti-sigma factors (37). Although zinc-free RsrA can bind to σR, its affinity may be decreased and all zinc ligands are essential for RsrA activity in vivo (3, 26, 37, 53). Therefore, under conditions of zinc deprivation, RsrA may accumulate in a zinc-free state, with reduced affinity for σR, allowing σR to interact with RNA polymerase and activate members of the σR regulon. The σR-RsrA system is the major controller of enzymes involved in thiol-disulfide metabolism, including the thioredoxin system, which maintains cellular thiols in their normal reduced state. The increased expression of such enzymes during zinc deprivation may be beneficial because the very juxtaposition of cysteine zinc ligands may increase the likelihood of intramolecular disulfide bond formation when zinc is absent. Thus, the increased expression of enzymes that can catalyze thiol-disulfide exchange may play an important role in the reassembly of zinc metalloproteins during zinc depletion. There is accumulating evidence for a link between zinc homeostasis and the oxidative stress response in bacteria. For example, in B. subtilis, the deletion of the low-affinity zinc importer ZosA leads to increased sensitivity to the thiol oxidant diamide and hydrogen peroxide (14). Interestingly, rather than being controlled by a zinc-responsive regulator, ZosA is regulated by PerR, a redox-responsive repressor.
Although the control of rpmG3-rpmJ2 by σR may be on account of the ability of the σR-RsrA system to sense zinc, it is also possible that the production of 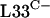 and
and 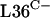 provides a selective advantage during disulfide stress. The C+ versions of these proteins may be susceptible to oxidation, and the production of alternative redox-insensitive forms may compensate for this susceptibility. Such an arrangement is reminiscent of the induction of alternative superoxide-resistant forms of fumarase and aconitase in E. coli (17, 27, 51). Further studies are needed to investigate whether
provides a selective advantage during disulfide stress. The C+ versions of these proteins may be susceptible to oxidation, and the production of alternative redox-insensitive forms may compensate for this susceptibility. Such an arrangement is reminiscent of the induction of alternative superoxide-resistant forms of fumarase and aconitase in E. coli (17, 27, 51). Further studies are needed to investigate whether 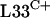 and
and 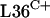 , and possibly other C+ R proteins, are sensitive to disulfide stress.
, and possibly other C+ R proteins, are sensitive to disulfide stress.
Finally, the results of this study are in close agreement with those of an accompanying study by Shin et al. (47) into the function of S. coelicolor Zur. These investigators also noted that Zur negatively regulates znuA and C− R-protein gene expression and additionally showed that Zur positively regulates its own expression. It was also found that σR activity was induced by chelating agents, further supporting the possible role of the σR-RsrA system as a novel sensor of zinc deficiency.
Supplementary Material
Acknowledgments
We thank the University of Sussex and the Biotechnology and Biological Sciences Research Council for providing studentships to G.A.O. and B.P., respectively. This work was partially funded by Biotechnology and Biological Sciences Research Council grant BBC5038541.
We thank Philip Doughty for technical assistance and Nigel Robinson for useful discussions.
Footnotes
Published ahead of print on 30 March 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahn, B. E., J. Cha, E. J. Lee, A. R. Han, C. J. Thompson, and J. H. Roe. 2006. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59:1848-1858. [DOI] [PubMed] [Google Scholar]
- 2.Akanuma, G., H. Nanamiya, Y. Natori, N. Nomura, and F. Kawamura. 2006. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J. Bacteriol. 188:2715-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, J. B., J. H. Park, M. Y. Hahn, M. S. Kim, and J. H. Roe. 2004. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J. Mol. Biol. 335:425-435. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Blindauer, C. A., M. D. Harrison, A. K. Robinson, J. A. Parkinson, P. W. Bowness, P. J. Sadler, and N. J. Robinson. 2002. Multiple bacteria encode metallothioneins and SmtA-like zinc fingers. Mol. Microbiol. 45:1421-1432. [DOI] [PubMed] [Google Scholar]
- 7.Brocklehurst, K. R., J. L. Hobman, B. Lawley, L. Blank, S. J. Marshall, N. L. Brown, and A. P. Morby. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31:893-902. [DOI] [PubMed] [Google Scholar]
- 8.Brodersen, D. E., and P. Nissen. 2005. The social life of ribosomal proteins. FEBS J. 272:2098-2108. [DOI] [PubMed] [Google Scholar]
- 9.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 10.Chakraburtty, R., and M. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fallon, A. M., C. S. Jinks, G. D. Strycharz, and M. Nomura. 1979. Regulation of ribosomal protein synthesis in Escherichia coli by selective mRNA inactivation. Proc. Natl. Acad. Sci. USA 76:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaballa, A., and J. D. Helmann. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45:997-1005. [DOI] [PubMed] [Google Scholar]
- 15.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil, R., F. J. Silva, J. Pereto, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol Rev. 68:518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruer, M. J., and J. R. Guest. 1994. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140:2531-2541. [DOI] [PubMed] [Google Scholar]
- 18.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, J. S., S. Y. Oh, K. F. Chater, Y. H. Cho, and J. H. Roe. 2000. H2O2-sensitive Fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275:38254-38260. [DOI] [PubMed] [Google Scholar]
- 20.Hard, T., A. Rak, P. Allard, L. Kloo, and M. Garber. 2000. The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold. J. Mol. Biol. 296:169-180. [DOI] [PubMed] [Google Scholar]
- 21.Hong, H. J., M. I. Hutchings, J. M. Neu, G. D. Wright, M. S. Paget, and M. J. Buttner. 2004. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52:1107-1121. [DOI] [PubMed] [Google Scholar]
- 22.Huckle, J. W., A. P. Morby, J. S. Turner, and N. J. Robinson. 1993. Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol. Microbiol. 7:177-187. [DOI] [PubMed] [Google Scholar]
- 23.Kelemen, G. H., K. A. Plaskitt, C. G. Lewis, K. C. Findlay, and M. J. Buttner. 1995. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 17:221-230. [DOI] [PubMed] [Google Scholar]
- 24.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 25.Larson, J. L., and C. L. Hershberger. 1986. The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid 15:199-209. [DOI] [PubMed] [Google Scholar]
- 26.Li, W., A. R. Bottrill, M. J. Bibb, M. J. Buttner, M. S. Paget, and C. Kleanthous. 2003. The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J. Mol. Biol. 333:461-472. [DOI] [PubMed] [Google Scholar]
- 27.Liochev, S. I., and I. Fridovich. 1992. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc. Natl. Acad. Sci. USA 89:5892-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maaloe, O., and N. O. Kjeldgaard. 1966. Control of macromolecular synthesis: a study of DNA, RNA, and protein synthesis in bacteria. Benjamin, New York, NY.
- 29.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palu, G. Riccardi, and R. Manganelli. 2007. Global analysis of Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189:730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova, K. S., V. A. Ponomarev, and E. V. Koonin. 2001. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2:RESEARCH 0033. [DOI] [PMC free article] [PubMed]
- 31.Moore, P. B., and T. A. Steitz. 2003. The structural basis of large ribosomal subunit function. Annu. Rev. Biochem. 72:813-850. [DOI] [PubMed] [Google Scholar]
- 32.Nanamiya, H., G. Akanuma, Y. Natori, R. Murayama, S. Kosono, T. Kudo, K. Kobayashi, N. Ogasawara, S. M. Park, K. Ochi, and F. Kawamura. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52:273-283. [DOI] [PubMed] [Google Scholar]
- 33.Ortiz de Orue Lucana, D., and H. Schrempf. 2000. The DNA-binding characteristics of the Streptomyces reticuli regulator FurS depend on the redox state of its cysteine residues. Mol. Gen. Genet. 264:341-353. [DOI] [PubMed] [Google Scholar]
- 34.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 35.Outten, C. E., D. A. Tobin, J. E. Penner-Hahn, and T. V. O'Halloran. 2001. Characterization of the metal receptor sites in Escherichia coli Zur, an ultrasensitive zinc(II) metalloregulatory protein. Biochemistry 40:10417-10423. [DOI] [PubMed] [Google Scholar]
- 36.Owen, G. A. 2005. Stress-induced ribosomal proteins of Streptomyces coelicolor A3(2). PhD dissertation. University of Sussex, Brighton, United Kingdom.
- 37.Paget, M. S., J. B. Bae, M. Y. Hahn, W. Li, C. Kleanthous, J. H. Roe, and M. J. Buttner. 2001. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol. Microbiol. 39:1036-1047. [DOI] [PubMed] [Google Scholar]
- 38.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paget, M. S., G. Hintermann, and C. P. Smith. 1994. Construction and application of streptomycete promoter probe vectors which employ the Streptomyces glaucescens tyrosinase-encoding gene as reporter. Gene 146:105-110. [DOI] [PubMed] [Google Scholar]
- 40.Paget, M. S., J. G. Kang, J. H. Roe, and M. J. Buttner. 1998. SigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17:5776-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 42.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 100:9912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patzer, S. I., and K. Hantke. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321-24332. [DOI] [PubMed] [Google Scholar]
- 44.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 45.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38:749-770. [DOI] [PubMed] [Google Scholar]
- 46.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 47.Shin, J.-H., S.-Y. Oh, S.-J. Kim, and J.-H. Roe. 2007. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 189:4070-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 49.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 50.Tsiboli, P., D. Triantafillidou, F. Franceschi, and T. Choli-Papadopoulou. 1998. Studies on the Zn-containing S14 ribosomal protein from Thermus thermophilus. Eur. J. Biochem. 256:136-141. [DOI] [PubMed] [Google Scholar]
- 51.Varghese, S., Y. Tang, and J. A. Imlay. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J. Bacteriol. 185:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasak, M., and D. W. Hasler. 2000. Metallothioneins: new functional and structural insights. Curr. Opin. Chem. Biol. 4:177-183. [DOI] [PubMed] [Google Scholar]
- 53.Zdanowski, K., P. Doughty, P. Jakimowicz, L. O'Hara, M. J. Buttner, M. S. Paget, and C. Kleanthous. 2006. Assignment of the Zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry 45:8294-8300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.