Abstract
The Caulobacter crescentus DNA adenine methyltransferase CcrM and its homologs in the α-Proteobacteria are essential for viability. CcrM is 34% identical to the yhdJ gene products of Escherichia coli and Salmonella enterica. This study provides evidence that the E. coli yhdJ gene encodes a DNA adenine methyltransferase. In contrast to an earlier report, however, we show that yhdJ is not an essential gene in either E. coli or S. enterica.
Eubacterial genomes can encode DNA methyltransferases that are not part of restriction-modification systems, also referred to as orphan or solitary DNA methyltransferases. The best studied of these enzymes are two adenine methyltransferases: Dam, present in the γ subdivision of the Proteobacteria, and CcrM (for “cell cycle-regulated methyltransferase”), found in species of the α subdivision. These methyltransferases are known to be involved in the control of many cellular events, including gene expression, cell cycle regulation, and DNA repair (reviewed in references 7 and 14). Where it has been examined, CcrM is essential for viability, whereas Dam and its homologs are essential in only a few species. In Escherichia coli and Salmonella enterica, Dam methyltransferase mutants have pleiotropic defects but are viable.
The E. coli yhdJ gene, or b3262, was annotated as a putative methyltransferase-encoding gene (10), and the predicted protein contains conserved methyltransferase motifs for S-adenosyl-l-methionine (SAM) binding and catalysis. The order of these domains places YhdJ in the beta group of methyltransferases, along with CcrM and its homologs (12). E. coli YhdJ has identity with the adenine methyltransferases M. AvaIII (55.8%) and CcrM from Caulobacter crescentus (34.3%). Like CcrM but unlike M. AvaIII, yhdJ does not appear to be part of a restriction-modification system, as there is no gene encoding the former in close proximity.
The existence of an essential DNA methyltransferase in E. coli could have significant implications for our understanding of DNA replication and cell cycle control. The recently published collection of in-frame E. coli deletions contains a yhdJ mutant, suggesting that YhdJ is not essential (1), whereas a previous report concluded that a gene deletion could not be obtained and that yhdJ expression was essential for viability (6). In this study, we therefore reexamine the role and function of the yhdJ gene product.
The yhdJ gene is not essential, and its overexpression does not alter cell morphology.
To address whether yhdJ is essential, the yhdJ coding sequence (except for the first and last codons) was replaced (9) with a chloramphenicol resistance gene (cat) in E. coli strain AB1157 (4), resulting in isolate GM8494. The ΔyhdJ::cat mutation was transduced with P1vir (8) from GM8494 into E. coli WA802 (15) and designated GM8613. E. coli strain BW25113 and its yhdJ deletion derivative JW5543, from the E. coli gene knockout collection (1), were also analyzed. The yhdJ gene was also disrupted in S. enterica serovar Typhimurium LT2 by the procedure of Datsenko and Wanner (3). Disruption of yhdJ in all strains was confirmed by Southern blot analysis and/or PCR (data not shown). All constructs lacking yhdJ were viable, and no significant growth deficiency was observed based on doubling times in Luria broth (LB) (for example, the doubling times of BW25113 and its ΔyhdJ::Kmr [JW5543] derivative [1] were 49 min and 52 min, respectively). No difference in cell morphology was observed between the yhdJ mutants and otherwise isogenic wild-type strains. We used the WA802 strain background because, in a previous study, a yhdJ deletion could not be constructed in this background (6). Thus, yhdJ is not an essential gene in E. coli or S. enterica.
For the following experiments, two derivatives were constructed from the E. coli plasmid pCA24N, which carries yhdJ-his6-gfp cloned behind an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (5). pMV237 is pCA24N without the NotI fragment that contains gfp, and in pMV238, the yhdJ-his6 insert has been removed by deleting the SfiI fragment from pMV237, resulting in an “empty” vector.
Based on observations by Kossykh and Lloyd (6), strains overexpressing YhdJ should show aberrant growth and morphology. Hence, we examined whether induction of ydhJ expression from a plasmid in JW5543 (ΔyhdJ::Kmr) (1) caused morphological changes. As controls, we used the same strain under noninducing conditions, as well as the yhdJ-less plasmid pMV238. However, we did not observe any change in cell morphology as a result of induction of YhdJ. The doubling times of wild-type and YhdJ-overexpressing strains were very similar, within 5% of each other (results not shown).
YhdJ is an adenine DNA methyltransferase.
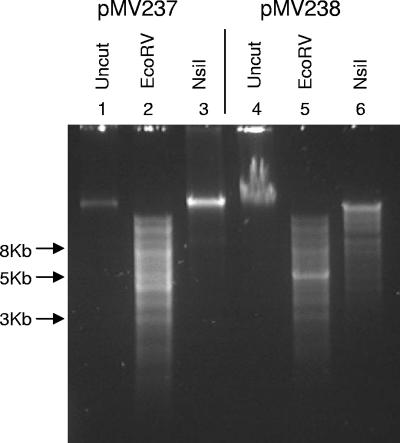
A previous report indicated that YhdJ methylation confers protection to NsiI digestion (6). To further examine the enzymatic activity of YhdJ, methylation of this putative target sequence was examined by determining DNA susceptibility to NsiI digestion, both in vivo and in vitro. The NsiI recognition sequence is 5′-ATGCAT-3′. If the 5′ adenine is methylated on the top and bottom strands, cleavage by NsiI is reduced by 50%. In contrast, if the 3′ adenine is methylated on both DNA strands, cleavage is completely inhibited (http://rebase.neb.com/cgi-bin/msget?NsiI) (13). NsiI digestion patterns of chromosomal DNA isolated from stationary-phase cultures of the wild-type and yhdJ deletion strains JW5543 and GM8494 were identical, with no protection from digestion (results not shown). However, when the same analysis was performed on total DNA isolated from a culture of BW25113 carrying pMV237(yhdJ) grown with 1 mM IPTG, protection from NsiI cleavage occurred (Fig. 1, lane 3). Protection from NsiI digestion did not occur in the strain that carried pMV238 (empty vector) (Fig. 1, lane 6). These results indicate that the level of expression in a wild-type isolate grown under the conditions used here is too low for methylation to be detected by this method. However, YhdJ overexpression can lead to methylation at the NsiI recognition sequence.
FIG. 1.
Overexpression of yhdJ protects DNA from NsiI digestion. Total DNA was isolated from BW25113/pMV237(6xhis-yhdJ) (lanes 1 to 3) and BW25113/pMV238 (vector) (lanes 4 to 6) and incubated without enzyme (lanes 1 and 4), with EcoRV (lanes 2 and 5) as a positive control, and with NsiI (lanes 3 and 6).
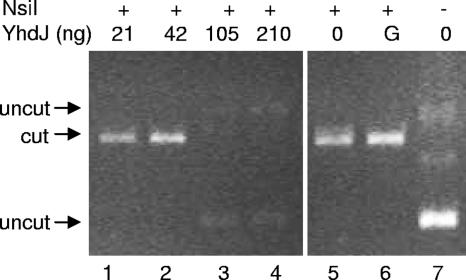
To confirm that protection of NsiI digestion is conferred by YhdJ, plasmid DNA was methylated in vitro with partially purified His-YhdJ. Strain DH5α(pMV237) was grown in LB with 1 mM IPTG added at an optical density at 600 nm of 0.3. Cells were harvested 3 h later, and His-YhdJ was partially purified with His-GraviTrap (GE Healthcare). The final fraction was dialyzed against 50 mM Tris (pH 8), 1 mM EDTA, 1 mM dithiothreitol (DTT), and 100 mM NaCl and stored at −20°C in 50% glycerol. In vitro methylation was carried out on pACYC177, which has two NsiI target sequences. The methylation reaction consisted of YhdJ-His, 40 ng of pACYC177, 50 mM Tris (pH 8), 1 mM EDTA, 1 mM DTT, 100 mM NaCl, and 50 μM SAM. Methylation was carried out at 37°C for 2 h and was stopped by incubation at 65°C. NsiI (6 units) and 10 mM MgCl2 were added, and DNA was incubated for 3 h at 37°C. The results (Fig. 2) show that cleavage by NsiI is inhibited in a concentration-dependent manner by YhdJ-His (compare lanes 1 and 2 with lanes 3 and 4), indicating that one of the adenines in the recognition sequence 5′-ATGCAT-3′ is modified (http://rebase.neb.com/cgi-bin/msget?NsiI).
FIG. 2.
In vitro YhdJ-dependent methylation protects pACYC177 from NsiI cleavage. Plasmid DNA was incubated with partially purified YhdJ as indicated (lanes 1 to 4) and then digested with NsiI (lanes 1 to 6). “G” (lane 6) indicates the addition of YhdJ storage buffer; lane 7 is undigested plasmid.
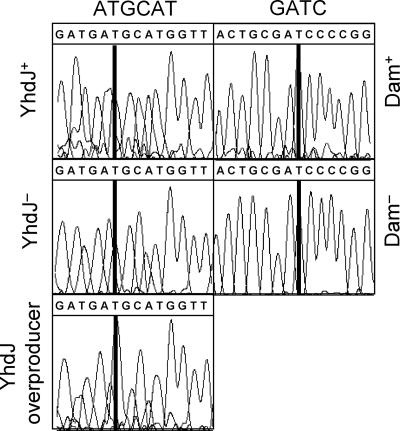
Methylation of adenine can be shown directly by automated dye terminator sequencing, since the modification leads to an increase in the signal of the complementary thymidine (2, 11). To examine YhdJ-dependent modification, several strains were constructed. The kanamycin cassette was deleted from JW5543 (BW25113 yhdJ::Kmr) (1) as described (3), resulting in MV1061. Strain MV1074 is MV1061(ΔyhdJ) with pACYC177 and pMV237(6xhis-yhdJ), and MV1081 is MV1061(ΔyhdJ) with pACYC177 and pMV238 (vector). Plasmid DNA was isolated from these strains and from BW25113 containing pACYC177 (MV1070) grown in LB in the presence of 3 mM IPTG, and the region of pACYC177 flanking the NsiI sites was sequenced. Examination of the sequencing chromatograms permitted the detection of N6-methyladenine, as described by Rao and Buckler-White (11). Sections of a representative trace showing an NsiI sequence are presented in Fig. 3. Also shown as controls are GATC sequences, the methylation targets for Dam (14), in plasmid pACYC177 isolated from isogenic Dam+ and Dam− hosts (SV5421 and SV5422, respectively). The relative increase in the signal of the 5′ thymidine residue specifically in DNA incubated with YhdJ-His indicates that the adenine on the opposing strand (the 3′ adenine of the 5′-ATGCAT-3′ NsiI recognition sequence) is methylated. Similar results were obtained with the second NsiI sequence in pACYC177 (not shown).
FIG. 3.
The 3′ adenine of the NsiI recognition sequence is modified by YhdJ. The traces of 5′-ATGCAT-3′ sequence (left) on plasmid pACYC177, isolated from MV1070 (top), MV1081 (middle), and MV1074 (bottom) (see the text), are shown. Also shown are 5′-GATC-3′ sequence traces (right) from plasmid pACYC177, isolated from SV5421 (top) and SV5422 (bottom).
YhdJ is a nonessential DNA adenine methyltransferase.
Our study provides two lines of evidence, based on DNA digestion and DNA sequencing, indicating that YhdJ is an adenine methyltransferase. However, in contrast to the earlier report, our analysis suggests that YhdJ methylates the second, or 3′, adenine of the sequence 5′-ATGCAT-3′ (Fig. 2 and 3). Since the two NsiI sequences in pACYC177 are not flanked by identical nucleotides, this is likely to be the same as, or to at least contain, the YhdJ recognition sequence. DNA methyltransferase enzymes are highly specific for their target sequence (http://rebase.neb.com/cgi-bin/msget?NsiI); thus, it is unlikely that another target sequence exists, even though this cannot be completely ruled out. The absence of detectable YhdJ-dependent methylation in bacterial cells (Fig. 1) suggests that YhdJ is unlikely to play a significant role in bacterial physiology or cell cycle regulation, nor does it seem that yhdJ is an essential gene (6), since deletions could be made in various genetic backgrounds of both E. coli and S. enterica. Furthermore, there was no difference in synchronization between dnaC2(Ts) strains carrying either pMV237 or pMV238 (results not shown). This suggests that YhdJ overproduction does not affect initiation of DNA replication, which is a key feature of the cell cycle. Thus, yhdJ is distinct in these relevant features from the CcrM family of DNA methyltransferases (reviewed in reference 14). Therefore, to avoid confusion, YhdJ should not be referred to as M.EcoKCcrM (6).
Acknowledgments
We thank Richard Roberts (NEB) for helpful comments and Renata Kaminska for help with cell synchronization studies.
This research was supported by the BBSRC (grant BB/C502849/1) (M.V.D.W.), the Spanish Ministry of Education and Science and the European Regional Fund (grants GEN2003-20234-CO6-03 and BIO2004-03455-CO2-02) (J.C.), and National Institutes of Health grant GM63790 (M.G.M.).
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed]
- 2.Bart, A., M. W. van Passel, K. van Amsterdam, and A. van der Ende. 2005. Direct detection of methylation in genomic DNA. Nucleic Acids Res. 33:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeWitt, S. K., and E. A. Adelberg. 1962. The occurrence of a genetic transposition in a strain of Escherichia coli. Genetics 47:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitagawa, M., T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, and H. Mori. 2005. Complete set of ORF clones of Escherichia coli ASKA library. A complete set of E. coli K-12 ORF archive: unique resources for biological research. DNA Res. 12:291-299. [DOI] [PubMed] [Google Scholar]
- 6.Kossykh, V. G., and R. S. Lloyd. 2004. A DNA adenine methyltransferase of Escherichia coli that is cell cycle regulated and essential for viability. J. Bacteriol. 186:2061-2067. (Retraction, 186:6340.) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Lobner-Olesen, A., O. Skovgaard, and M. G. Marinus. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8:154-160. [DOI] [PubMed] [Google Scholar]
- 8.Marinus, M. G. 1973. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol. Gen. Genet. 127:47-55. [DOI] [PubMed] [Google Scholar]
- 9.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 10.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao, B. S., and A. Buckler-White. 1998. Direct visualization of site-specific and strand-specific DNA methylation patterns in automated DNA sequencing data. Nucleic Acids Res. 26:2505-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reisenauer, A., L. S. Kahng, S. McCollum, and L. Shapiro. 1999. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts, R. J., T. Vincze, J. Posfai, and D. Macelis. 2005. REBASE—restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 33:D230-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wion, D., and J. Casadesus. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4:183-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood, W. B. 1966. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol. 16:118-133. [DOI] [PubMed] [Google Scholar]