Abstract
Anaerobic growth of Pseudomonas aeruginosa PAO1 was affected by quorum sensing. Deletion of genes that produce N-acyl-l-homoserine lactone signals resulted in an increase in denitrification activity, which was repressed by exogenous signal molecules. The effect of the las quorum-sensing system was dependent on the rhl quorum-sensing system in regulating denitrification.
Bacteria regulate their metabolism to adapt to various conditions by sensing environmental signals. Under anoxic conditions, many bacteria are able to use N-oxides as terminal electron acceptors. Pseudomonas aeruginosa is a denitrifying bacterium capable of anaerobic growth by utilizing N-oxides such as nitrate (NO3−) and nitrite (NO2−). Denitrification is induced under low-oxygen conditions when N-oxides are also present (1, 9, 11).
Recent studies on bacteria have revealed new types of environmental signals known as cell-to-cell communication signals (25). P. aeruginosa is reported to control gene expression globally in response to cell density by utilizing N-acyl-l-homoserine lactone (AHL) signals. This cell-density-dependent regulation is termed quorum sensing (5). P. aeruginosa possesses at least two quorum-sensing systems: the LasR-LasI (las) and RhlR-RhlI (rhl) systems (20). LasI directs the synthesis of the AHL signal N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) (17, 18), and RhlI directs the synthesis of another AHL signal, N-butyryl-l-homoserine lactone (C4-HSL) (19). The transcription regulatory proteins, LasR (6) and RhlR (16), are specifically activated by 3-oxo-C12-HSL and C4-HSL, respectively. Previous studies have indicated that production of virulence factors, such as protease, exotoxin A, rhamnolipids, and siderophores, is regulated by quorum sensing in P. aeruginosa (7, 12, 26), suggesting that quorum sensing is important in the pathogenesis of infection with this bacterium.
Recent transcriptomic and proteomic studies indicate that quorum sensing is a global regulation system in P. aeruginosa (3, 23, 28). From these findings, it can be presumed that quorum-sensing systems have ecologically important roles in addition to the control of pathogenesis for the bacterium. For example, recent work suggests that quorum sensing regulates the activities of denitrification enzymes. In a recent study by Yoon et al. (31), the authors reported that levels of denitrifying enzyme activities of anaerobically grown P. aeruginosa cells are higher for an rhlR mutant than for its parent strain in an in vitro system. We were interested in further characterizing the potential anaerobic regulation of denitrification by the quorum-sensing system. Here we present a comprehensive analysis of the impact of quorum sensing on the denitrification pathway under anaerobic conditions by using in vivo and in vitro analyses.
Effect of quorum sensing on denitrification activity.
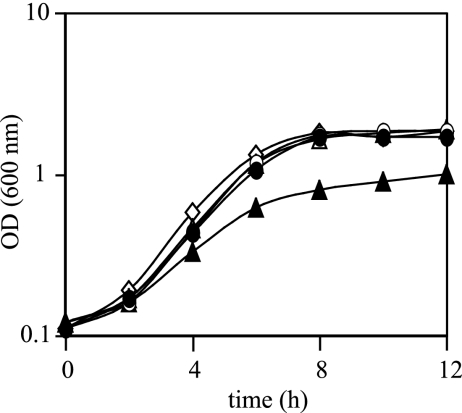
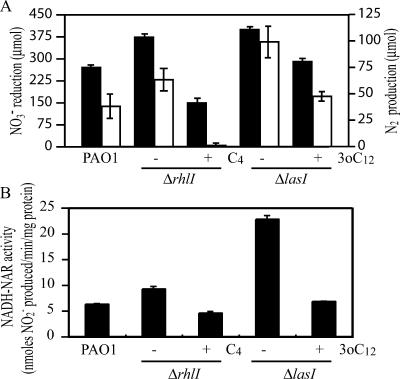
P. aeruginosa PAO1 was cultured anaerobically in 17-ml Hungate tubes containing 5 ml Luria-Bertani (LB) medium supplemented with 100 mM KNO3 with shaking at 200 rpm at 37°C. The tubes were sealed with rubber stoppers, and the air was replaced with argon. The growth of ΔrhlI and ΔlasI mutants, which are deficient in producing C4-HSL and 3-oxo-C12-HSL, respectively, was almost identical to that of the parental strain, PAO1. However, when C4-HSL was added to the ΔrhlI mutant, the turbidity at 600 nm was decreased approximately 50% when cells entered the stationary phase (Fig. 1). No effect on growth was observed with addition of 3-oxo-C12-HSL to the ΔlasI mutant. To further investigate the effect of quorum sensing on denitrification, the amounts of NO3− reduced and N2 produced during a 12-h incubation were measured. NO3− was measured by the brucine method (15), and N2 was measured by a gas chromatograph (GC-8AIT; Shimadzu) equipped with a Molecular Sieve 5A column. The quorum-sensing mutants reduced more NO3− and produced more N2 than PAO1, a pattern that was reversed by addition of 3-oxo-C12-HSL and C4-HSL (Fig. 2A). To investigate whether the observed effect of quorum sensing on NO3− reduction is linked with the NO3− respiratory chain, the cell membrane was collected and NO3−-respiring activity was measured by using NADH as an electron donor. Cells were washed twice with 0.01 M potassium phosphate buffer, pH 7.0, containing 10% glycerol and were sonicated in the same buffer. The sonicated cells were centrifuged for 10 min at 2,000 × g and 4°C to remove unbroken cells and were then centrifuged at 104,000 × g and 4°C for 60 min. Pellets were resuspended in the same buffer and collected as membrane fractions. A 500-μl volume of reaction mixture for the NADH-dependent NO3− reductase (NAR) activity assay contained 1 mM NADH, 10 mM KNO3, and the membrane fraction of anaerobically cultured P. aeruginosa in 0.01 M potassium phosphate buffer (pH 7.0)-10% glycerol. The air of the reaction mixture was replaced with N2, and incubation was carried out at 30°C for 30 min. The reaction was started by addition of NADH and was stopped by boiling at 100°C for 15 min. NO2− production was measured according to the method of Nicholas and Nason (15). To ensure that NO3−-respiring activity was measured in this assay, rotenone, which is an inhibitor of complex I in the respiratory chain, was added. NO3− reduction was repressed to approximately 70%, as has been observed for other Pseudomonas species (21). The NO3−- respiring activity in the ΔrhlI mutant was 1.4-fold higher than that in PAO1 and was repressed 50% when the ΔrhlI mutant was cultured with C4-HSL (Fig. 2B). The ΔlasI mutant showed the highest activity (3.5-fold higher than that of PAO1), which could be repressed 70% by the addition of 3-oxo-C12-HSL in the culture. Taken together, these results indicate that the rhl and las quorum-sensing systems repress denitrification activity. Even though the ΔrhlI and ΔlasI mutants had higher denitrification activity than PAO1, no significant difference in growth could be observed between the mutants and PAO1. This may be due to the accumulation of toxic levels of nitric oxide (NO) in the quorum-sensing mutants (31), which may repress growth.
FIG. 1.
Effects of AHL signals on anaerobic growth. Cells were grown anaerobically in LB medium supplemented with 100 mM NO3−. Symbols: open diamonds, PAO1; open triangles, ΔrhlI mutant; solid triangles, ΔrhlI mutant with 10 μM C4-HSL; open circles, ΔlasI mutant; solid circles, ΔlasI mutant with 1 μM 3-oxo-C12-HSL. Three independent experiments were conducted, and representative data are shown.
FIG. 2.
Effects of AHL signals on denitrification. (A) Effects of AHL signals on NO3− reduction and N2 production. The amounts of NO3− reduced and N2 produced over 12 h were measured in PAO1 and in ΔrhlI and ΔlasI mutants. Solid bars, NO3− reduced; open bars, N2 produced. C4-HSL and 3-oxo-C12-HSL were added to the cultures at final concentrations of 10 μM and 1 μM, respectively. Data are means ± standard deviations from three independent experiments. (B) Effects of AHL signals on NO3−-respiring activity. NADH-dependent NAR activity was measured in cells grown anaerobically for 12 h. C4-HSL at 10 μM and 3-oxo-C12-HSL at 1 μM were added to the cultures of ΔrhlI and ΔlasI mutants, respectively. The experiment was performed in triplicate.
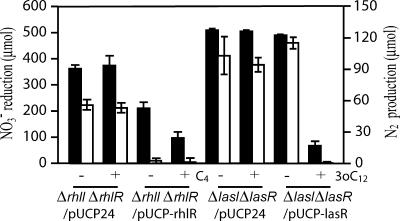
To investigate the involvement of the transcription regulatory proteins RhlR and LasR in the regulation of denitrification, C4-HSL or 3-oxo-C12-HSL was added to an ΔrhlI ΔrhlR or ΔlasI ΔlasR double mutant, respectively. As a result, denitrification was not decreased by either of the AHLs when their binding proteins were deficient (ΔrhlI ΔrhlR/pUCP24, ΔlasI ΔlasR/pUCP24), and the regulation was restored by complementing each mutant with its appropriate wild-type copy (ΔrhlI ΔrhlR/pUCP-rhlR, ΔlasI ΔlasR/pUCP-lasR) (Fig. 3), indicating that denitrification regulation by C4-HSL and 3-oxo-C12-HSL is dependent on their cognate transcription regulatory proteins. Interestingly, denitrification activity was decreased even in the absence of C4-HSL when rhlR was complemented and was more repressed when C4-HSL was added, as could be observed in the NO3− reduction data (Fig. 3). RhlR has been reported to regulate rhlAB oppositely in the presence and absence of C4-HSL (14); however, in this study, denitrification was repressed by RhlR both in the presence and in the absence of C4-HSL. Therefore, the C4-HSL-RhlR complex and RhlR alone may regulate denitrification through different pathways, although it is still a question whether RhlR represses denitrification under physiological conditions, since RhlR was overexpressed in this experiment.
FIG. 3.
Effects of RhlR and LasR transcriptional regulators on denitrification regulation. The amounts of NO3− reduced and N2 produced over 12 h were measured. Solid bars, NO3− reduced; open bars, N2 produced. C4-HSL and 3-oxo-C12-HSL were added to the cultures at final concentrations of 10 μM and 1 μM, respectively. rhlR or lasR was inserted into a pUCP24 plasmid (pUCP-rhlR or pUCP-lasR) and transformed into a ΔrhlI ΔrhlR or ΔlasI ΔlasR double mutant for complementation. pUCP24 (29) was transformed into a ΔrhlI ΔrhlR or ΔlasI ΔlasR mutant as a control. Data are means ± standard deviations from three independent experiments.
Regulation of denitrification by rhl and las quorum-sensing systems.
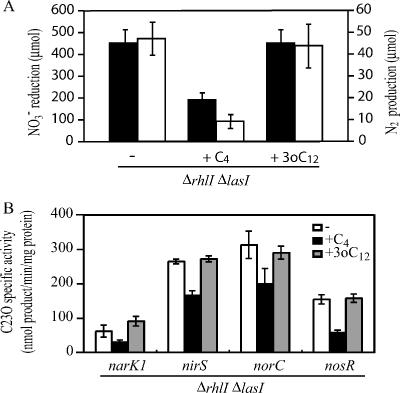
The rhl and las quorum-sensing systems are reported as a hierarchy, in which the las quorum-sensing system regulates the rhl quorum-sensing system (20). To study whether the rhl and las quorum-sensing systems regulate denitrification concertedly or not, C4-HSL and 3-oxo-C12-HSL were added to the culture of a ΔrhlI ΔlasI double mutant. The amounts of NO3− reduced and N2 produced during a 12-h incubation were measured. Addition of C4-HSL decreased the denitrification activity of the ΔrhlI ΔlasI mutant; however, 3-oxo-C12-HSL did not (Fig. 4A). To study whether denitrification is regulated at the transcriptional level, transcriptional fusion plasmids were constructed. During NO3− reduction to N2, NAR, NO2− reductase (NIR), NO reductase (NOR), and N2O reductase (NOS) are involved, and the genes encoding each of those enzymatic activities are organized into distinct operons (1, 2, 10, 24). The promoter regions of narK1, nirS, norC, and nosR were fused to a xylE gene on a pMEX9 plasmid, which was constructed by replacing a lacZα gene of pME4510 (22) with xylE. Promoter regions of denitrifying genes were amplified by PCR using appropriate oligonucleotide primers. The amplified DNA fragments were digested with appropriate restriction enzymes and then inserted into a multicloning site of pMEX9. Each plasmid constructed, as well as a promoterless pMEX9 plasmid used as a reference, was transformed into a ΔrhlI ΔlasI mutant, and catechol 2,3-dioxygenase (C23O) activity was measured during the late-exponential phase (culture for 6 h). C23O activity was measured as described previously (13), and the promoter-dependent C23O activity was divided by the basal C23O activity, which was measured by using a ΔrhlI ΔlasI mutant transformed with pMEX9. Transcription of the denitrifying genes in the ΔrhlI ΔlasI mutant was repressed by C4-HSL; however, 3-oxo-C12-HSL had no effect (Fig. 4B). Thus, these results indicate that the denitrification regulation by the las quorum-sensing system is dependent on the rhl quorum-sensing system and that each denitrifying enzyme is regulated at the transcriptional level. The repression of the narK1, nirS, and norC promoters by quorum sensing is in accordance with the previous result, which showed that NAR, NIR, and NOR activities are higher in an rhlR mutant than in its parental strain (31).
FIG. 4.
Denitrification regulation by rhl and las quorum-sensing systems. (A) Effect of 10 μM C4-HSL and 1 μM 3-oxo-C12-HSL on a ΔrhlI ΔlasI double mutant. The amounts of NO3− reduced and N2 produced over 12 h were measured. Solid bars, NO3− reduced; open bars, N2 produced. Data are means ± standard deviations from three independent experiments. (B) Effects of 10 μM C4-HSL and 1 μM 3-oxo-C12-HSL on the transcription of denitrifying genes in a ΔrhlI ΔlasI mutant. C23O activity was measured in cells cultured for 6 h (late-exponential phase). Data are means ± standard deviations from three independent experiments.
In conclusion, the denitrification activity in P. aeruginosa is regulated by C4-HSL and 3-oxo-C12-HSL. Regulation by the las quorum-sensing system was dependent on the rhl quorum-sensing system, suggesting a hierarchical regulation from the las to the rhl quorum-sensing system in denitrification regulation. However, regulation by the las quorum-sensing system may not be as simple, since NO3−-respiring activity was higher in the ΔlasI mutant than in the ΔrhlI mutant, indicating that other, rhl-independent factors are involved. Although the mechanism of denitrification regulation by quorum sensing remains to be investigated, this is the first study, to our knowledge, to directly demonstrate the role of quorum sensing in controlling bacterial respiration.
Quorum sensing has been reported to affect butanediol fermentation in Serratia species (27) and the growth rate in Rhizobium species, although the mechanisms remain obscure (8, 30). A study by Wagner et al. using DNA microarray analysis (28) reported that exogenous C4-HSL and 3-oxo-C12-HSL affect the transcription of denitrifying genes in P. aeruginosa PAO1 under aerobic conditions. In addition, the results of Yoon et al. (31) indicate that quorum sensing affects biofilm formation under anaerobic conditions. Furthermore, it was reported that a denitrification gene, nirS, is expressed in aerobically grown P. aeruginosa biofilms (4), suggesting that regulation of denitrification by quorum sensing may be important in both aerobically and anaerobically grown biofilms. These studies, along with our data concerning the regulation of respiration by quorum sensing, may provide new insight into how bacteria regulate their energy metabolism during biofilm development.
Acknowledgments
We thank Kiwamu Minamisawa, Yuichi Suwa, Sherry L. Kuchma, Daniel. P. MacEachran, and E. Peter Greenberg for helpful comments on the manuscript and Emiko Matsuzaka and Norihisa Okada for technical advice in determining denitrification activities. We also thank Michael A. Kertesz for providing the pME4510 plasmid and Herbert P. Schweizer for providing the pUCP24 plasmid.
This study was partially supported by a grant to N.N. from the Industrial Technology Research Grant Program 2003-2005 of the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Arai, H., T. Kodama, and Y. Igarashi. 1999. Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 170:19-24. [DOI] [PubMed] [Google Scholar]
- 2.Arai, H., M. Mizutani, and Y. Igarashi. 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149:29-36. [DOI] [PubMed] [Google Scholar]
- 3.Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5:1350-1369. [DOI] [PubMed] [Google Scholar]
- 4.Barraud, N., D. J. Hassett, S. H. Hwang, S. A. Rice, S. Kjelleberg, and J. S. Webb. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188:7344-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gambello, M. J., S. Kaye, and B. H. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185:809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ka, J. O., J. Urbance, R. W. Ye, T. Y. Ahn, and J. M. Tiedje. 1997. Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria. FEMS Microbiol. Lett. 156:55-60. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki, S., H. Arai, T. Kodama, and Y. Igarashi. 1997. Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa: sequencing and identification of a locus for heme d1 biosynthesis. J. Bacteriol. 179:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Körner, H., and W. G. Zumft. 1989. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 55:1670-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 13.Maseda, H., I. Sawada, K. Saito, H. Uchiyama, T. Nakae, and N. Nomura. 2004. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:1320-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medina, G., K. Juarez, B. Valderrama, and G. Soberon-Chavez. 2003. Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J. Bacteriol. 185:5976-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas, D. J. D., and A. Nason. 1957. Determination of nitrate and nitrite. Methods Enzymol. 3:981-984. [Google Scholar]
- 16.Ochsner, U. A., A. K. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 18.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radcliffe, B. C., and D. J. Nicholas. 1970. Some properties of a nitrate reductase from Pseudomonas denitrificans. Biochim. Biophys. Acta 205:273-287. [DOI] [PubMed] [Google Scholar]
- 22.Rist, M., and M. A. Kertesz. 1998. Construction of improved plasmid vectors for promoter characterization in Pseudomonas aeruginosa and other gram-negative bacteria. FEMS Microbiol. Lett. 169:179-183. [DOI] [PubMed] [Google Scholar]
- 23.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma, V., C. E. Noriega, and J. J. Rowe. 2006. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toder, D. S., M. J. Gambello, and B. H. Iglewski. 1991. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol. Microbiol. 5:2003-2010. [DOI] [PubMed] [Google Scholar]
- 27.Van Houdt, R., P. Moons, M. Hueso Buj, and C. W. Michiels. 2006. N-Acyl-l-homoserine lactone quorum sensing controls butanediol fermentation in Serratia plymuthica RVH1 and Serratia marcescens MG1. J. Bacteriol. 188:4570-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson, A., V. Danino, F. Wisniewski-Dye, J. K. Lithgow, and J. A. Downie. 2002. N-Acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 184:4510-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]