Abstract
Phthiocerol dimycocerosates (DIM) and phenolglycolipids (PGL) are functionally important surface-exposed lipids of Mycobacterium tuberculosis. Their biosynthesis involves the products of several genes clustered in a 70-kb region of the M. tuberculosis chromosome. Among these products is PpsD, one of the modular type I polyketide synthases responsible for the synthesis of the lipid core common to DIM and PGL. Bioinformatic analyses have suggested that this protein lacks a functional enoyl reductase activity domain required for the synthesis of these lipids. We have identified a gene, Rv2953, that putatively encodes an enoyl reductase. Mutation in Rv2953 prevents conventional DIM formation and leads to the accumulation of a novel DIM-like product. This product is unsaturated between C-4 and C-5 of phthiocerol. Consistently, complementation of the mutant with a functional pks15/1 gene from Mycobacterium bovis BCG resulted in the accumulation of an unsaturated PGL-like substance. When an intact Rv2953 gene was reintroduced into the mutant strain, the phenotype reverted to the wild type. These findings indicate that Rv2953 encodes a trans-acting enoyl reductase that acts with PpsD in phthiocerol and phenolphthiocerol biosynthesis.
Mycobacterium tuberculosis, the etiological agent of tuberculosis, remains one of the leading causes of mortality and morbidity worldwide. The mycobacterial cell envelope is a complex structure that plays a major role in both the virulence and the ability of the tubercle bacillus to resist the hostile environments encountered during the infection process. A unique feature of this cell envelope is its high lipid content (40 to 60% [dry weight]) that confers an exceptionally low permeability on mycobacteria (9, 13). Among the surface-exposed constituents of the cell envelope of M. tuberculosis, two structurally related families of lipids, the phthiocerol dimycocerosates (DIM) and the glycosylated phenolphthiocerol dimycocerosates, also called phenolglycolipids (PGL), have been extensively studied because of the key role they play in the pathogenesis of tuberculosis and the tubercle bacillus-host interactions (4, 8, 10, 12, 24). Consistently, DIM and PGL are produced by a limited group of slow-growing mycobacterial species, most of which are pathogenic for humans (10).
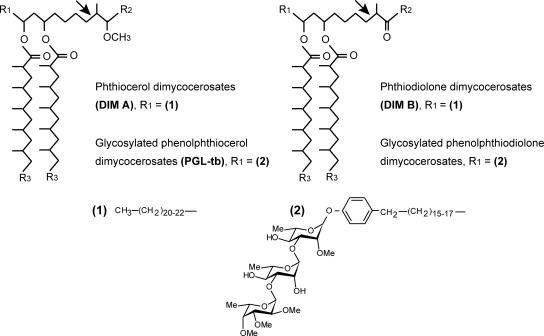
DIM are composed of a mixture of long-chain β-diols that are esterified by multimethyl-branched fatty acids called mycocerosic acids (Fig. 1). PGL consists of a lipid core similar to that of DIM, except that the phthiocerol chain is terminated by a glycosylated phenolic moiety (Fig. 1). More than 20 genes are required for the formation and translocation of DIM and PGL (for a review, see reference 21). These genes are clustered in a 73-kbp region of the chromosome of M. tuberculosis. The biosynthesis pathway of the lipid core common to DIM and PGL involves a group of six type I polyketide synthases (Pks) (18). These enzymes are large multienzymatic proteins that synthetize polyketides by decarboxylative condensation between an acyl chain and an extender unit (5, 16). The minimal core of a typical type I Pks consists of three domains, the ketosynthase (KS), the acyl transferase (AT), and the acyl carrier protein (ACP) domains, which are responsible for chain elongation. Additional modifying domains, such as a β-ketoreductase (KR) domain, a dehydratase (DH) domain, and an enoyl reductase (ER) domain, may subsequently modify the β-ketothioester intermediate generated by the core domains.
FIG. 1.
Structure of phthiocerol dimycocerosates and related compounds in M. tuberculosis. R2 is CH3 or C2H5; R3 is (CH2)16-18-CH3. Arrows show the position of the double bond reduced by the Rv2953 protein during phthiocerol and phenolphthiocerol biosynthesis.
A group of five modular type I Pks, PpsA through PpsE, was shown to be involved in the formation of phthiocerol and phenolphthiocerol chains (2). These Pks catalyze the elongation of either C22-C24 fatty acids or p-hydroxyphenylalkanoic acid by successive additions of three malonyl-CoA and two methylmalonyl-CoA extender units. Using the structure of these β-diols, Azad et al. (2) predicted the domain arrangement for each type I Pks encoded by the ppsABCDE genes. These domain assignments, except that for the PpsD protein, have been confirmed by bioinformatic studies with appropriate computational tools and also by biochemical studies (30, 33). Indeed, it had been proposed that PpsD contains the six domains, the KS, AT, DH, ER, KR, and ACP domains, but subsequent bioinformatic analyses failed to detect the ER domain (33), raising the possibility that a specific independent enzyme might be involved in the reduction of the double bond putatively left by PpsD during phthiocerol and phenolphthiocerol synthesis.
In this paper, we provide evidence that PpsD does not contain an ER domain and that Rv2953 encodes an independent reductase involved in the reduction of the double bond left in phthiocerol and phenophthiocerol chains by PpsD.
MATERIALS AND METHODS
Bacterial strains, growth media, and culture conditions.
The Escherichia coli DH5α strain used for cloning and M. tuberculosis strains were cultured as previously described (26).
General DNA techniques.
Molecular cloning and restriction endonuclease digestions were performed by standard procedures. Mycobacterial genomic DNA was isolated from 5-ml saturated cultures according to the method of Belisle and Sonnenberg (3).
Bioinformatic analysis of the Rv2953 protein.
We searched for sequence similarities between the Rv2953 and other proteins using the BLASTP program from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Secondary structure prediction and comparisons with known protein structures were carried out using the HHpred server at the Max-Planck Institute for Developmental Biology (http://toolkit.tuebingen.mpg.de/hhpred). HHpred builds a hidden Markov model (HMM) profile from a query sequence and compares it with a database of HMMs representing annotated protein families or domains with known structures (28). An HMM-HMM comparison using the HHpred server with the Rv2953 sequence as the query sequence was performed against the PDB70 database.
Construction of an Rv2953-disrupted mutant of M. tuberculosis H37Rv and a complementation vector.
An Rv2953 mutant of M. tuberculosis H37Rv containing a disrupted Rv2953::Km gene on the chromosome was constructed by allelic exchange using the Ts/sacB procedure (22, 26). A DNA fragment overlapping the Rv2953 gene was amplified by PCR from genomic DNA using oligonucleotides 2953A and 2953B (Table 1) and cloned, after the insertion of a Km resistance cassette between the ClaI-NruI restriction sites, into the mycobacterial thermosensitive suicide plasmid pPR27 (22). The resulting plasmid was transferred by electrotransformation into M. tuberculosis, and an allelic exchange at the Rv2953 locus was screened by PCR analysis of the genomic DNA from several Km- and sucrose-resistant colonies by using a set of specific primers (2953C, 2953D, 2953E, res1, and res2 [Table 1]). One clone with a pattern corresponding to the disruption of Rv2953 was selected and named PMM80.
TABLE 1.
Oligonucleotides used in this study
| Gene | Primer | Oligonucleotide sequence (5′-3′) |
|---|---|---|
| Rv2953 | 2953A | GCTCTAGAGTTTAAACATCAACCTGTACCACCGC |
| 2953B | GCTCTAGAGTTTAAACGGAACGGCATTTTCACAGC | |
| 2953C | GTCATGGCTAGGAACGCTAC | |
| 2953D | CTCATTGTTTCCGAGTAGCG | |
| 2953E | ACGTGTCAGCAGCTCGATAG | |
| 2953F | ACGGCATATGAGCCCAGCTGAGCGCGA | |
| 2953G | CAAAAGCTTAGCTCAGCCTGGTCGTTCC | |
| res | res1 | GCTCTAGAGCAACCGTCCGAAATATTATAAA |
| res2 | GCTCTAGATCTCATAAAAATGTATCCTAAATCAAATATC |
To construct pC2953 (for complementation), the Rv2953 gene was PCR amplified from M. tuberculosis H37Rv genomic DNA using oligonucleotides 2953F and 2953G (Table 1). The PCR product was digested with NdeI and HindIII and inserted between the NdeI and HindIII sites of pMV361eHyg, a pMV361 derivative harboring the pBlaF* promoter from pMIP12 instead of the original phsp60 promoter and carrying an hyg resistance marker (19, 29). This vector was transferred into PMM80 cells, and the transformants were selected on 7H11 agar plates supplemented with oleic acid-albumin-dextrose-catalase, Km, and Hyg.
Biochemical analyses of DIM and PGL.
DIM and PGL were obtained from mycobacterial cells as described in reference 26. For mass spectrometry and nuclear magnetic resonance (NMR) analyses, DIM and M. tuberculosis PGL were further purified by chromatography on a Florisil column (4, 7). Thin-layer chromatography (TLC), matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry, and NMR spectroscopy analyses were performed as previously described (26).
RESULTS
Identification of a gene putatively encoding the missing enoyl reductase of PpsD.
The ER domain of PpsC can be easily identified by bioinformatic analysis (33). In contrast, the PpsD sequence has no significant similarity with type I Pks ER domains. In an attempt to identify another enzyme which may fulfill this function, we searched the M. tuberculosis genome for a gene conserved in mycobacteria producing DIM, encoding a protein with similarities to reductase, and mapping within the DIM locus, which is the locus where all the other genes involved in DIM and PGL synthesis map. We identified a single, previously uncharacterized, candidate gene, Rv2953.
Rv2953 maps immediately downstream from Rv2952, which is involved in the methylation of the phthiocerol and phenolphthiocerol chains in mycobacterial species producing DIM, and Rv2953 orthologs are found in Mycobacterium marinum, Mycobacterium bovis, and even Mycobacterium leprae, a mycobacterial species which has undergone massive gene decay. Protein-protein BLAST and translated database similarity searches using the Rv2953 protein sequence as the query sequence revealed that the Rv2953 protein has similarities with a number of conserved prokaryotic proteins of unknown function. Many of these share similarities with saccharopine dehydrogenase, also known as saccharopine reductase, an oxidoreductase involved in the biosynthesis of lysine via the α-aminoadipate pathway in higher fungi (32). The three-dimensional structures of saccharopine reductases from the plant pathogen Magnaporthe grisea and from Saccharomyces cerevisiae have recently been identified (1, 14). Each subunit of this homodimer consists of three structural domains; the first domain contains a Rossmann fold variant binding NADPH (14). The primary sequence of the Rv2953 protein has a low similarity with that of saccharopine reductase from M. grisea (15% identity; 33% similarity), but an HMM-HMM comparison using the HHpred program (28) revealed a strong correlation between the predicted secondary structure of the Rv2953 protein and the secondary structure of saccharopine reductase (see Fig. S1 in the supplemental material). This result indicates that the two proteins may adopt similar three-dimensional structures. In addition, many saccharopine reductase residues responsible for binding NADPH are either conserved or replaced with similar amino acids in the Rv2953 protein (see Fig. S1 in the supplemental material). Together these bioinformatic analyses strongly suggest that the Rv2953 protein has a Rossmann fold and binds NADPH, consistent with the function of the Rv2953 protein as a reductase.
Thus, Rv2953 fulfilled our criteria for encoding the missing enoyl reductase of PpsD. However, the biosynthetic role of the Rv2953 protein remained to be demonstrated experimentally.
Disruption of the Rv2953 gene in M. tuberculosis H37Rv and biochemical characterization of the Rv2953 mutant.
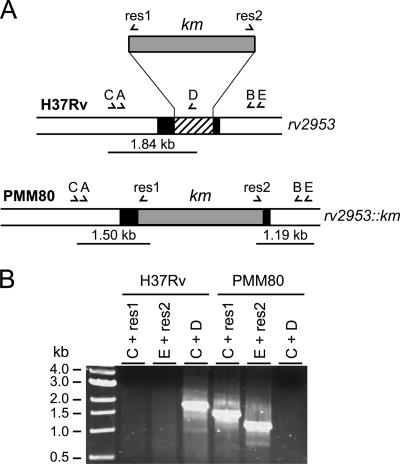
To establish whether the Rv2953 protein is involved in DIM and PGL biosynthesis, we constructed an M. tuberculosis H37Rv mutant by exchanging the wild-type allele of Rv2953 with a Km resistance cassette-disrupted allele (Fig. 2A). One clone, named PMM80, exhibiting an amplification pattern consistent with an allelic exchange at the Rv2953 locus, was retained for further analyses (Fig. 2B).
FIG. 2.
Construction and characterization of the M. tuberculosis H37Rv Rv2953::Km mutant strain. (A) Schematic diagram of the genomic organization of the Rv2953 locus in the wild-type strain of M. tuberculosis H37Rv and the PMM80 (Rv2953::Km) mutant. The black box represents the Rv2953 gene, and the hatched box represents the fragment deleted during the construction of the knockout mutant. The Km resistance cassette used for targeted disruption is represented by a gray box. Names of primers are indicated by letters (A, 2953A; B, 2953B; C, 2953C; D, 2953D; E, 2953E; res1; and res2). Positions are indicated by arrowheads below each genetic structure, and the expected sizes for PCR products are indicated. (B) PCR analysis of the recombinant strain PMM80 using various combinations of specific primers.
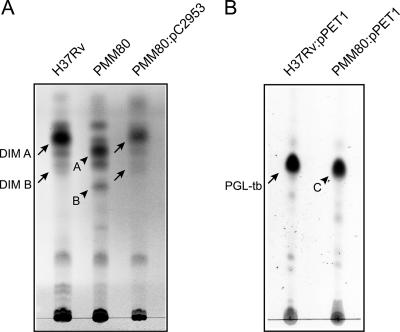
To determine the effects of the mutation in gene Rv2953, we then examined DIM production in the PMM80 mutant strain. TLC analyses showed that Rv2953 disruption resulted in the accumulation of compounds A and B (Fig. 3A) that exhibited lower mobilities than those of DIM A and DIM B, a related lipid of the DIM family harboring a keto moiety at the terminal end of the β-diol chain (Fig. 1) (10). This result suggested that the DIM produced by the Rv2953::Km mutant strain was structurally modified.
FIG. 3.
TLC analyses of lipids extracted from M. tuberculosis H37Rv and its isogenic Rv2953::Km mutant strain (PMM80). (A) TLC analysis of DIM from the M. tuberculosis wild-type strain, the PMM80 mutant, and the PMM80::pC2953 strain. Lipid extracts dissolved in CHCl3 were separated with petroleum ether-diethylether (90:10, vol/vol), and DIM were visualized by spraying the TLC plate with 10% phosphomolybdic acid in ethanol, followed by heating. Positions of DIM A and DIM B (arrows) and of products A and B (arrowheads) are indicated. (B) TLC analysis of glycolipids extracted from the M. tuberculosis wild-type and PMM80 mutant strains complemented with pPET1. Lipids were dissolved in CHCl3 and run in CHCl3/CH3OH (95:5, vol/vol). Glycoconjugates were visualized by spraying the TLC plate with 0.2% anthrone (wt/vol) in concentrated H2SO4, followed by heating. Positions of M. tuberculosis PGL (PGL-tb; arrow) and product C (arrowhead) are indicated.
Because DIM A and M. tuberculosis PGL share a common lipid domain, we also studied the role of the Rv2953 protein in the biosynthesis of the phenolphthiocerol moiety of this glycolipid. We previously demonstrated that M. tuberculosis H37Rv is unable to produce PGL due to the presence of a frameshift mutation in the pks15/1 gene and that the production of PGL can be restored by introducing a functional pks15/1 gene (7). Accordingly, both M. tuberculosis H37Rv wild-type and the PMM80 mutant strains were transformed with pPET1, a plasmid with a functional M. bovis BCG pks15/1 gene (7). Lipids were extracted, and the glycolipids were analyzed. The PMM80::pPET1 mutant produced a major glycoconjugate (product C) (Fig. 3B, lane 2) exhibiting a slightly lower TLC mobility than that of PGL produced from the wild-type strain carrying pPET1, strongly suggesting that the Rv2953 protein is also involved in the production of the conventional PGL in M. tuberculosis.
Structural analyses of DIM and PGL derivatives from M. tuberculosis Rv2953-disrupted mutant.
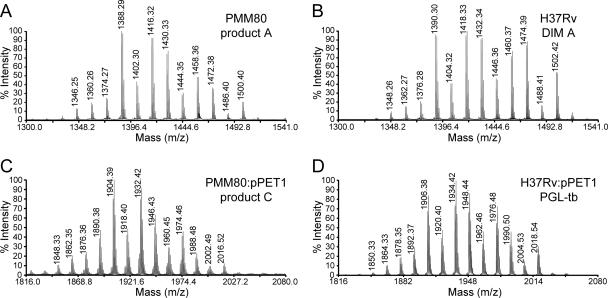
To describe the DIM-like compounds produced by the PMM80 mutant, products A and B were purified and analyzed by MALDI-TOF mass spectrometry. The mass spectrum of product A showed a series of pseudomolecular ion (M + Na+) peaks whose major homologues appeared at m/z ratios of 1388, 1416, 1430, and 1458 (Fig. 4A). Product B contained a series of pseudomolecular ion (M + Na+) peaks 16 atomic mass units lower than those for product A (data not shown). These values for products A and B were consistent with compound A bearing a methoxy group, whereas compound B contains a keto group instead. However, these values were 2 mass units lower than those observed in the spectra of DIM A (major peaks at m/z 1390, 1418, 1432, 1460, and 1474) (Fig. 4B) and DIM B (major peaks at m/z 1374, 1402, 1416, and 1444) (data not shown) from the wild-type strain. These data suggest that the PMM80 mutant produced lipids differing from DIM A and DIM B by the presence of an additional double bond. The transformation of this mutant strain with pC2953, a plasmid containing an intact Rv2953 gene, restored the wild-type phenotype (Fig. 3A). In addition, the mass spectra of the lipids purified from the complemented PMM80::pC2953 strain were superimposable to those of DIM A and DIM B from the wild-type H37Rv strain (data not shown). These data confirmed that the disruption of Rv2953 was solely responsible for the structural modification of DIM in the PMM80 mutant strain.
FIG. 4.
MALDI-TOF mass spectra of purified product A from the PMM80 mutant strain (panel A), of purified DIM A from M. tuberculosis wild-type H37Rv (panel B), of purified glycolipid (product C) from the PMM80::pPET1 strain (panel C), and of purified PGL from M. tuberculosis H37Rv::pPET1 (PGL-tb; panel D).
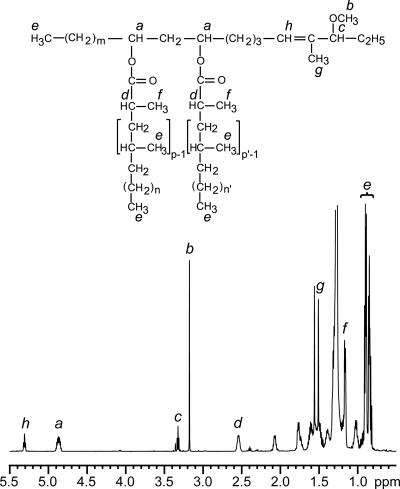
We also used NMR to analyze the A and B lipid products accumulated in PMM80. The 1H-NMR spectrum of product A from PMM80 and the structure deduced from the structural analysis are shown in Fig. 5. Most of the proton signal resonances that typify DIM A (4) were detected: (i) the multiplet centered at 4.83 p.p.m. (signal a), attributable to the resonances of the methine protons of the esterified β-diol; (ii) resonances of several terminal methyl protons seen at 0.8 to 1.0 p.p.m. (signal e), consistent with the presence of multimethyl-branched fatty acyl residues (11); (iii) resonances at 2.55 p.p.m. (signal d) and 1.15 p.p.m. (signal f) that correspond to the resonances of the methine protons at the α position of the fatty acyl residues and to those of the methyl groups located on this α carbon, respectively; and (iv) proton resonances of the methoxyl group observed at 3.16 p.p.m. (signal b). Interestingly, the proton resonance of the methine proton of the carbon bearing this methoxyl group, expected at 2.85 p.p.m. in the spectrum of DIM A (4), was deshielded and observed at 3.31 p.p.m. (signal c) because of the proximity of the double bond. Additional resonances were also observed at 1.50 p.p.m. and 5.30 p.p.m. and correspond to the proton resonance of the methyl group in the α position of the double bond (signal g) and to that of the double bond (signal h), respectively. These resonances were assigned by two-dimensional homonuclear 1H-1H correlation spectroscopy and heteronuclear 1H-13C single-bond correlation (heteronuclear single-quantum coherence spectroscopy) experiments, and the terminal motif of compound A containing the double bond was unambiguously established by a proton-detected 1H-13C heteronuclear multiple-bond correlation experiment (data not shown).
FIG. 5.
The 1H-NMR spectrum of product A from the PMM80 mutant strain. The structure of the analyzed compound deduced from the various structural analyses is shown above the spectrum (p, p' = 3 to 5; n, n' = 16 to 18; m = 20 to 22). Letters (a to h) shown above the formula indicate the protons, and those above the spectrum indicate the corresponding signal resonances.
These data clearly demonstrated that product A is a phthiocerol dimycocerosate derivative unsaturated between C-4 and C-5. Similar NMR analyses of product B, purified from PMM80, indicated that this lipid is a DIM B-like compound with a double bond between C-4 and C-5 (data not shown).
The TLC results with wild-type or mutant strains made competent for PGL production also strongly suggested that a similar structural change had occurred in the PGL-like substance produced by the PMM80::pPET1 mutant strain. This glycoconjugate was purified from bacterial cells and analyzed by MALDI-TOF mass spectrometry. Major pseudomolecular ion (M + Na+) peaks were seen at m/z ratios of 1904, 1932, and 1946 (Fig. 4C). These mass peaks were 2 mass units lower than those in the mass spectrum of M. tuberculosis PGL from the H37Rv::pPET1 strain (Fig. 4D), strongly supporting the concept that the major glycolipid produced by the PMM80::pPET1 mutant strain is M. tuberculosis PGL with an additional unsaturated bond. These various biochemical and structural analyses show that mutation in gene Rv2953 had similar effects on DIM and M. tuberculosis PGL biosynthesis, leading to the accumulation of unsaturated lipids.
DISCUSSION
The lipid core common to DIM and PGL is synthesized by a group of five modular type I Pks: PpsA through PpsE (30). It had been suggested that PpsD contains an ER domain responsible for the reduction of the double bond between the C-4 and C-5 during phthiocerol and phenolphthiocerol biosynthesis (2, 20). We and other groups (21, 33), however, failed to detect an ER domain by in silico analyses. This suggests that this reduction step might be catalyzed by an unidentified enzyme. We propose that PpsD generates an α,β-unsaturated thioester intermediate that is reduced by a trans enoyl reductase activity, encoded by the Rv2953 gene. This statement is supported by several key findings. Orthologs of Rv2953 are found in all analyzed mycobacterial species producing DIM (i.e., M. bovis, M. leprae, Mycobacterium ulcerans, and M. marinum), consistent with the involvement of the protein in the biosynthesis of phthiocerol. Moreover, we found that an Rv2953::Km mutant of H37Rv produces new lipids corresponding to DIM A and DIM B derivatives containing double bonds between C-4 and C-5. The presence of a functional pks15/1 gene in this strain consistently resulted in the synthesis of a major glycosylated phenolphthiocerol dimycocerosate containing a double bond. This phenotype was not due to a polar effect on downstream genes as the transfer of a functional Rv2953 gene, carried on a mycobacterial plasmid, fully reversed the biochemical phenotype. These findings demonstrate that (i) PpsD, which is thought to be involved in the synthesis of this fragment of the phthiocerol and phenolphthiocerol chain, has no functional ER domain and (ii) the product of Rv2953 is involved in the reduction of the double bond left by the PpsD synthase. An alternative model is that the Rv2953 protein has no reductase activity but may recruit an additional enzyme that fulfils this function. However, bioinformatic analyses of the Rv2953 protein sequence are consistent with this enzyme being an enoyl reductase and therefore do not support this alternative proposal. Enoyl reductases are NADH- or NADPH-dependent enzymes which have been categorized in two superfamilies: the short-chain dehydrogenase/reductase (SDR) superfamily and the medium-chain dehydrogenase/reductase superfamily (15, 23, 25). Most SDR enzymes have an N-terminal-binding motif G-X-G-X-X-G required for the binding of the cofactor (15, 23). However, short-chain ERs are members of the divergent family of SDRs, and they exhibit sequences that deviate from the typical motif encountered in most SDRs (23). Interestingly, the Rv2953 protein contains a G-A-T-G-F-S-G sequence (amino acids 14 to 20) at the N-terminal part of the protein. Additionally, the three-dimensional structures of several enoyl reductases have been established and they all exhibit a Rossmann fold (Protein Data Bank [http://www.rcsb.org/pdb/home/home.do]). The Rv2953 protein sequence was predicted to fold in the same way as saccharopine reductase, an enzyme with a Rossmann fold (14). Furthermore, many key residues involved in the binding of NADPH are either conserved or replaced with similar amino acids in the Rv2953 protein. Therefore, these bioinformatic analyses of the Rv2953 protein suggest that it has a Rossmann fold structure and binds NADH or NADPH, consistent with it being an enoyl reductase.
To our knowledge, this is the first report of a type I Pks requiring an additional accessory enzyme for polyketide biosynthesis in mycobacteria, although direct interactions between type I Pks and discrete enzymes have been reported for AT, KR, DH, and ER domains in other microorganisms (6, 17, 27, 31). For instance, in Aspergillus terreus, the lovastatin nonaketide synthase, a type I Pks that lacks a functional ER activity, interacts with LovC, a putative ER, for the correct assembly of dihydromonacolin L during lovastatin production (17). LovC probably acts on the growing polyketide chain bound to lovastatin nonaketide synthase. How the Rv2953 protein acts during DIM and M. tuberculosis PGL biosynthesis is still unclear. One possibility is that the Rv2953 protein interacts directly with PpsD during the synthesis of (phenol)phthiocerol and provides the ER activity in trans to PpsD to reduce the α,β-unsaturated thioester intermediate generated by this Pks. Alternatively, the Rv2953 protein might reduce the double bond after synthesis of the whole lipid chain by the five modular Pks, PpsA through PpsE. This reaction may occur before or after esterification of the chain with the mycocerosic acids. Further investigations are necessary to confirm our model and to determine the timing of the reduction step by the Rv2953 protein with respect to the synthesis of the phthiocerol chain.
Supplementary Material
Acknowledgments
We are grateful to Françoise Laval (mass spectrometry) and Anne Lemassu (NMR spectroscopy) for their valuable assistance.
The NMR spectrometers were financed by the CNRS, the University Paul Sabatier, the Région Midi-Pyrénées, and the European Structural Funds (FEDER). Roxane Siméone is a recipient of a European Commission fellowship.
Footnotes
Published ahead of print on 27 April 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andi, B., P. F. Cook, and A. H. West. 2006. Crystal structure of the his-tagged saccharopine reductase from Saccharomyces cerevisiae at 1.7-Å resolution. Cell Biochem. Biophys. 46:17-26. [DOI] [PubMed] [Google Scholar]
- 2.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 3.Belisle, J. T., and M. G. Sonnenberg. 1998. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 101:31-44. [DOI] [PubMed] [Google Scholar]
- 4.Camacho, L. R., P. Constant, C. Raynaud, M. A. Lanéelle, J. A. Triccas, B. Gicquel, M. Daffé, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 5.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 6:R319-R325. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, Y. Q., G. L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constant, P., E. Perez, W. Malaga, M. A. Lanéelle, O. Saurel, M. Daffé, and C. Guilhot. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277:38148-38158. [DOI] [PubMed] [Google Scholar]
- 8.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 9.Daffé, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 10.Daffé, M., and M. A. Lanéelle. 1988. Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 134:2049-2055. [DOI] [PubMed] [Google Scholar]
- 11.Daffé, M., and P. Servin. 1989. Scalar, dipolar-correlated and J-resolved 2D-NMR spectroscopy of the specific phenolic mycoside of Mycobacterium tuberculosis. Eur. J. Biochem. 185:157-162. [DOI] [PubMed] [Google Scholar]
- 12.Goren, M. B., O. Brokl, and W. B. Schaefer. 1974. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect. Immun. 9:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarlier, V., and H. Nikaido. 1990. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J. Bacteriol. 172:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson, E., J. J. Steffens, Y. Lindqvist, and G. Schneider. 2000. Crystal structure of saccharopine reductase from Magnaporthe grisea, an enzyme of the alpha-aminoadipate pathway of lysine biosynthesis. Structure 8:1037-1047. [DOI] [PubMed] [Google Scholar]
- 15.Kallberg, Y., U. Oppermann, H. Jornvall, and B. Persson. 2002. Short-chain dehydrogenases/reductases (SDRs). Eur. J. Biochem. 269:4409-4417. [DOI] [PubMed] [Google Scholar]
- 16.Keating, T. A., and C. T. Walsh. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3:598-606. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy, J., K. Auclair, S. G. Kendrew, C. Park, J. C. Vederas, and C. R. Hutchinson. 1999. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 284:1368-1372. [DOI] [PubMed] [Google Scholar]
- 18.Kolattukudy, P. E., N. D. Fernandes, A. K. Azad, A. M. Fitzmaurice, and T. D. Sirakova. 1997. Biochemistry and molecular genetics of cell-wall lipid biosynthesis in mycobacteria. Mol. Microbiol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 19.Le Dantec, C., N. Winter, B. Gicquel, V. Vincent, and M. Picardeau. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183:2157-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minnikin, D. E., L. Kremer, L. G. Dover, and G. S. Besra. 2002. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9:545-553. [DOI] [PubMed] [Google Scholar]
- 21.Onwueme, K. C., C. J. Vos, J. Zurita, J. A. Ferreras, and L. E. Quadri. 2005. The dimycocerosate ester polyketide virulence factors of mycobacteria. Prog. Lipid Res. 44:259-302. [DOI] [PubMed] [Google Scholar]
- 22.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson, B., Y. Kallberg, U. Oppermann, and H. Jornvall. 2003. Coenzyme-based functional assignments of short-chain dehydrogenases/reductases (SDRs). Chem.-Biol. Interact. 143-144:271-278. [DOI] [PubMed] [Google Scholar]
- 24.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84-87. [DOI] [PubMed] [Google Scholar]
- 25.Riveros-Rosas, H., A. Julian-Sanchez, R. Villalobos-Molina, J. P. Pardo, and E. Pina. 2003. Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur. J. Biochem. 270:3309-3334. [DOI] [PubMed] [Google Scholar]
- 26.Siméone, R., P. Constant, W. Malaga, C. Guilhot, M. Daffé, and C. Chalut. 2007. Molecular dissection of the biosynthetic relationship between phthiocerol and phthiodiolone dimycocerosates and their critical role in the virulence and permeability of Mycobacterium tuberculosis. FEBS J. 274:1957-1969. [DOI] [PubMed] [Google Scholar]
- 27.Simunovic, V., J. Zapp, S. Rachid, D. Krug, P. Meiser, and R. Muller. 2006. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. Chembiochem 7:1206-1220. [DOI] [PubMed] [Google Scholar]
- 28.Söding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, Jr., and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 30.Trivedi, O. A., P. Arora, A. Vats, M. Z. Ansari, R. Tickoo, V. Sridharan, D. Mohanty, and R. S. Gokhale. 2005. Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell 17:631-643. [DOI] [PubMed] [Google Scholar]
- 31.Ward, S. L., Z. Hu, A. Schirmer, R. Reid, W. P. Revill, C. D. Reeves, O. V. Petrakovsky, S. D. Dong, and L. Katz. 2004. Chalcomycin biosynthesis gene cluster from Streptomyces bikiniensis: novel features of an unusual ketolide produced through expression of the chm polyketide synthase in Streptomyces fradiae. Antimicrob. Agents Chemother. 48:4703-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, H., B. Andi, J. Qian, A. H. West, and P. F. Cook. 2006. The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem. Biophys. 46:43-64. [DOI] [PubMed] [Google Scholar]
- 33.Yadav, G., R. S. Gokhale, and D. Mohanty. 2003. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328:335-363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.