Abstract
The obligate intracellular nature of chlamydiae presents challenges to the characterization of its phages, which are potential tools for a genetic transfer system. An assay for phage infectivity is described, and the infectious properties of phage Chp2 were determined.
Chlamydia and Chlamydophila spp. are pathogenic obligate intracellular parasitic bacteria. The lack of a DNA transfer system has hindered the use of genetic approaches which could elucidate the molecular mechanisms of pathogenesis, microbial control, and cellular physiology. Although chlamydiaphages could be adapted for this purpose (3, 5, 6), host intracellular replication presents technical challenges to phage characterization. Several novel assays were developed to determine the specific infectivity of chlamydiaphage Chp2 and its host Chlamydophila abortus. The phage is highly infectious, with specific infectivity of 0.5 to 1.0. In contrast, the specific infectivity of C. abortus is 2 orders of magnitude lower than that of its phage.
Infectious virion assays rely on host cell viability and, in this case, the ability of the bacterial host to infect a eukaryotic cell. To infect Chlamydophila abortus with phage Chp2, a member of the Microviridae, the bacterial host must first be propagated and isolated from eukaryotic cells. Although preparations typically yield a mixture of elementary bodies (EBs) and reticulate bodies (RBs), the latter of which are not infectious (8), RBs are osmotically fragile (4) and were lysed during preparation. However, not all EBs are infectious in laboratory procedures. Thus, to determine the specific infectivity of Chp2, the specific infectivity of C. abortus B577 (inclusion-forming units [ifu]/total cell count) had to be determined. The titer of ifu represents the number of infectious and/or viable EBs in a sample under the chosen laboratory conditions.
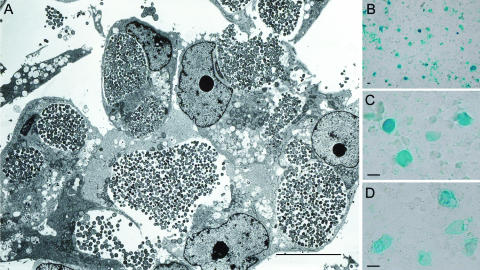
C. abortus forms densely packed inclusions during infection (Fig. 1A), which makes it amenable for the development of an accurate immunostaining titration protocol. EBs of the host, a bacteriophage-free strain of C. abortus (B577), were titrated in BGMK cells seeded at 5.0 × 104 cells/well on 96-well microtiter tissue-culture-quality plates. At 72 h postinfection, the time to completion of the C. abortus developmental cycle, cells were fixed with methanol for 20 min at −20°C before incubation with a chlamydia-specific monoclonal antibody at 4.0°C for 16 h. After unbound antibody was removed, samples were incubated with a second-affinity anti-mouse antibody conjugated with β-galactosidase (Calbiochem). β-Galactosidase was chosen as the conjugate over fluorescent conjugates to facilitate the assay for a variety of reasons, including ease of use and cost. Moreover, stained-inclusion procedures can be conducted with a standard light microscope, allowing observation of an entire well, as opposed to fields, which could yield more reliable counts. For staining, 100 μl of a staining solution [5.0 mM K3Fe(CN)6, 5.0 mM K4Fe(CN)6·3H2O, 2.0 mM MgCl2·6H2O, 0.25 M 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)] was added per well and incubated for 4.0 h at 37°C. The chromogenic substrate X-Gal generates blue-stained C. abortus inclusions, which were easily and reliably recognized using the light microscope (Fig. 1B and C). The titer of infectious EBs in this preparation was approximately 1.7 × 107 ifu/ml.
FIG. 1.
The morphology of C. abortus inclusions. (A) Transmission electron micrograph illustrating the morphology of BGMK cells infected at a high multiplicity of infection by C. abortus strain B577 at 72 h postinfection. Inclusions were densely packed and contained a mixture of mature EBs and RBs. (B) Cells infected with C. abortus stained with a chlamydia-specific monoclonal antibody (MAb 29) at 72 h postinfection at low magnification, demonstrating a microscopic field. (C and D) Chp2-infected C. abortus inclusions stained with MAb 29 at high magnification (C) and stained with a chlamydiaphage-specific monoclonal antibody (MAb 55) (D). The scale bars represent 20 μm.
To determine the total number of bacterial cells in samples regardless of infectivity, the concentration of C. abortus genomes was calculated by quantitative PCR. By this assay the number of genomes was found to be 6.9 × 109 genomes/ml (Fig. 2). Thus, approximately 1 out of 400 C. abortus bacterial cells was infectious in this preparation, assuming one genome per bacterium. A similar specific infectivity for a C. trachomatis strain that also requires centrifugation for cell infection has been previously reported (7). To determine whether free chlamydial genomes were influencing the quantitative PCR results, a particle count of EBs (infectious and noninfectious) was also performed in the presence of concentration-quantified 100-nm-diameter latex spheres (data not shown). The EB concentration from this assay was approximately 7.3 × 109 bacterial cells/ml, a value in good agreement with the results of the quantitative PCR assay. While a small percentage of aggregates must exist in any chlamydial preparation, none were observed in multiple fields assayed by electron microscopy (EM) during EB particle counting. Thus, eukaryotic cells infected by an aggregate of EBs are unlikely to significantly affect the results.
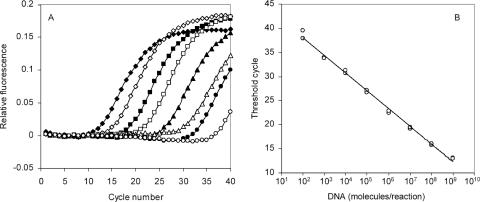
FIG. 2.
Real-time PCR analysis of C. abortus uvrD standard DNA. (A) Amplification plot of the C. abortus uvrD assay using serial, 10-fold dilutions of the puvrD plasmid as the template. ⧫, 109 plasmid DNA molecules/reaction; ⋄, 108; ▪, 107; □, 106; ▴, 105; ▵, 104; •, 103; ○, 102. The amplification plots are representative and illustrate, for each template concentration, the change in fluorescence as the PCR cycle number increases incrementally. (B) Standard curve derived from data presented in panel A. The cycle threshold (CT) for each reaction is the cycle number at which a fixed threshold value of relative fluorescence is attained in the amplification plot. Standard curves of the CT versus the number of DNA molecules/reaction were used to calculate unknowns. Standard reactions were performed in duplicate.
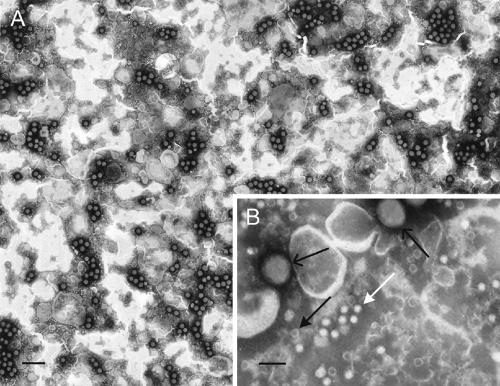
Partially purified chlamydiaphage preparations contain infectious virions, procapsids, and packaging intermediates (1). The presence of packaging intermediates, with unpackaged replicative-form DNA, complicates infectious particle titer determination by quantitative PCR. To circumvent this problem, the titer of Chp2 virions was determined by EM counting with latex spheres of a known concentration. Ratios of virions and procapsids to latex spheres can be calculated, as each particle has a distinctive morphology (Fig. 3). Latex spheres are relatively large structures (diameter, ∼100 nm) compared to procapsids and virions (diameter, ∼20 nm). Virions and procapsids can be further differentiated by their appearance. Procapsids, devoid of genomes, stain with a darkened center, while stain does not penetrate virions (Fig. 3B). The titers determined by this technique were approximately 3.0 × 1012 virions/ml and 1.5 × 1012 procapsids/ml.
FIG. 3.
Electron micrographs of chlamydiaphages. (A) Typical low-magnification field of latex spheres mixed with Chp2. The scale bar represents 500 nm. (B) A higher-magnification field showing procapsids (black arrow, solid head), virions (white arrow), and latex spheres (black arrow, line head). The scale bar represents 100 nm.
To determine the number of virion-like staining particles that were infectious, a phage-specific primary monoclonal antibody was used to stain phage-infected C. abortus inclusions. For these assays, BGMK cells were infected with C. abortus B577 at a multiplicity of infection of 1.0 ifu/eukaryotic cell. A 10-fold dilution series of semipurified Chp2 was preincubated with C. abortus for 30 min prior to infecting BGMK cells. Cells were stained with a phage-specific primary monoclonal antibody and then with a β-galactosidase-conjugated second-affinity antibody as described above. Inclusions formed by bacteriophage-infected C. abortus stained as blue compact structures, which had a sharply defined outline and therefore were easy to count directly by light microscopy (Fig. 1D).
By this assay, which was performed multiple times with little variation, the titer of infectious particles was approximately 5.8 × 109 chlamydiaphage-stained inclusions/ml, a value significantly at variance with the EM-determined particle count of approximately 3.0 × 1012 virion-staining particles/ml. However, only 1 in 400 (0.25%) C. abortus B577 EBs is infectious. The presence of noninfectious EBs affects the determination of viable phage titers. Chp2 is known to bind noninfectious RBs and killed EBs with the same efficiency with which it binds to live EBs (2).
Thus, the low specific infectivity of the host (2.5 × 10−3 ifu/bacterial cell) should be taken into account when determining the true specific infectivity of the phage. For every event of attachment to an infectious EB, there are approximately 400 events involving replication-incompetent bacterial host cells. Thus, a titer of approximately 5.8 × 109 chlamydiaphage-stained inclusions/ml reflects approximately 2.3 × 1012 infectious particles/ml, a value close to the EM-determined titer of approximately 3.0 × 1012, indicating that Chp2 is highly infectious.
Acknowledgments
We are grateful to Sylvia Everson and Patrick Pead for technical support and to Priscilla Wyrick for useful discussions.
This work was funded by project grant 074062 from the Wellcome Trust (I.N.C.), NSF MCB 0542978 (B.A.F.), and USDA-Hatch funds to the University of Arizona.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Clarke, I. N., L. T. Cutcliffe, J. S. Everson, S. A. Garner, P. R. Lambden, P. J. Pead, M. A. Pickett, K. L. Brentlinger, and B. A. Fane. 2004. Chlamydiaphage Chp2, a skeleton in the ϕX174 closet: scaffolding protein and procapsid identification. J. Bacteriol. 186:7571-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Everson, J. S., S. A. Garner, B. Fane, B. L. Liu, P. R. Lambden, and I. N. Clarke. 2002. Biological properties and cell tropism of Chp2, a bacteriophage of the obligate intracellular bacterium Chlamydophila abortus. J. Bacteriol. 184:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garner, S. A., J. S. Everson, P. R. Lambden, B. A. Fane, and I. N. Clarke. 2004. Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila pecorum. Virus Genes 28:207-214. [DOI] [PubMed] [Google Scholar]
- 4.Hatch, T. P., I. Allan, and J. H. Pearce. 1984. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J. Bacteriol. 157:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsia, R. C., L. M. Ting, and P. M. Bavoil. 2000. Microvirus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis: isolation and molecular characterization. Microbiology 146:1651-1660. [DOI] [PubMed] [Google Scholar]
- 6.Liu, B. L., J. S. Everson, B. Fane, P. Giannikopoulou, E. Vretou, P. R. Lambden, and I. N. Clarke. 2000. Molecular characterization of a bacteriophage (Chp2) from Chlamydia psittaci. J. Virol. 74:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peeling, R. W., and R. C. Brunham. 1991. Neutralization of Chlamydia trachomatis: kinetics and stoichiometry. Infect. Immun. 59:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913-925. [DOI] [PubMed] [Google Scholar]