Abstract
Salmonella enterica modifies its lipopolysaccharide (LPS), including the lipid A portion, to adapt to its environments. The lipid A 3-O-deacylase PagL exhibits latency; deacylation of lipid A is not usually observed in vivo despite the expression of PagL, which is under the control of a two-component regulatory system, PhoP-PhoQ. In contrast, PagL is released from latency in pmrA and pmrE mutants, both of which are deficient in aminoarabinose-modified lipid A, although the biological significance of this is not clear. The attachment of aminoarabinose to lipid A decreases the net anionic charge at the membrane's surface and reduces electrostatic repulsion between neighboring LPS molecules, leading to increases in bacterial resistance to cationic antimicrobial peptides, including polymyxin B. Here we examined the effects of the release of PagL from latency on resistance to polymyxin B. The pmrA pagL and pmrE pagL double mutants were more susceptible to polymyxin B than were the parental pmrA and pmrE mutants, respectively. Furthermore, introduction of the PagL expression plasmid into the pmrA pagL double mutant increased the resistance to polymyxin B. In addition, PagL-dependent deacylation of lipid A was observed in a mutant in which lipid A could not be modified with phosphoethanolamine, which partly contributes to the PmrA-dependent resistance to polymyxin B. These results, taken together, suggest that the release of PagL from latency compensates for the loss of resistance to polymyxin B that is due to a lack of other modifications to LPS.
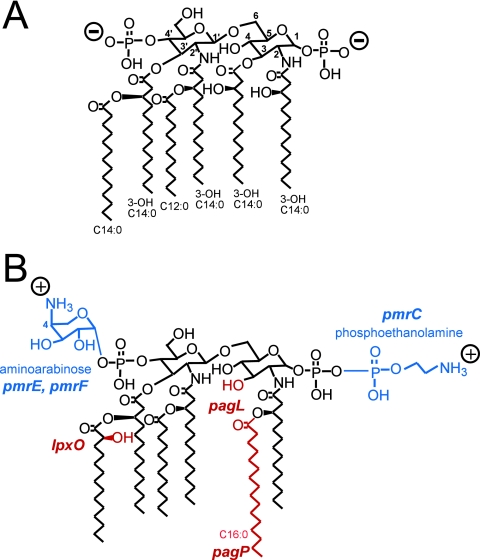
The outer membrane of pathogenic gram-negative bacteria, including Salmonella enterica, functions as a barrier to harmful host-derived compounds, such as antimicrobial peptides, gastric secretions, bile, and reactive oxygen. The protection is a consequence of strong lateral interaction between lipopolysaccharide (LPS) molecules, which occupy exclusive areas of the outer leaflet of the outer membrane (reviewed in reference 30). LPS consists of a hydrophobic membrane anchor portion known as lipid A and a nonrepeating core oligosaccharide coupled to a distal polysaccharide (O antigen) that extends from the bacterial surface (reviewed in reference 33). The prototype lipid A structure synthesized by different enteric bacteria, such as S. enterica serovar Typhimurium and Escherichia coli, is relatively conserved (Fig. 1A). In response to environmental conditions, S. enterica covalently modifies its lipid A through palmitoylation, deacylation, the formation of a 2-hydroxymyristate group (hydroxylation), and the addition of 4-amino-4-deoxy-l-arabinose (aminoarabinose) or phosphoethanolamine (7, 33) (Fig. 1B). Similar modifications occur in other gram-negative bacteria, including E. coli (44), Yersinia pestis (22), and Pseudomonas aeruginosa (8), and the modifications are thought to be conserved among gram-negative bacteria for adaptation to their environments.
FIG. 1.
PhoP-PhoQ- and PmrA-PmrB-regulated lipid A modifications in S. enterica serovar Typhimurium. (A) Prototype lipid A of S. enterica serovar Typhimurium. (B) Modified lipid A of S. enterica serovar Typhimurium. The phosphate residues and acyl chains of lipid A of S. enterica can be derivatized in a PhoP-PhoQ- or PmrA-PmrB-regulated manner (reviewed in reference 7). Aminoarabinose and/or phosphoethanolamine groups (shown in blue) can be attached to phosphate residues, under the control of PmrA-PmrB (18, 45). Minor species were present in which the locations of the aminoarabinose and phosphoethanolamine groups were reversed or in which both phosphates were modified with the same substituent (45). Both the pmrF operon and pmrE are necessary for the PmrA-PmrB-regulated attachment of aminoarabinose to lipid A (15, 45). pmrC mediates the PmrA-PmrB-regulated attachment of phosphoethanolamine to lipid A (27). The addition of the palmitate chain is catalyzed by PagP (3, 19), the formation of the 2-hydroxymyristate group requires LpxO (13), and the deacylation at position 3 of lipid A is catalyzed by PagL (39) (shown in red). The pagL and pagP genes are regulated by PhoP-PhoQ (2), and the lpxO gene is partly regulated by PhoP-PhoQ (12, 13). PhoP-PhoQ also activates PmrA-PmrB; therefore, the aminoarabinose and phosphoethanolamine modifications occur under PhoP-PhoQ-activating conditions (18, 44).
In S. enterica serovar Typhimurium, the two-component regulatory system PhoP-PhoQ induces the expression of genes involved in modifying lipid A (18). PhoQ is a sensor histidine kinase that responds to environmental conditions, including those within mammalian tissues, which are mimicked by magnesium-limited growth medium (9, 10, 14, 28). In response to specific environmental signals, PhoQ phosphorylates PhoP, leading to the activation of pagL and pagP encoding lipid A 3-O-deacylase and lipid A palmitoyltransferase, respectively (2, 3, 19, 39). Both PagL and PagP are outer membrane enzymes (3, 39), indicating that these enzymes modify mature LPS molecules that are transported to the outer membrane. Lipid A 3-O-deacylation and palmitoylation reduce the ability of lipid A to activate the host Toll-like receptor 4, suggesting that these modifications help pathogens to avoid innate immune recognition (23).
Activation of PhoP-PhoQ leads to the activation of a second two-component regulatory system, PmrA-PmrB (16, 21), which promotes the attachment of aminoarabinose and phosphoethanolamine to phosphate groups on lipid A (Fig. 1). The modification with aminoarabinose is essential for resistance to cationic antimicrobial peptides including polymyxin B (15, 16, 27), which is a cyclic antimicrobial lipopeptide produced by the soil bacterium Paenibacillus polymyxa (32). Cationic polymyxin B binds initially to the anionic surface of gram-negative bacteria, in particular to LPS (40), and then invades hydrophobic membranes. The modification of lipid A with aminoarabinose reduces the net anionic charge at this position and the electrostatic repulsion between neighboring LPS molecules (30), and these changes are essential for resistance to polymyxin B (15, 27, 31). In a previous study, PmrA-PmrB-regulated lipid A modifications were required for maximal virulence in BALB/c mice infected with S. enterica via the oral route (17), suggesting that the changes to the membrane, which were represented by resistance to polymyxin B, were involved in the virulence of salmonellae.
Previous studies demonstrated that deacylated lipid A species were not detected despite the presence of the lipid A deacylase PagL in S. enterica (24, 39); therefore, PagL is thought to be latent in the outer membrane under these conditions (24). In contrast, PagL-dependent deacylation of lipid A was detected in the pmrA, pmrE, and pmrF mutants, which are deficient in aminoarabinose-modified lipid A (Fig. 1B) (24). These results suggest that the modification with aminoarabinose is involved in the latency of PagL, although its biological significance is not clear (24).
In this study, we examined the effects of PagL-dependent deacylation of lipid A using pmrA pagL and pmrE pagL double mutants. The PagL-null strains were more susceptible to polymyxin B than were the parental strains. These results indicate that the release of PagL from latency compensates for the loss of inducible polymyxin B resistance that is dependent on the modification of lipid A with aminoarabinose.
MATERIALS AND METHODS
Materials.
All chemicals were of reagent grade or better. Restriction endonucleases and DNA-modifying enzymes were from New England Biolabs (Beverly, MA) and TAKARA BIO (Ohtsu, Japan). Polymyxin B was from Sigma-Aldrich (St. Louis, MO). Oligonucleotides were prepared commercially by Invitrogen Japan (Tokyo, Japan) and Texas Genomics Japan (Tokyo, Japan). Prestained molecular weight standards were from Apro Science (Naruto, Japan).
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. enterica serovar Typhimurium strain 14028s (American Type Culture Collection, Manassas, VA) was used as the wild-type strain in this study.
TABLE 1.
S. enterica serovar Typhimurium strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium | ||
| ATCC 14028s | Wild type | ATCC |
| JSG421 | pmrA::Tn10d | 16 |
| KCS041 | pmrE1::Tn10d | 24 |
| KCS178 | ΔpmrC::Kan | This work |
| KCS180 | ΔpmrC | This work |
| KCS202 | ΔpagL::Kan | This work |
| KCS203 | pmrA::Tn10d ΔpagL::Kan | This work |
| KCS204 | pmrE1::Tn10d ΔpagL::Kan | This work |
| KCS205 | ΔpmrC ΔpagL::Kan | This work |
| KCS208 | pmrA::Tn10d ΔpagL | This work |
| KCS209 | pmrE1::Tn10d ΔpagL | This work |
| KCS210 | ΔpmrC ΔpagL | This work |
| KCS216 | ΔpagL | This work |
| Plasmid | ||
| pWKS30 | Low-copy-number cloning vector | 41 |
| pWLP23 | pWKS30 containing pagL including 79-bp upstream region | 39 |
| pBAD24 | Inducible expression plasmid | 20 |
| pKK28 | pBAD24 containing pmrC-coding region | This work |
| pWLP21 | pBluescript KS II+ containing pagL | 39 |
| pKK7 | pEt15b containing pagL, which bears His6 epitope instead of 1 to 18 amino acid residues at N terminus | This work |
Bacteria were grown at 37°C with aeration in N-minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.28% (vol/vol) glycerol, 0.1% (wt/vol) Casamino Acids, 2 mg of thiamine/liter, and 0.1 M Tris/HCl (pH 7.4 or 5.8)] supplemented with 10 μM MgCl2 (10, 24, 27, 29). Ampicillin (100 μg/ml) was used for the cultivation of strains transformed with the plasmid pBAD24 (20) and its derivatives, and ampicillin (10 μg/ml) was used for the cultivation of strains transformed with the low-copy-number vector pWKS30 (41) and its derivatives. Bacterial colonies were picked out and grown overnight at 37°C in 1 to 10 ml of growth medium. The overnight cultures were diluted 1:10 with fresh growth medium and then grown at 37°C for 24 h. After the cultivation, stationary-phase cells were used for further analysis.
Bacterial genetic and molecular biology techniques.
Phage P22-mediated transduction was performed as described previously (6). Plasmid DNA was introduced into bacterial strains by electroporation using E. coli Pulser (Bio-Rad, Hercules, CA) following the manufacturer's instructions. Recombinant DNA techniques were performed according to standard protocols (37).
Construction of nonpolar pagL or pmrC deletion mutants.
Nonpolar deletion mutant strains were generated according to the method established by Datsenko and Wanner (5). A DNA fragment containing the kanamycin resistance cassette of pKD4 (5) was amplified by PCR with EX Taq DNA polymerase (TAKARA BIO). Primers KK85 (GCAAGGGCAACAAGCATCAGATCTCTTTTGCTGCGGGAGAAAGTATAAGAGTGTAGGCTGGAGCTGCTTC) and KK86 (TATGCCCTGAATTTTTATCCGTAAGTGATCCATTCGAGAAATGCCGGATACATATGAATATCCTCCTTAG) were designed to delete 350 bp of the 558-bp PagL-coding DNA region. Primers KK71 and KK72, which were identical to primers 2635 and 2636 (27), respectively, were used for the generation of a nonpolar pmrC-deletion mutant (ΔpmrC). The PCR products were introduced into S. enterica serovar Typhimurium strain 14028s carrying the plasmid pKD46 (5), which encodes Red recombinase. Generation of the ΔpagL::kan allele in the resulting strain, KCS202, and the ΔpmrC::kan allele in KCS178 was confirmed by PCR analysis. Kanamycin resistance cassettes were eliminated from the ΔpagL::kan and ΔpmrC::kan strains by using the plasmid pCP20 (5), and the elimination was confirmed by PCR analysis.
Construction of PmrC expression plasmid.
The pmrC-coding region was amplified from genomic DNA of strain 14028s by PCR with Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The primers used were KK33 (GATTGGATCCGTCGCGTTTGTGTATTGCATCTGG) and KK35 (ATCATCCCATGGTAATGGACGCATCAACATGTTAAAGCG). The amplified DNA fragment was cloned into NcoI and HindIII sites of pBAD24 (20) under the control of the arabinose PBAD promoter, and the resulting expression construct was named pKK28. The insert in the plasmid construct was verified by sequencing.
Preparation of lipid A.
The lipid A used for mass spectrometry was purified as described previously (43). In brief, cells collected from 25 ml of culture were resuspended in 500 μl of Tri-reagent (Molecular Research Center Inc., Cincinnati, OH). After incubation for 30 min at room temperature, 100 μl of chloroform was added. After 15 min, the mixture was centrifuged, and the aqueous phase was recovered. LPS was extracted three times by the addition of 500 μl of water to the organic phase, and the aqueous phase containing LPS was dried up with a vacuum concentrator. Five hundred microliters of 10 mM sodium acetate buffer (pH 4.5) containing 1% sodium dodecyl sulfate (SDS) was add to the dried LPS, and then the LPS was hydrolyzed to remove sugar chains from lipid A by incubation at 100°C for 1 h (35) followed by drying. The dried lipid A was washed once with 0.02 N HCl in 95% ethanol, and three times with 95% ethanol. The washed lipid A was dried up with a vacuum concentrator and then used for mass spectrometric analysis.
Alternatively, the lipid A used for mass spectrometry (insets in Fig. 2A, B, E, and H) was prepared from LPS as described previously (4). In brief, LPS purified from 25 ml of cell culture by using a LPS extraction kit (iNtRON Biotechnologies Inc., Seongnam-Si, Korea) was hydrolyzed for 3 h at 95°C in 150 μl of 100 mM sodium acetate buffer (pH 4.5). Then, 600 μl of a chloroform/methanol mixture (1:2, vol/vol), 200 μl of chloroform, and 100 μl of phosphate-buffered saline were added in succession, and the lipid A fraction (chloroform phase) was dried under a stream of nitrogen gas.
FIG. 2.
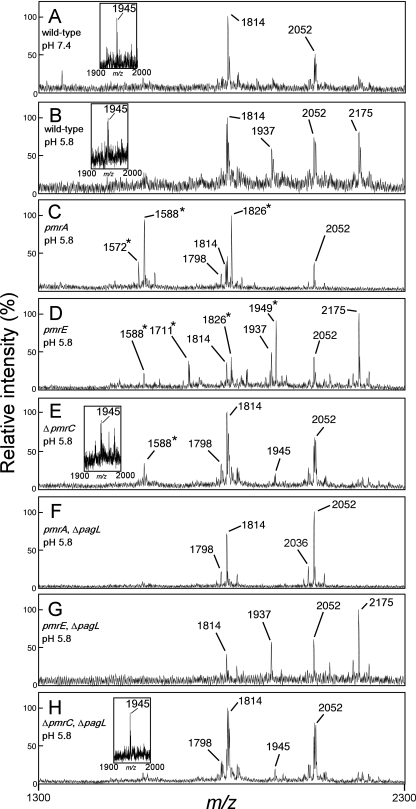
MALDI-TOF mass spectrometry of lipid A purified from S. enterica serovar Typhimurium strains. The wild-type (ATCC 14028s), pmrA (JSG421), pmrE (KCS041), ΔpmrC (KCS180), ΔpagL (KCS216), pmrA ΔpagL (KCS208), pmrE ΔpagL (KCS209), and ΔpmrC ΔpagL (KCS210) strains were cultivated in growth medium at pH 7.4 (A) or 5.8 (B to H). The m/z values of lipid A species are shown, and those that represent deacylated lipid A species are denoted by asterisks. Insets in panels A, B, E, and H show results of MALDI-TOF mass spectrometry of lipid A using 2-5-dihydroxybenzoic acid matrices. The structural interpretations of lipid A species are summarized in Table 2.
Mass spectrometry.
Dried lipid A was dissolved in 20 mg/ml 5-chloro-2-mercaptobenzothiazole matrices in chloroform/methanol (1:1, vol/vol). Alternatively, it was dissolved in chloroform/methanol (1:2, vol/vol) and then mixed with 2-5-dihydroxybenzoic acid matrices (77 mg/ml in methanol) at a ratio of 1:1 (insets in Fig. 2A, B, E, and H). The mixtures were allowed to dry at room temperature on the sample plate prior to analysis. Spectra were obtained in the negative reflection mode using a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) Voyager-DE STR mass spectrometer (Applied Biosystems Japan, Tokyo, Japan). Each spectrum was the average of 200 shots. Structural interpretations of lipid A species detected by mass spectrometry in this study are summarized in Table 2.
TABLE 2.
Structural interpretations of lipid A species detected by mass spectrometry in this study
| m/z | Lipid A modificationsa |
|---|---|
| 1572 | Deacylation |
| 1588 | Deacylation and hydroxylation |
| 1668 | Deacylation and hydroxylation (pyrophosphate lipid A)b |
| 1711 | Deacylation, hydroxylation, and phosphoethanolamine attachment |
| 1798 | (Unmodified) |
| 1810 | Deacylation and palmitoylation |
| 1814 | Hydroxylation |
| 1826 | Deacylation, palmitoylation, and hydroxylation |
| 1894 | Hydroxylation (pyrophosphate lipid A)b |
| 1921 | Phosphoethanolamine attachment |
| 1937 | Hydroxylation and phosphoethanolamine attachment |
| 1945 | Aminoarabinose attachment and hydroxylation |
| 1949 | Deacylation, palmitoylation, hydroxylation, and phosphoethanolamine attachment |
| 2036 | Palmitoylation |
| 2052 | Palmitoylation and hydroxylation |
| 2132 | Hydroxylation and palmitoylation (pyrophosphate lipid A)b |
| 2159 | Palmitoylation and phosphoethanolamine attachment |
| 2175 | Palmitoylation, hydroxylation, and phosphoethanolamine attachment |
Anti-PagL antibodies.
The His6-tagged recombinant PagL protein was used as an antigen with which to produce polyclonal antibody. The recombinant protein was prepared as follows. The pagL-coding region was amplified from the plasmid pWLP21 (39) with Pfu Turbo DNA polymerase. The primers used for PCR were KK14 (GATTGGATCCGTCGCGTTTGTGTATTGCATCTGG) and KK15 (CGCGGATCCTCAGAAATTATAACTAATTGA). A PagL expression plasmid was designed to express a partial PagL protein, which lacks 18 amino acid residues at the N terminus. The amplified DNA fragment was cloned into NdeI and BamHI sites of pET15b (Novagen, Darmstadt, Germany), which was designed to produce a recombinant protein carrying a His6 epitope at the N terminus. The resulting PagL expression construct pKK7 was introduced into E. coli strain BL21(DE3). The transformant grown in Luria-Bertani medium containing ampicillin (100 μg/ml) was cultivated for 4 h in the presence of 1 mM isopropyl β-d-thiogalactoside to induce the expression of the recombinant PagL protein. The His6-tagged recombinant PagL protein was purified from E. coli lysate with Ni-nitrilotriacetic agarose (QIAGEN) in accordance with the manufacturer's instructions. Immunization of a rabbit and preparation of serum were performed by Charles River Japan (Yokohama, Japan). The antibody against PagL protein was affinity purified as described previously (25) using the purified recombinant PagL protein.
SDS-polyacrylamide gel electrophoresis and Western blotting.
Proteins were fractionated by SDS-12.5% polyacrylamide gel electrophoresis under reducing conditions (26). Proteins separated on the gel were stained with Coomassie blue. For the Western blot analysis, proteins separated on the gel were electroblotted onto a nitrocellulose membrane in 25 mM Tris-192 mM glycine-0.02% SDS-20% methanol at 22 V/cm for 60 min. The blot was then incubated with affinity-purified anti-PagL antibodies and subsequently with anti-rabbit immunoglobulin G linked to horseradish peroxidase. Cross-reactive proteins were detected with ECL Western blotting detection reagents (GE Healthcare Bio-Sciences, Piscataway, NJ).
Polymyxin B killing assay.
Harvested cells from 5 ml of culture were washed two times with 1 ml of Luria-Bertani medium and then suspended with Luria-Bertani medium at an optical density at 600 nm of 0.12. The bacterial suspension (475 μl) was mixed with 25 μl of water or a polymyxin B solution and stood at 37°C. After 1 h, the cells were diluted 1:10,000 with Luria-Bertani medium and the diluted suspension (50 μl) was plated on a Luria-Bertani agar plate to determine the number of CFU. The dilution eliminated the killing effect of polymyxin B on the plates (data not shown). Percent survival was calculated as follows: (CFU of polymyxin B-treated culture/CFU of water-treated culture) × 100.
RESULTS
PagL-dependent lipid A deacylation was observed in Salmonella pmrA and pmrE mutant strains grown in mild acid medium but not in the wild-type strain.
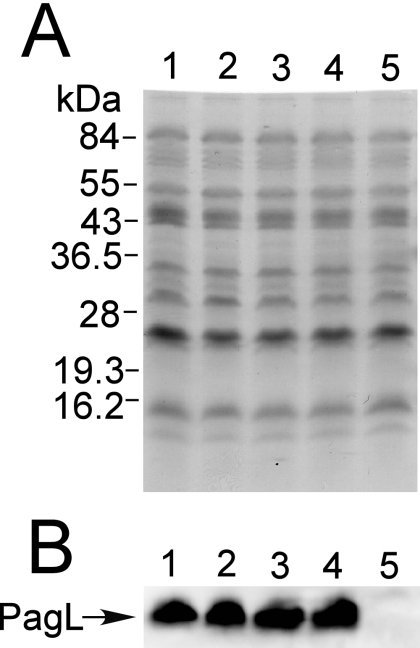
Previously, Kawasaki et al. showed that PagL-dependent 3-O-deacylation of lipid A was clearly observed in mutant strains of S. enterica serovar Typhimurium deficient in aminoarabinose-modified lipid A, including the pmrA, pmrE, and pmrF strains, when grown in defined minimal magnesium-limited neutral medium (pH 7.4), but not in the wild-type strain (24). We examined whether or not aminoarabinose-dependent latency of PagL is also observed in S. enterica grown in defined minimal magnesium-limited mild acid medium (pH 5.8), which is often used to mimic host microenvironments, such as phagolysosomes of macrophages (1, 10, 12, 27, 38, 42). In a MALDI-TOF mass spectrometric analysis, deacylated species were not detected in the lipid A prepared from the wild-type strain grown in the mild acid medium or in the neutral medium (Fig. 2A and B). In addition, significant amounts of lipid A species modified with phosphoethanolamine (m/z 1937 and 2175) were detected in the wild-type strain cultivated in the mild acid medium but not in that cultivated in the neutral medium (Fig. 2A and B). These observations were consistent with the finding that the wild-type Salmonella strain modifies its lipid A with phosphoethanolamine in defined minimal magnesium-limited mild acid medium (pH 5.8) (27). Aminoarabinose-modified lipid A (m/z 1945) was clearly detected in the wild-type strain (insets of Fig. 2A and B) but not in the pmrA and pmrE mutant strains (data not shown). On the other hand, deacylated lipid A species (m/z 1572, 1588, 1711, 1826, and 1949) were detected in the pmrA and pmrE mutant strains cultivated in the mild acid medium (Fig. 2C and D) as well as in the neutral medium (data not shown) (24). 3-O-deacylated lipid A species were not observed in the nonpolar pagL deletion (ΔpagL) mutant strains (pmrA ΔpagL and pmrE ΔpagL) cultivated in the mild acid medium (Fig. 2F and G), indicating that the deacylation observed in the pmrA and pmrE mutants is dependent on PagL. Furthermore, PagL protein levels were similar among the wild-type, pmrA, and pmrE strains grown in the mild acid medium (Fig. 3). These results indicate that PagL is latent in S. enterica strains grown in the mild acid medium as well as in the neutral medium.
FIG. 3.
Expression levels of PagL protein were similar among Salmonella wild-type, pmrA, pmrE, and ΔpmrC strains grown in the mild acid medium. Ten-microgram samples of membrane proteins prepared from strains cultivated in mild acid medium (pH 5.8) containing 10 μM MgCl2 were subjected to SDS-polyacrylamide gel (12.5%) electrophoresis and analyzed with staining (A) or by Western blotting (B). Lanes: 1, wild-type strain; 2, pmrA strain (JSG421); 3, pmrE strain (KCS041); 4, ΔpmrC strain (KCS180); 5, ΔpagL strain (KCS216).
PagL mutant strains are more susceptible to polymyxin B than the parental strains.
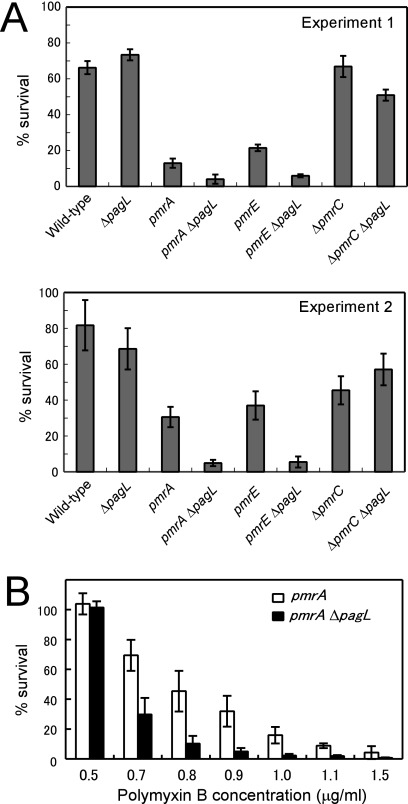
In S. enterica, pmrA-pmrB-dependent attachment of aminoarabinose to lipid A, which decreases the net charge of LPS and the electrostatic repulsion between neighboring LPS molecules, is responsible for the resistance to the cationic antimicrobial peptide polymyxin B (15, 31). It is plausible that the release of PagL from latency observed in the pmrA and pmrE mutant strains, which lack aminoarabinose-modified lipid A, increases the resistance of salmonellae to polymyxin B. Mutant strains pmrA ΔpagL and pmrE ΔpagL grown in the mild acid medium were more susceptible to polymyxin B (1.0 μg/ml) than were the parental pmrA and pmrE strains, respectively, suggesting that PagL-dependent deacylation of lipid A in the pmrA and pmrE mutant strains increased resistance to polymyxin B (Fig. 4A). To confirm the involvement of the PagL-dependent deacylation in the resistance, the pmrA and pmrA ΔpagL mutant strains grown in the mild acid medium were treated with various concentrations of polymyxin B. As shown in Fig. 4B, the pmrA strain was more resistant to polymyxin B than was pmrA ΔpagL. These results suggest that the PagL-dependent deacylation of lipid A in the pmrA strain increased resistance to polymyxin B. Mutant strains pmrA and pmrE were more susceptible to polymyxin B than was the wild-type strain (Fig. 4A), and these observations are consistent with the previous finding that pmrA and pmrE are essential for developing resistance to polymyxin B (15).
FIG. 4.
The pagL mutants were more susceptible to polymyxin B than were the parental mutants. (A) The wild-type (14028s), ΔpagL (KCS216), pmrA (JSG421), pmrA ΔpagL (KCS208), pmrE (KCS041), pmrE ΔpagL (KCS209), ΔpmrC (KCS180), and ΔpmrC ΔpagL (KCS210) strains grown in the growth medium (pH 5.8) were treated with 1 μg/ml of polymyxin B. Percent survival was determined as described in Materials and Methods. The results shown are for two independent sets of experiments (experiments 1 and 2). (B) The pmrA and pmrA ΔpagL mutant strains cultivated in the growth medium (pH 5.8) were treated with the indicated concentrations of polymyxin B. Percent survival is the average of triplicate measurements, and error bars indicate standard deviations.
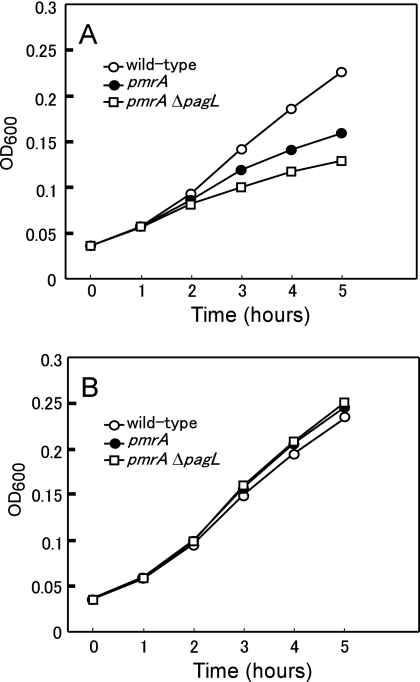
To further confirm that the PagL-dependent deacylation of lipid A increases resistance to polymyxin B in the mutant strains, the wild-type, pmrA mutant, and pmrA ΔpagL mutant strains were grown in the presence or absence of 1 μg/ml of polymyxin B. In the presence of the polymyxin B, the growth rate of the pmrA ΔpagL strain was lower than that of the pmrA strain, which was in turn lower than that of the wild-type strain (Fig. 5A). In contrast, the pmrA ΔpagL and pmrA strains had a rate of growth similar to that of the wild-type strain in the absence of polymyxin B (Fig. 5B), indicating that the pmrA ΔpagL strain is more susceptible to polymyxin B than is the parental pmrA strain. These results, taken together, indicate that PagL-dependent deacylation of lipid A in the pmrA and pmrE strains increases their resistance to the cationic antimicrobial peptide polymyxin B.
FIG. 5.
Growth of S. enterica serovar Typhimurium strains in the presence of polymyxin B. The wild-type (14028s), pmrA (JSG421), and pmrA ΔpagL (KCS208) strains cultivated in the growth medium (pH 5.8) as described in Materials and Methods were diluted 1:10 with fresh growth medium (pH 5.8). Then, the cells were grown at 37°C in the presence (A) or absence (B) of 1 μg/ml of polymyxin B. The results shown are representative of at least two independent experiments. OD600, optical density at 600 nm.
Introduction of the PagL expression construct into a pagL mutant increased polymyxin B resistance.
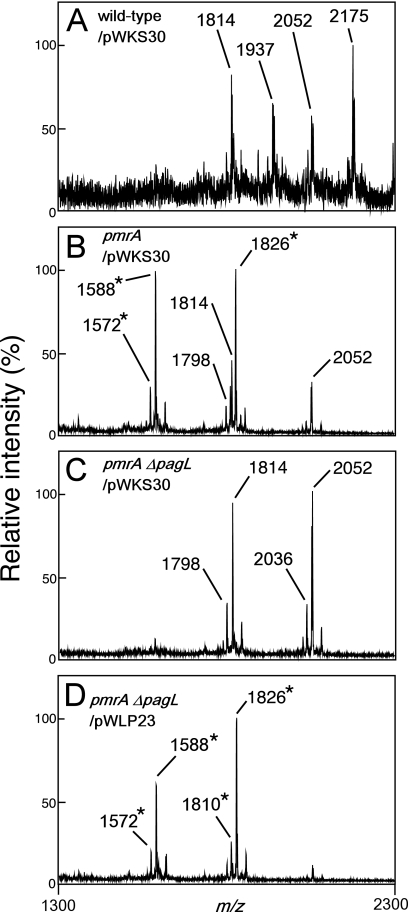
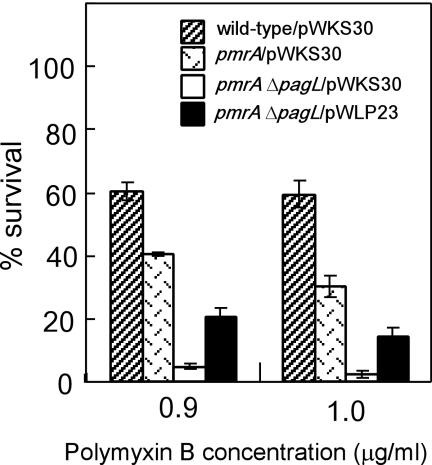
We introduced a PagL expression plasmid construct into the pmrA ΔpagL strain in order to examine whether or not expression of PagL in the pmrA ΔpagL mutant increased the resistance to polymyxin B. At first, we examined lipid A species prepared from pmrA ΔpagL transformed with pWLP23, a derivative of the low-copy-number vector pWKS30 containing the pagL gene. MALDI-TOF mass spectrometry revealed that almost all lipid A molecules (m/z 1572, 1588, 1810, and 1826) were 3-O-deacylated in pmrA ΔpagL/pWLP23 (Fig. 6D). In contrast, nondeacylated lipid A species (m/z 1798, 1814, and 2052) as well as deacylated lipid A species were apparently observed in the pmrA strain transformed with the control vector pWKS30 (pmrA/pWKS30) (Fig. 6B), indicating that the level of deacylated lipid A molecules was higher in pmrA ΔpagL/pWLP23 than in pmrA/pWKS30. Compared with pmrA ΔpagL/pWKS30, pmrA ΔpagL/pWLP23 was resistant to polymyxin B treatment (Fig. 7), indicating that the deacylation increased the resistance. It is noteworthy that pmrA ΔpagL/pWLP23 was more susceptible to polymyxin B treatment than was pmrA/pWKS30 (Fig. 7), indicating that the susceptibility of the pmrA ΔpagL strain was not complemented completely on introduction of the PagL expression plasmid construct pWLP23. Since the level of deacylated lipid A was lower in pmrA/pWKS30 than in pmrA ΔpagL/pWLP23 as described above, it appears that not only the existence of deacylated lipid A molecules in the outer membrane but also the balance of modified lipid A species or the amount of deacylated lipid A might be important for the susceptibility.
FIG. 6.
MALDI-TOF mass spectrometry of lipid A purified from pagL-null mutant strains transformed with a PagL expression plasmid. Wild-type (ATCC 14028s), pmrA (JSG421), and pmrA ΔpagL (KCS208) strains transformed with a PagL expression construct pWLP23 or the control vector pWKS30 were grown in the growth medium (pH 5.8), and their lipid A was analyzed with a MALDI-TOF mass spectrometer. The m/z values of lipid A species are shown, and those that represent deacylated lipid A species are denoted by asterisks. The structural interpretations of lipid A species are summarized in Table 2.
FIG. 7.
Introduction of PagL expression plasmids increased the resistance of the pagL-null mutant strain to polymyxin B. Wild-type (ATCC 14028s), pmrA (JSG421), and pmrA ΔpagL (KCS208) strains transformed with the control vector (pWKS30) or PagL expression construct (pWLP23) were cultivated in the growth medium (pH 5.8) and then treated with 0.9 or 1.0 μg/ml of polymyxin B. Percent survival is the average of triplicate measurements, and the error bars indicate standard deviations.
Modification of lipid A with phosphoethanolamine is partly involved in the latency of PagL.
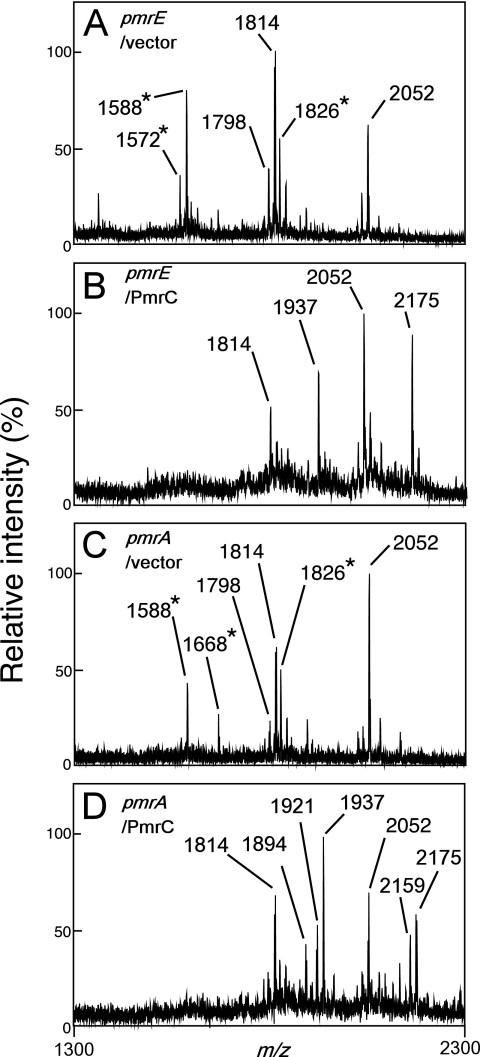
The attachment of phosphoethanolamine to lipid A, a modification mediated by the PmrA-PmrB-activated gene product PmrC, partly contributes to the pmrA-dependent resistance to polymyxin B of S. enterica serovar Typhimurium (27). Interestingly, a 3-O-deacylated lipid A species (m/z 1588) was observed in the pmrC mutant cultivated in the mild acid medium (Fig. 2E), suggesting that this modification of lipid A is partly involved in the latency of PagL. Although the effect of pmrC's deletion on the resistance to polymyxin B was not observed (Fig. 4A), the lack of phosphoethanolamine-modified lipid A might be enough to release PagL from latency in the membrane. To confirm the involvement of the modification of lipid A with phosphoethanolamine in the latency of PagL, a PmrC expression plasmid construct was introduced into the pmrE and pmrA mutants. Lipid A was modified in the mutants transformed with PmrC expression plasmid, and no 3-O-deacylation was observed (Fig. 8). These results, taken together, indicate that the attachment of phosphoethanolamine as well as aminoarabinose to lipid A is involved in the latency of PagL.
FIG. 8.
The attachment of phosphoethanolamine inhibited the deacylation of lipid A. The pmrE (KCS041) or pmrA (JSG421) strain transformed with pBAD24 (control vector) or pKK28 (PmrC) was cultivated in growth medium (pH 7.4) containing 0.2% (wt/vol) arabinose to induce PmrC expression under the control of the PBAD promoter, and then its lipid A was analyzed with a MALDI-TOF mass spectrometer. The m/z values of lipid A species are shown, and those that represent deacylated lipid A species are denoted by asterisks. The structural interpretations of lipid A species are summarized in Table 2.
DISCUSSION
Previously, Kawasaki et al. demonstrated that the Salmonella lipid A deacylase PagL is dormant in the outer membrane and that the modification of lipid A with aminoarabinose is involved in the latency (24), although its biological significance was not apparent. In this paper, we showed that the release of PagL from latency compensates for the loss of the aminoarabinose-dependent resistance to polymyxin B.
Gram-negative pathogenic bacteria such as S. enterica serovar Typhimurium adapt to host microenvironments through changes at the cellular surface, including changes to LPS. The PmrA-PmrB-regulated attachment of aminoarabinose to lipid A alters the net charge at the surface, which reduces electrostatic repulsion between neighboring LPS molecules. These changes may help S. enterica to adapt to host microenvironments because they increase resistance to cationic antimicrobial peptides, including polymyxin B, although the machinery responsible for the resistance remains to be elucidated. In this study, we demonstrated that PagL-dependent lipid A deacylation increases polymyxin B resistance in mutants lacking aminoarabinose-modified lipid A. In addition, introduction of a PagL expression construct into the pmrA ΔpagL strain increased resistance to polymyxin B. These results, taken together, indicate that lipid A deacylation is involved in polymyxin B resistance under specific conditions. However, the pmrA ΔpagL strain transformed with the PagL expression construct was more susceptible to polymyxin B than the pmrA strain transformed with the control vector. We speculated that the introduction of a PagL expression construct induced deacylation of lipid A, but the quality of the deacylation was not controlled; the quantity of deacylated lipid A molecules and the balance of lipid A modifications in the bacterial membrane were not well controlled by the transformation, although we used the pagL promoter region and a low-copy-number vector for the expression construct. The uncontrolled deacylation might not be sufficient for complementation. The deacylation of lipid A helps salmonellae to resist antimicrobial peptides including polymyxin B under certain conditions, but the molecular machinery that increases the resistance is complex and remains to be elucidated.
We demonstrated that a lack of aminoarabinose-dependent resistance to polymyxin B is compensated for by the induction of lipid A 3-O-deacylation. These observations suggest that bacterial lipid A modifications were flexible: a defect in a specific type of modification was functionally compensated by another type of modification. Similar flexibility has been reported previously. Zhou et al. showed that phosphoethanolamine-modified lipid A species are much less abundant than aminoarabinose-modified ones in wild-type S. enterica serovar Typhimurium (45). In contrast, phosphoethanolamine-modified lipid A species accumulate at high levels in pmrA-constitutive strains that harbor a null mutation in either pmrE or pmrF, which is essential for the modification of lipid A with aminoarabinose (45). Since the modification of lipid A with phosphoethanolamine as well as aminoarabinose is involved in pmrA-regulated resistance to polymyxin B (15, 27), induction of the phosphoethanolamine-type modification might be beneficial for salmonellae that lack aminoarabinose-modified lipid A to increase their resistance to cationic antimicrobial peptides including polymyxin B. Although the molecular machinery underlying this regulation remains to be elucidated, Salmonella lipid A-modifying enzymes might directly sense the conditions and then regulate their activity in order to adapt to their environment.
Recently, Tommassen and coworkers reported the crystal structure of P. aeruginosa PagL and identified the active site of PagL (11, 36). They speculated that PagL is active as a single molecule and that dimerization might silence it (36). Changes in the outer membrane, including the modification of lipid A with aminoarabinose, might affect the structure of the outer membrane protein PagL and regulate its activity. Recently, a lipid A 3′-O-deacylase, LpxR, was identified in S. enterica serovar Typhimurium (34). Interestingly, LpxR is similar to PagL in that it is an outer membrane lipase and latent; LpxR deacylates lipid A in vitro, but 3′-O-deacylated lipid A species have not been reported for this organism (34). Elucidation of the latency of an outer membrane enzyme might provide new insight into bacterial membrane remodeling, which is very important for pathogenic bacterial adaptation to host tissues.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, by the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and by the Research Foundation for Pharmaceutical Sciences.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Alpuche Aranda, C. M., J. A. Swanson, W. P. Loomis, and S. I. Miller. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA 89:10079-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belden, W. J., and S. I. Miller. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, R. E., H. S. Gibbons, T. Guina, M. S. Trent, S. I. Miller, and C. R. Raetz. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 19:5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett, P. J., M. N. Burtnick, D. S. Snyder, J. G. Shannon, P. Azadi, and F. C. Gherardini. 2007. Burkholderia mallei expresses a unique lipopolysaccharide mixture that is a potent activator of human Toll-like receptor 4 complexes. Mol. Microbiol. 63:379-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 7.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 8.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 9.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 10.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 11.Geurtsen, J., L. Steeghs, J. T. Hove, P. van der Ley, and J. Tommassen. 2005. Dissemination of lipid A deacylases (pagL) among gram-negative bacteria: identification of active-site histidine and serine residues. J. Biol. Chem. 280:8248-8259. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons, H. S., S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2005. Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol. Microbiol. 55:425-440. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons, H. S., S. Lin, R. J. Cotter, and C. R. Raetz. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, a new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J. Biol. Chem. 275:32940-32949. [DOI] [PubMed] [Google Scholar]
- 14.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 16.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 19.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, A., and E. A. Groisman. 2004. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 18:2302-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara, K., H. Tsukano, H. Watanabe, B. Lindner, and M. Matsuura. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki, K., R. K. Ernst, and S. I. Miller. 2004. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J. Biol. Chem. 279:20044-20048. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki, K., R. K. Ernst, and S. I. Miller. 2005. Inhibition of Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. J. Bacteriol. 187:2448-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki, K., O. Kuge, Y. Yamakawa, and M. Nishijima. 2001. Purification of phosphatidylglycerophosphate synthase from Chinese hamster ovary cells. Biochem. J. 354:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lee, H., F. F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, D. L., and E. P. Kennedy. 1971. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J. Biol. Chem. 246:3042-3049. [PubMed] [Google Scholar]
- 30.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nummila, K., I. Kilpelainen, U. Zahringer, M. Vaara, and I. M. Helander. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271-278. [DOI] [PubMed] [Google Scholar]
- 32.Paulus, H., and E. Gray. 1964. The Biosynthesis of polymyxin B by growing cultures of Bacillus polymyxa. J. Biol. Chem. 239:865-871. [PubMed] [Google Scholar]
- 33.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, C. M., A. A. Ribeiro, S. C. McGrath, R. J. Cotter, C. R. Raetz, and M. S. Trent. 2006. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J. Biol. Chem. 281:21974-21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosner, M. R., J. Tang, I. Barzilay, and H. G. Khorana. 1979. Structure of the lipopolysaccharide from an Escherichia coli heptose-less mutant. I. Chemical degradations and identification of products. J. Biol. Chem. 254:5906-5917. [PubMed] [Google Scholar]
- 36.Rutten, L., J. Geurtsen, W. Lambert, J. J. Smolenaers, A. M. Bonvin, A. de Haan, P. van der Ley, M. R. Egmond, P. Gros, and J. Tommassen. 2006. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 103:7071-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 38.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trent, M. S., W. Pabich, C. R. Raetz, and S. I. Miller. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 276:9083-9092. [DOI] [PubMed] [Google Scholar]
- 40.Vaara, M., and P. Viljanen. 1985. Binding of polymyxin B nonapeptide to gram-negative bacteria. Antimicrob. Agents Chemother. 27:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 42.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 43.Yi, E. C., and M. Hackett. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651-656. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Z., S. Lin, R. J. Cotter, and C. R. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-l-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]