Abstract
The Escherichia coli dnaN159 allele encodes a mutant form of the β-sliding clamp (β159) that is impaired for interaction with the replicative DNA polymerase (Pol), Pol III. In addition, strains bearing the dnaN159 allele require functional Pol I for viability. We have utilized a combination of genetic and biochemical approaches to characterize the role(s) played by Pol I in the dnaN159 strain. Our findings indicate that elevated levels of Pol I partially suppress the temperature-sensitive growth phenotype of the dnaN159 strain. In addition, we demonstrate that the β clamp stimulates the processivity of Pol I in vitro and that β159 is impaired for this activity. The reduced ability of β159 to stimulate Pol I in vitro correlates with our finding that single-stranded DNA (ssDNA) gap repair is impaired in the dnaN159 strain. Taken together, these results suggest that (i) the β clamp-Pol I interaction may be important for proper Pol I function in vivo and (ii) in the absence of Pol I, ssDNA gaps may persist in the dnaN159 strain, leading to lethality of the dnaN159 ΔpolA strain.
The Escherichia coli dnaN-encoded β-sliding clamp functions as a homodimer and is “loaded” onto primed DNA by the multisubunit DnaX clamp loader complex (5, 21). Once loaded, the β clamp slides freely along duplex DNA and functions to tether a variety of different proteins involved in replication and repair to the DNA (4, 6, 7, 9, 19, 23, 24, 26, 28, 41-43). The dnaN159 allele encodes a mutant form of the β-sliding clamp (β159) that bears two amino acid substitutions: G66E (glycine at position 66 replaced with glutamic acid) and G174A (14, 35, 38). The G174A substitution appears to impair the ability of a hydrophobic cleft in β to interact with the eubacterial clamp-binding motif (QL[S/D]LF) that is conserved in most partner proteins reported to interact with the β clamp (8, 38). E. coli strains bearing the dnaN159 allele display temperature-sensitive growth (14, 35, 38) and altered DNA polymerase (Pol) usage (29, 38-40). These phenotypes appear to result, at least in part, from impaired interactions of the mutant β159 clamp protein with the replicative DNA polymerase, Pol III (38). As a result of the impaired β159-Pol III interaction, E. coli strains bearing the dnaN159 allele display increased utilization of the three SOS-regulated DNA polymerases, Pol II (polB), Pol IV (dinB), and Pol V (umuDC) (29, 38-40). In contrast to these three Pols, which under certain conditions impede growth of the dnaN159 strain (29, 39, 40), presumably by impairing DNA replication, the catalytic DNA polymerase activity of Pol I (polA) is essential for viability of the dnaN159 strain (38). Importantly, Pol I function is not required by the isogenic dnaN+ strain (38), indicating that Pol I plays one or more essential roles in the dnaN159 strain. Since Pol I is a multifunctional protein that participates in DNA replication, as well as numerous DNA repair pathways, several possibilities exist, including Okazaki fragment maturation (21, 31) and single-stranded DNA (ssDNA) gap repair (16, 22).
Since Pol III replication is abated in strains bearing the dnaN159 allele (14, 35, 38), due to the impaired ability of β159 to interact with the α catalytic subunit of Pol III (38), we hypothesized that the requirement for Pol I function in the dnaN159 strain might stem from its ability to augment Pol III function in DNA replication. The experiments discussed in this report were designed to test this hypothesis. Our findings indicate that elevated levels of Pol I partially suppress the temperature-sensitive growth phenotype of the dnaN159 strain. In addition, we provide evidence that ssDNA gap repair is impaired in the dnaN159 strain. Moreover, we determined that the β clamp confers a modest degree of processivity upon Pol I replication in vitro, while the mutant β159 clamp protein is impaired for this function, suggesting that the β clamp-Pol I interaction is important for ssDNA gap repair in vivo. Finally, we provide evidence that the fidelity of lagging-strand replication is impaired in the dnaN159 strain, consistent with altered Okazaki fragment maturation in the dnaN159 strain. Taken together, these findings suggest that (i) the β clamp-Pol I interaction may be important for proper Pol I function in vivo and (ii) in the absence of Pol I, ssDNA gaps may persist in the dnaN159 strain, leading to lethality.
MATERIALS AND METHODS
E. coli strains, plasmid DNAs, and bacteriological techniques.
All E. coli strains used in this study are derivatives of either RW118 [thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) hisG4(Oc) rpsL31 xylA5 mtl-1 argE3(Oc) thi-1 sulA211] (15), CJ278 [Δ(gal-bio) thi-1 relA1 spoT1 ΔpolA::kan] (18), or EC3144 and EC3138 {ara Δ(lac-proB)XIII attB::lacI+Z[GGG→ GAG]Y+A+} (10). The strains were constructed using P1vir-mediated transduction (30), and the presence of the desired allele was confirmed by either diagnostic PCR or automated nucleotide sequence analysis of the PCR-amplified allele, as described previously (38). Transduction of the dnaN+ and dnaN159 alleles was achieved by selection for the closely linked (∼80%) tnaA300::Tn10 allele. E. coli strains were made competent for transformation with plasmid DNA by rubidium chloride treatment (36). All E. coli strains were grown in either LB or M9 medium (30), as indicated. When necessary, the following antibiotics were added at the indicated concentrations: ampicillin, 150 mg/ml; chloramphenicol, 20 mg/ml; kanamycin, 60 mg/ml; rifampin (Rif), 100 mg/ml; and tetracycline, 10 mg/ml.
Plasmid pRM100 is a pWSK29 (44) derivative that contains the complete polA+ gene (coding sequence and promoter) and was described previously (29). Plasmid pRM104 is a pRM100 derivative that expresses the Klenow fragment of Pol I (lacking amino acids 1 to 322) from the native polA promoter. It was constructed by introducing NcoI restriction sites at nucleotide positions −2 through +4 (where +1 corresponds to the A in the ATG initiation codon) and +965 through +970 of the polA coding sequence using the Quickchange mutagenesis kit (Stratagene) with primers polA-Klenow-1 (5′-CAC GGA CAC CAT GGT TCA GAT CCC-3′) and polA-Klenow-2 (5′-GGG ATC TGA ACC ATG GTG TCC GTG-3′) and with polA-Klenow-3 (5′-GTG ACG GCC ATG GTG ATT TCT TAT GAC-3′) and polA-Klenow-4 (5′-GTC ATA AGA AAT CAC CAT GGC CGT CAC-3′), respectively (the NcoI sites are underlined). After confirming the correct nucleotide sequence (Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, NY), the double-mutant plasmid was digested with NcoI, gel purified, and ligated, resulting in a plasmid expressing the Klenow fragment of Pol I from the native polA+ promoter.
Plasmids pRM105 and pRM106 express untagged Pol I and N-terminally His10-tagged Klenow, respectively. Plasmid pRM105 was cloned by PCR amplification of the polA+ gene from RW118 genomic DNA using primers polA-forward (5′-CCG ACA CAT ATG GTT CAG ATC CCG CAG AAT CC-3′) and polA-reverse (5′-GGG ACA CCT AGG TTA GTG CGC CTG ATC CCA G-3′). Primer polA-forward introduces an NdeI site overlapping the ATG initiation codon (underlined). The PCR fragment was cloned into plasmid pCR-BluntII-Topo (Invitrogen), and the complete nucleotide sequence of the polA+ reading frame was verified by automated nucleotide sequence analysis (Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, NY). The polA+ coding sequence was then subcloned into the overexpression vector pET11a (Novagen) using NdeI and BamHI (the site was present in the multiple cloning site of pCR-BluntII-Topo) restriction, placing it under the control of the T7 promoter. Plasmid pRM106 was created by PCR amplifying the Klenow fragment of Pol I from the polA+ gene, using primers Klenow-forward (5′-CCA GAA GTG ACG GCA CAT ATG ATT TCT TAT GAC AAC TAC-3′, which introduces an NdeI restriction site [underlined] in place of residues 322 and 333) and polA-reverse (see above). The resulting PCR fragment was cloned similarly to polA+, except that the overexpression vector pET16b (Novagen) was used to introduce an N-terminal His10 tag. As for pRM105, the sequence of the Klenow coding sequence was confirmed by automated nucleotide sequence analysis (Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, NY).
UV sensitivity and mutation frequency measurements.
Sensitivity to UV light (254 nm) was measured using a germicidal lamp (General Electric) as described previously (38). Briefly, cultures were grown at 30°C in M9 minimal medium supplemented with glucose (0.2%) and Casamino Acids (0.5%) until they reached mid-exponential growth (an optical density at 595 nm of ∼0.6). The cultures were then transferred to a sterile 100-mm glass petri dish and irradiated with the indicated UV dose. Appropriate dilutions of each sample were plated onto LB plates supplemented with the appropriate antibiotics. Colonies were counted after overnight incubation at 30°C, and survival was expressed as percent viability relative to control samples that were mock UV irradiated.
Spontaneous-mutation frequencies were determined as previously described (29). Briefly, cultures were grown overnight at 30°C in LB medium and appropriate dilutions were plated onto LB plates or LB plates supplemented with Rif (100 μg/ml). The mutation frequency was calculated by dividing the number of Rifr colonies observed by the number of viable cells.
Protein purification and in vitro primer extension assays.
Purification of β, β159, and the γ3δδ′ clamp loader complex will be described elsewhere (R. W. Maul, S. K. Scouten-Ponticelli, J. M. Duzen, and M. D. Sutton, submitted for publication). Recombinant Pol I and N-terminally His10-tagged Klenow were purified as described previously (17). Primer SP20 (5′-ACG CCT GTA GCA TTC CAC AG-3′) was gel purified (Sigma Genosys) and 5′-end labeled using polynucleotide kinase and [γ32P]ATP as described previously (1). SP20 was annealed to M13mp18 ssDNA (New England Biolabs) in a reaction mixture (100 μl) containing 15 pmol 32P-labeled SP20, 5 pmol M13mp18 ssDNA, and annealing buffer (25 mM Tris-HCl [pH 7.0], 100 mM NaCl, 15 mM MgCl2) by incubation at 95°C for 3 minutes, followed by cooling to room temperature for 1 hour. To remove unannealed SP20, the annealing reaction mixture was passed over an S-400 microspin column (GE Healthcare) following the manufacturer's recommendations.
Primer extension reaction mixtures (10 μl) were assembled on ice and contained replication assay buffer (20 mM Tris-HCl [pH 7.5], 8.0 mM MgCl2, 0.1 mM EDTA, 5 mM dithiothreitol, 1 mM ATP, 5% glycerol, 0.8 μg/ml bovine serum albumin) supplemented with 0.133 mM deoxynucleoside triphosphates (dNTPs), 2 μM ssDNA-binding protein, and 2.5 nM 32P-labeled SP20/M13mp18 ssDNA template. Reactions were initiated by addition of the indicated β clamp protein (40 nM) and γ3δδ′ clamp loader complex (10 nM). Following a 5-min preincubation at 30°C to allow loading of the clamp, Pol I or Klenow was added as indicated in the figure legends. After 10 min at 30°C, the reactions were quenched with 20 μl of quench buffer (95% formamide, 20 mM EDTA, 0.1% bromophenol blue), and 5 μl of each quenched reaction mixture was electrophoresed through an 8.0% urea-polyacrylamide gel. Replication products were visualized using a phosphorimaging screen K and personal FX imaging instrumentation (Bio-Rad).
For quantitation of replication products, reactions (20 μl) were performed as described above using either unlabeled SP20/M13mp18 as a template or 250 ng of a multiply nicked form of the double-stranded DNA (dsDNA) plasmid pBluescript KS II, together with a [3H]dTTP-dNTP mixture (133 μM; 101 cpm/pmol). Reactions were quenched with 15% trichloroacetic acid, 100 mM sodium pyrophosphate. Acid-insoluble products were collected on 2.4-cm glass fiber filters (VWR) by vacuum filtration, and incorporation of [3H]dTTP was quantitated by liquid scintillation counting, as described previously (34). The results shown represent the average of triplicates. The error bars represent the standard deviations. The nicked pBluescript template was generated by incubating 10 μg of pBluescript KS II dsDNA with 0.002 units of DNase I (Promega) for 30 min at room temperature. The reaction was quenched by extraction with a 50% phenol-chloroform suspension, followed by ethanol precipitation.
RESULTS AND DISCUSSION
Modest overexpression of Pol I partially suppresses the temperature-sensitive growth phenotype of the dnaN159 strain.
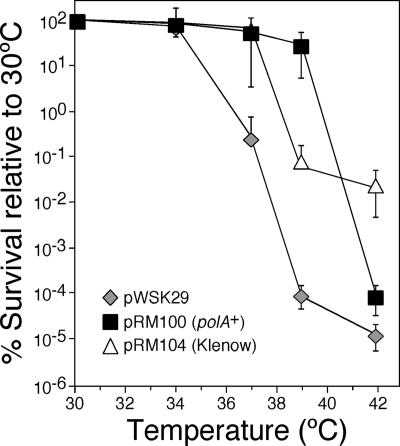
In contrast to Pol II, Pol IV, and Pol V, which confer lethality upon the dnaN159 strain under certain conditions (29, 38, 39), the catalytic polymerase activity of Pol I is required for viability of the dnaN159 strain (38). Since Pol III function is abated in the dnaN159 strain (14, 35, 38), due to an impaired β159-Pol III interaction (38), we hypothesized that Pol I might act to augment Pol III function during DNA replication in the dnaN159 strain. As a test of this hypothesis, we measured growth at different temperatures of the dnaN159 strain MS105 bearing either the low-copy-number plasmid pWSK29 (44) or pRM100 (29), a pWSK29 derivative containing E. coli Pol I under the control of its native promoter. Consistent with our previous findings (29), strain MS105 bearing pWSK29 was severely impaired for growth at temperatures above 34°C (Fig. 1). In contrast, strain MS105 bearing the Pol I-expressing plasmid pRM100 grew normally at temperatures up to and including 39°C (Fig. 1), consistent with the idea that Pol I augments Pol III function during DNA replication in the dnaN159 strain.
FIG. 1.
Elevated levels of Pol I partially suppress the temperature-sensitive growth phenotype of the dnaN159 strain. Strain MS105 [thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) hisG4(Oc) rpsL31 xylA5 mtl-1 argE3(Oc) thi-1 sulA211 lexA51(Def) tnaA300::Tn10 dnaN159] was transformed with either pWSK29 (control), pRM100 (polA+), or pRM104 (Klenow). We used strain MS105 for this experiment because it displays a more pronounced temperature-sensitive phenotype than many other dnaN159 strains, thus increasing the range in which we could observe an effect by Pol I or Klenow. Temperature sensitivity was measured by plating out dilutions of overnight cultures grown at 30°C onto M9 minimal medium, followed by incubation at the indicated temperature overnight. The number of CFU observed at each indicated temperature is expressed as a percentage of that observed for the same strain at 30°C, which was set equal to 100%. The error bars indicate standard deviations. MS105 bearing pWSK29 had an average titer of 5.1 × 108 viable cells/ml, MS105 bearing pRM100 had an average titer of 1.5 × 108 viable cells/ml, and MS105 bearing pRM104 had an average titer of 0.1 × 107 viable cells/ml.
E. coli Pol I contains three distinct activities: (i) a 5′-to-3′ template-directed DNA polymerase activity, (ii) a 3′-to-5′ exonuclease proofreading activity, and (iii) a FEN1-like 5′-to-3′ exonuclease activity important for DNA repair functions, as well as Okazaki fragment maturation (21). Since growth of the dnaN159 strain requires only the polymerase activity of Pol I (38), which, together with the 3′-to-5′ exonuclease proofreading activity, is contained within the Klenow fragment (residues 323 to 928 of Pol I), we asked whether plasmid-expressed Klenow fragment was sufficient to support growth of the dnaN159 strain at elevated temperatures. To examine this, we utilized a pRM100 derivative named pRM104 that expresses the Klenow fragment of Pol I from the endogenous polA promoter. In contrast to Pol I, plasmid-expressed Klenow fragment was only able to support growth at temperatures up to and including 37°C (Fig. 1). In addition, the titer of this strain was ∼100-fold lower than those of the other strains examined (see the legend to Fig. 1). This finding suggests that elevated levels of the Klenow fragment are deleterious to the dnaN159 strain. Taken together, these results indicate that both the DNA polymerase and the FEN1-like 5′-to-3′ exonuclease activities of Pol I are required for maximal suppression of temperature-sensitive growth of the dnaN159 strain.
The dnaN159 strain is impaired for ssDNA gap repair.
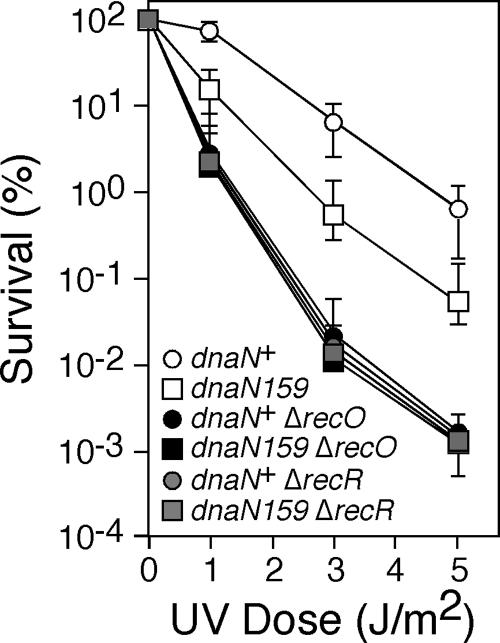
Our finding that the FEN1-like 5′-to-3′ exonuclease activity of Pol I is required for maximal suppression of temperature-sensitive growth in the dnaN159 strain (Fig. 1) suggests that temperature sensitivity is the result, at least in part, of the accumulation of ssDNA nicks and/or gaps (hereafter referred to jointly as ssDNA gaps for simplicity). Consistent with this idea, the dnaN159 strain is sensitive to UV light at the permissive temperature of 30°C, and this sensitivity is enhanced ∼3-fold at the semipermissive temperature of 37°C (38). Therefore, we hypothesized that the UV sensitivity of the dnaN159 strain would be epistatic with mutations that impair the RecFOR-mediated pathway of ssDNA gap repair, which is reported to utilize both Pol I and Pol III for gap filling (16, 37). Since the recF allele is located immediately downstream of dnaN (32, 33), we chose to construct isogenic E. coli strains bearing dnaN+ or dnaN159, together with either the recO+, ΔrecO1504::Tn5 (20), recR+, or ΔrecR252::Tn10∼9 (25) alleles, and subsequently compared their respective levels of sensitivity to UV irradiation. Consistent with our previous findings (38), the dnaN159 strain was ∼10-fold more sensitive to UV irradiation than was the isogenic dnaN+ strain (Fig. 2). This UV sensitivity was completely epistatic with both the ΔrecO1504::Tn5 and the ΔrecR252::Tn10∼9 alleles (Fig. 2), suggesting that the UV sensitivity of the dnaN159 strains is a result of impaired ssDNA gap repair. However, our finding that the dnaN159 strain was less sensitive to UV irradiation than were the isogenic dnaN+ ΔrecO1504::Tn5 and dnaN+ ΔrecR252::Tn10∼9 strains indicates that the dnaN159 strain is only partially impaired for ssDNA gap repair. Furthermore, our finding that the UV sensitivity of the dnaN159 strain was suppressed by (not epistatic with) inactivation of Pol IV [Δ(dinB-yafN)::kan] (38) suggests that Pol IV competes with Pol I and/or Pol III for access to ssDNA gaps to impede repair in the dnaN159 strain.
FIG. 2.
The UV sensitivity of the dnaN159 strain is epistatic with mutations that impair recFOR function. Survival following the indicated dose of UV irradiation was measured by plating serial dilutions as previously described (29). The strains examined are derivatives of RW118 [thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44(AS) galK2(Oc) hisG4(Oc) rpsL31 xylA5 mtl-1 argE3(Oc) thi-1 sulA211] (15): RM149 (dnaN+ ΔuvrB::cat), RM150 (dnaN159 ΔuvrB::cat), RM151 (dnaN+ ΔuvrB::cat ΔrecO1504::Tn5), RM152 (dnaN159 ΔuvrB::cat ΔrecO1504::Tn5), RM163 (dnaN+ ΔuvrB::cat ΔrecR252::Tn10∼9), and RM164 (dnaN159 ΔuvrB::cat ΔrecR252::Tn10∼9). The results shown represent the average of two independent experiments, each done in duplicate. The error bars represent the range.
The β159 clamp is impaired for stimulating Pol I and Klenow replication on both a singly primed ssDNA and a nicked dsDNA template in vitro.
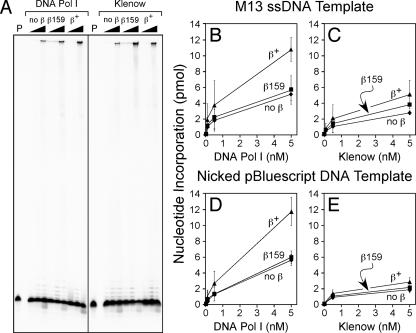
The β clamp is reported to interact with, and to confer processivity upon, the Klenow fragment of Pol I in vitro during replication of singly primed M13 ssDNA (24) (Fig. 3). However, due to the fact that a consensus clamp-binding motif has not yet been identified in Pol I, it remains unclear exactly how Pol I interacts with the β clamp. As a result, it is currently unknown whether the Pol I-β clamp interaction is required for proper Pol I function in vivo. We hypothesized that the ssDNA gap repair defect of the dnaN159 strain could be the result of an impaired ability of the β159 clamp protein to stimulate Pol I function, leading to a reduced capacity for ssDNA gap repair. As a test of this hypothesis, we cloned and purified recombinant forms of Pol I and Klenow for in vitro replication assays. In our first experiment, we utilized a M13 ssDNA template bearing a 32P-labeled primer. Using this assay, the wild-type β clamp conferred a modest degree of processivity upon both Pol I and Klenow, as judged by the increase in the abundance of full-length product near the top of the gel with 5 nM Pol I or Klenow in a reaction mixture containing the β clamp relative to those lacking the clamp (Fig. 3A). Our finding that the wild-type β clamp conferred processivity upon Klenow confirms an earlier report (24). Although β159 was impaired for conferring processivity upon Pol I as measured by this assay, it did confer modest processivity upon Klenow (Fig. 3A).
FIG. 3.
The wild-type β clamp stimulates Pol I replication in vitro. (A) Reaction mixtures containing M13 ssDNA bearing a 5′-32P-labeled primer as a template, together with either 0.05, 0.5, or 5 nM of the indicated polymerase, as described in Materials and Methods. After being quenched, aliquots of each reaction mixture were electrophoresed through an 8% urea-polyacrylamide gel. The lanes labeled P represent control reactions lacking clamp and polymerase and serve to indicate the position of free primer. Replication products were visualized by phosphorimager analysis. The faint full-length bands visible in reactions containing β159 and 0.05 or 0.5 nM Pol I or Klenow are due to a trace polymerase contamination in the β159 preparation. Alternatively, replication utilizing either the same M13 ssDNA template lacking the 5′ 32P label (B and C) or a multiply nicked form of pBluescript KS II dsDNA (D and E), together with [3H]dTTP-dNTPs, was quantified by liquid scintillation spectroscopy of acid-insoluble products collected on 2.4-cm glass fiber filters as described in Materials and Methods. Reaction mixtures contained the indicated amounts (0.05, 0.5, or 5 nM) of Pol I (B and D) or the Klenow fragment of Pol I (C and E).
In order to quantify the extent to which the wild-type and β159 clamp proteins influenced Pol I and Klenow replication in vitro, we repeated the experiments described above using the same singly primed M13 ssDNA template, but instead of the 5′-32P-label, we utilized [3H]dTTP-dNTPs and measured nucleotide incorporation by liquid scintillation spectroscopy of acid-insoluble [3H]dTTP retained on glass fiber filters (34). Consistent with the results shown in Fig. 3A, addition of the wild-type β clamp and γ complex stimulated both Pol I and Klenow replication ∼2-fold at all levels of enzyme (0.05, 0.5, and 5 nM) examined (Fig. 3B and C). In contrast, β159 failed to stimulate Pol I replication under these conditions (Fig. 3B) and stimulated Klenow only moderately (Fig. 3C).
We next asked whether the β clamp might stimulate Pol I or Klenow more effectively on a nicked DNA substrate. For these experiments, we used a multiply nicked form of the double-stranded plasmid pBluescript KS II as the DNA template and again measured nucleotide incorporation by Pol I or Klenow by liquid scintillation spectroscopy. Although the wild-type β clamp stimulated Pol I replication ∼2-fold on the multiply nicked template, β159 failed to stimulate Pol I to a detectable level on this DNA template (Fig. 3D). Consistent with a need for the FEN1-like 5′-to-3′ exonuclease domain of Pol I for nick translation (21), the Klenow fragment inefficiently replicated the nicked dsDNA template regardless of whether the reaction mixture contained the wild-type β or β159 clamp protein (Fig. 3E). These biochemical findings, taken together with the genetic results discussed above (Fig. 2), suggest that ssDNA gap repair is abated in the dnaN159 strain due, at least in part, to the diminished ability of the mutant β159 clamp protein to stimulate Pol I function.
Pol I and Pol V compete with each other in the dnaN159 strain.
The results discussed above are consistent with a model in which the ssDNA gap repair defect of the dnaN159 strain is the result, at least in part, of the impaired ability of β159 to stimulate Pol I (Fig. 3) and Pol III (38). We hypothesized that ssDNA gap repair might be further abated in the dnaN159 strain by the actions of the SOS-regulated Pols via their ability to compete with Pol I and/or Pol III for access to ssDNA gaps. It was previously reported that spontaneous Pol V-dependent mutagenesis was enhanced in E. coli strains either bearing the ΔpolA allele or expressing the Klenow fragment of Pol I in place of the full-length Pol I (2). The ability of Pol V to contribute to spontaneous mutagenesis under these conditions presumably results from the ability of Pol V to gain access to ssDNA gaps in the absence of Pol I or in the presence of a Pol I mutant (Klenow) impaired for proper function. Based on our finding that Pol V plays a more prominent role in DNA replication in the dnaN159 strain in response to UV irradiation (38), we hypothesized that Pol V would likewise play a larger role in replication in the dnaN159 strain bearing mutations impairing Pol I function. Due to the fact that Pol V is error prone, we further reasoned that if Pol I and Pol V do in fact compete with each other for access to ssDNA gaps in the dnaN159 strain, then we should be able to manipulate the frequency of spontaneous mutagenesis by using different combinations of polA (Pol I) and umuDC (Pol V) alleles.
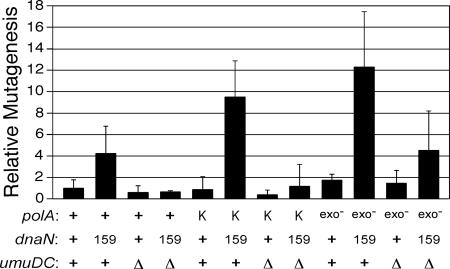
Although a dnaN159 ΔpolA strain is nonviable (38), it is possible to construct a dnaN159 strain bearing a ΔpolA allele on the chromosome that is complemented by the Klenow fragment of Pol I expressed from an F′ episome (38). We therefore utilized Rifr as a measure of the spontaneous mutation frequencies of isogenic dnaN+ and dnaN159 strains bearing either the umuD+C+ (Pol V) or ΔumuDC alleles together with the ΔpolA allele on their chromosome and expressing either the polA+ allele or the Klenow fragment of Pol I in order to gauge the respective effects of β159, Klenow, and Pol V. Although we observed little effect on the frequency of spontaneous mutagenesis in the dnaN+ strain expressing the Klenow fragment of Pol I relative to the same strain expressing Pol I (Fig. 4), the dnaN159 strains expressing Pol I or the Klenow fragment of Pol I displayed ∼4-fold- and ∼9-fold-higher spontaneous mutation frequencies, respectively, than did the dnaN+ Pol I control strain. Consistent with our hypothesis, the elevated frequency of spontaneous mutagenesis in these strains was completely dependent upon Pol V, as the mutation frequencies of the dnaN159 ΔumuDC strains expressing either Pol I or Klenow were comparable to that of the dnaN+ Pol I strain (Fig. 4). Our observation that the dnaN159 Klenow strain displayed a higher Pol V-dependent spontaneous-mutator phenotype than the isogenic dnaN159 Pol I strain correlates with our finding that the β clamp stimulated Pol I replication more robustly than it did Klenow (Fig. 3), consistent with the β clamp-Pol I interaction being important for proper Pol I function in vivo.
FIG. 4.
Spontaneous mutation frequencies in dnaN+ and dnaN159 strains expressing either Pol I or the Klenow fragment of Pol I. All strains used were derivatives of CJ278 [Δ(gal-bio) thi-1 relA1 spoT1 ΔpolA::kan] and expressed either the polA+ allele, the Klenow fragment of Pol I, or the polA-exo mutant allele from an F′ episome (18). The strains examined were RM137, dnaN+ (F′ polA+); RM138, dnaN159 (F′ polA+); RM139, dnaN+ ΔumuDC595::cat (F′ polA+); RM140, dnaN159 ΔumuDC595::cat (F′ polA+); RM141, dnaN+ (F′ Klenow); RM142, dnaN159 (F′ Klenow); RM143, dnaN+ ΔumuDC596::ermGT (F′ Klenow); RM144, dnaN159 ΔumuDC596::ermGT (F′ Klenow); RM145, dnaN+ (F′ polA-exo mutant); RM146, dnaN159 (F′ polA-exo mutant); RM147, dnaN+ ΔumuDC596::ermGT (F′ polA-exo mutant); RM148, dnaN159 ΔumuDC596::ermGT (F′ polA-exo mutant). The mutation frequency was calculated following growth at 30°C by dividing the number of Rifr colonies by the total number of viable cells, as described previously (29, 38, 40). Mutation frequencies are expressed relative to the dnaN+ polA+ strain (9.7 ± 7.5 Rifr CFU/109 total viable cells), which was set equal to 1.0, and are the averages of at least five independent determinations. The error bars represent the standard deviations. Abbreviations: +, polA+, dnaN+, or umuD+C+, as indicated; exo−, Pol I exo mutant; K, Klenow fragment; 159, dnaN159; Δ, ΔumuDC596::ermGT or ΔumuDC595::cat.
Evidence for a more prominent role for Pol I in DNA replication in the dnaN159 strain.
In addition to being impaired for stimulation of Pol I replication in vitro (Fig. 3), β159 is also impaired for interactions with the α catalytic subunit of Pol III (38). We hypothesized that the impaired ability of β159 to interact with Pol III might not only impair ssDNA gap repair, due to a role for Pol III in gap filling (11, 16), but in addition might actually contribute to the further formation of ssDNA gaps in the dnaN159 strain by decreasing the efficiency of Pol III replication. We further hypothesized that if the dnaN159 strain contains on average more ssDNA gaps than the isogenic dnaN+ strain, then Pol I should play a more prominent role in DNA replication in the dnaN159 strain due to its role, albeit impaired, in the repair of these gaps. We tested this hypothesis by measuring spontaneous-mutation frequencies in isogenic dnaN+ and dnaN159 strains expressing the polA-exo mutant allele from an F′ episome (18). The polA-exo mutant allele encodes D355A and E357A substitutions that essentially eliminate the 3′-to-5′ proofreading nuclease activity of Pol I, thereby reducing its polymerase fidelity ∼4-fold, allowing a measure of its contribution to DNA replication in vivo (3). As shown in Fig. 4, the dnaN159 Pol I exo mutant strain displayed an ∼12-fold-higher spontaneous-mutation frequency compared to an ∼2-fold increase for the dnaN+ Pol I exo mutant strain relative to the dnaN+ Pol I control, suggesting that Pol I plays a more prominent role in replication in the dnaN159 strain. In light of our results discussed above indicating that Pol V contributes to spontaneous mutagenesis in the dnaN159 strains expressing Pol I (Fig. 4), we questioned whether Pol V contributed to the mutation frequencies observed for the strains expressing the Pol I exo mutant. Inactivation of Pol V (ΔumuDC) had essentially no effect on the mutation frequency of the dnaN+ Pol I exo mutant and produced only an ∼3-fold reduction in the mutation frequency of the dnaN159 Pol I exo mutant (Fig. 4), suggesting that the elevated mutation frequencies observed were largely the result of Pol I replication errors. Thus, these findings suggest that Pol I plays a more prominent role in DNA replication in the dnaN159 strain than it does in the otherwise isogenic dnaN+ strain, consistent with the idea that the dnaN159 strain suffers from more numerous ssDNA gaps.
Lagging-strand replication is more severely impaired than leading-strand replication in the dnaN159 strain.
The lagging strand, by virtue of its discontinuous synthesis, contains ssDNA nicks between Okazaki fragments. These nicks are ultimately repaired by the combined actions of RNase H, Pol I, and DNA ligase as part of the Okazaki fragment maturation process (21). Given that the dnaN159 strain is impaired for repair of ssDNA gaps, we hypothesized that it would likewise be impaired for proper processing of ssDNA nicks located between adjacent Okazaki fragments on the lagging strand. As a test of this hypothesis, we asked whether the fidelity of lagging-strand replication was reduced in the dnaN159 strain.
Using a collection of isogenic E. coli strains bearing defined lacZ mutant alleles integrated in both orientations at the attB site (i.e., so that the lacZ coding sequence was replicated as part of either the leading or the lagging strand) to measure lacZ mutant→ lacZ+ reversion, the fidelity of lagging-strand replication was reported to be ∼2- to ∼6-fold higher (depending upon the lacZ mutant allele analyzed) than that of the leading strand (10, 13, 27). Using this same approach, we measured the respective fidelities of leading- and lagging-strand replication in four isogenic E. coli strains bearing the lacZ[GGG→ GAG] allele integrated in both orientations at attB together with a ΔmutL::cat mutation to inactivate mismatch repair (to allow a more accurate measure of replication errors) and either the dnaN+ or the dnaN159 allele.
Using the two isogenic dnaN+ strains bearing the lacZ[GGG→ GAG] allele in each orientation, we observed a 2.9-fold-lower spontaneous lacZ[GGG→ GAG]→ lacZ+ reversion frequency on the lagging strand (0.21 × 10−6 ± 0.05 × 10−6) than on the leading strand (0.60 × 10−6 ± 0.13 × 10−6) (Table 1), indicating that the lagging strand displayed 2.9-fold-higher fidelity than the leading strand, consistent with previous reports (10, 13, 27). This difference was specific to the respective orientations of the lacZ[GGG→ GAG] allele, as it was not observed when measuring Rifr arising from mutations within rpoB, which is present in the same native orientation in both strains (Table 1), also consistent with previous reports (10, 13, 27). In contrast, the fidelity of lagging-strand replication in the pair of dnaN159 strains was more severely impaired than that of the leading strand: the frequency of lacZ[GGG→ GAG]→ lacZ+ reversion on the leading strand was 2.3-fold higher in the dnaN159 strain (1.40 × 10−6 ± 0.47 × 10−6) than it was in the isogenic dnaN+ strain (0.60 × 10−6 ± 0.13 × 10−6), while that of the lagging strand in the dnaN159 strain (1.59 × 10−6 ± 1.29 × 10−6) was 7.6-fold higher than that of the isogenic dnaN+ strain (0.21 × 10−6 ± 0.05 × 10−6) (Table 1). Furthermore, the fidelities of the leading and lagging strands were comparable to each other in the dnaN159 strain (Table 1). The frequencies of Rifr for these two dnaN159 strains were also comparable to each other, although they were ∼2-fold higher than those observed in the isogenic dnaN+ strains (Table 1).
TABLE 1.
Leading- and lagging-strand mutation frequencies in dnaN+ and dnaN159 strains
| Straina | dnaN allele | Orientation of lacZ[GGG→ GAG] alleleb | Mutants/106 CFUc
|
|||
|---|---|---|---|---|---|---|
|
lacZ[GGG→ GAG]→ Lac+
|
Rifr
|
|||||
| Mutation frequency | Effect (n-fold) (leading/lagging) | Mutation frequency | Effect (n-fold) (leading/lagging) | |||
| JL100 | dnaN+ | Leading | 0.60 ± 0.13 | 2.9 | 4.87 ± 1.77 | 1.3 |
| JL101 | dnaN+ | Lagging | 0.21 ± 0.05 | 3.74 ± 0.22 | ||
| JL102 | dnaN159 | Leading | 1.40 ± 0.47 | 0.9 | 7.31 ± 1.95 | 1.1 |
| JL103 | dnaN159 | Lagging | 1.59 ± 1.29 | 6.59 ± 1.23 | ||
Strains JL100 to JL103 are derived from strains EC3144 and EC3138 (10) and differ only in their respective dnaN alleles (dnaN+ or dnaN159), as well as the orientation of the lacZ[GGG→ GAG] allele (leading or lagging) integrated at attB. Their remaining genotype is ara Δ(lac-proB)XIII attB::lacI+Z[GGG→ GAG]Y+A+ tnaA300::Tn10 ΔmutL::cat.
The lacZ[GGG→ GAG] allele is integrated at attB so that the coding strand is replicated as part of the leading strand (Leading) or as part of the lagging strand (Lagging).
Growth of strains bearing the lacZ[GGG→ GAG] allele on minimal media containing lactose as the sole carbon source requires a “true” reversion of the lacZ mutation, as no other mutation confers a Lac+ phenotype. Mutation frequencies were measured as described previously (10). The results shown represent the averages of at least three independent determinations ± the standard deviation.
In order to confirm that the predominant base substitution contributing to spontaneous mutations in the dnaN159 strain resulted from G→ A transitions (the mutation that was measured by lacZ[GGG→ GAG]→ lacZ+ reversion), as has been reported for the dnaN+ strain (12), we sequenced 22 independent Rifr mutants from the dnaN159 strain and compared the spectrum to those observed for 15 independent Rifr dnaN+ strains (Table 2). Based on this analysis, Rifr of all 15 dnaN+ strains examined resulted from a single GC→ AT transition mutation affecting one of five different nucleotide positions (Table 2). Of the 23 mutations identified in the dnaN159 strains, 17 (74%) corresponded to GC→ AT transition mutations, 2 corresponded to TA→ CG transition mutations, and 4 corresponded to CG→ AT transversion mutations (Table 2). Based on these results, we conclude that spontaneous mutations in both the dnaN+ and dnaN159 strains result largely from GC→ AT transition mutations. These results, taken together with our findings discussed above, indicate that the fidelity of lagging-strand replication is more severely impaired than that of the leading strand in the dnaN159 strain, suggesting that Okazaki fragment synthesis/maturation is impaired.
TABLE 2.
Nucleotide sequence analysis of the rpoB alleles from independent Rifr dnaN+ and dnaN159 isolates
| Nucleotide positionb |
rpoB mutations identified in straina:
|
|||
|---|---|---|---|---|
| JL100 (dnaN+)
|
JL103 (dnaN159)
|
|||
| Base substitution | No. of occurrences | Base substitution | No. of occurrences | |
| 1455 | ND | NA | TA→ CG | 1 |
| 1534 | ND | NA | TA→ CG | 1 |
| 1546 | GC→ AT | 4 | GC→ AT | 10 |
| 1576 | GC→ AT | 5 | CG→ AT | 4 |
| 1586 | GC→ AT | 1 | GC→ AT | 4 |
| 1592 | GC→ AT | 1 | GC→ AT | 3 |
| 1691 | GC→ AT | 4 | ND | NA |
Cultures of the indicated strains were spread on LB plates supplemented with Rif (100 μg/ml) and incubated overnight at 30°C. A single Rifr isolate from each plate was colony purified prior to PCR amplification of the rpoB gene, as described previously (12). The nucleotide change(s) in rpoB alleles leading to Rifr in independent Rifr isolates of strains JL100 (dnaN+; 15 isolates) and JL103 (dnaN159; 22 isolates) were determined by automated nucleotide sequence analysis (Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, NY), as described previously (38). Each Rifr dnaN+ isolate examined contained a single rpoB mutation, while one of the Rifr dnaN159 isolates contained two mutations (TA→ CG at position 1455 and GC→ AT at position 1586). Abbreviations: ND, none detected; NA, not applicable.
The position of each mutation identified is indicated, with position 1 referring to the A in the ATG initiation codon of rpoB.
Summary and conclusions.
The results discussed above suggest that Pol I plays an important role in both DNA replication (Fig. 1) and ssDNA gap repair in the dnaN159 strain (Fig. 2). In addition, our findings discussed above (Fig. 4), taken together with those discussed previously (29, 38, 39), suggest that the SOS-regulated Pol II (polB), Pol IV (dinB), and Pol V (umuDC) are able to compete with Pol I and Pol III for access to ssDNA gaps in the dnaN159 strain, particularly on the lagging strand (Table 1). This apparent competition is presumably attributable in large part to impaired interactions of β159 with Pol I (Fig. 3) and Pol III (38). Finally, the observation that the β clamp confers processivity upon Pol I in vitro, and that β159 is impaired for this activity (Fig. 3), suggests that the β clamp might act to manage the actions of Pol I in vivo, as has been suggested previously for the lagging strand (24). Further work utilizing mutant forms of Pol I deficient for interaction with the β clamp will help to establish whether Pol I must interact with the β clamp in order to catalyze ssDNA gap repair and/or Okazaki fragment maturation in vivo.
Acknowledgments
We thank Mary Berlyn (E. coli Genetic Stock Center, Yale University), Justin Courcelle (Portland State University), Iwona Fijalkowska (Polish Academy of Sciences), Cathy Joyce (Yale University), and Martin Marinus (University of Massachusetts Medical School) for generously providing us with E. coli strains and the members of our laboratory for helpful discussions.
This work was supported by a Public Service Health grant GM066094 (M.D.S.) and NIH Undergraduate Minority Administrative Supplement GM066094-S1 (M.D.S. and R.B.).
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Ausubel, F. M. 1988. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 2.Bates, H., S. K. Randall, C. Rayssiguier, B. A. Bridges, M. F. Goodman, and M. Radman. 1989. Spontaneous and UV-induced mutations in Escherichia coli K-12 strains with altered or absent DNA polymerase I. J. Bacteriol. 171:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bebenek, K., C. M. Joyce, M. P. Fitzgerald, and T. A. Kunkel. 1990. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 265:13878-13887. [PubMed] [Google Scholar]
- 4.Becherel, O. J., R. P. Fuchs, and J. Wagner. 2002. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair 1:703-708. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, L. B. 2006. Dynamics of loading the Escherichia coli DNA polymerase processivity clamp. Crit. Rev. Biochem. Mol. Biol. 41:179-208. [DOI] [PubMed] [Google Scholar]
- 6.Bonner, C. A., P. T. Stukenberg, M. Rajagopalan, R. Eritja, M. O'Donnell, K. McEntee, H. Echols, and M. F. Goodman. 1992. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J. Biol. Chem. 267:11431-11438. [PubMed] [Google Scholar]
- 7.Burgers, P. M., A. Kornberg, and Y. Sakakibara. 1981. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:5391-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalrymple, B. P., K. Kongsuwan, G. Wijffels, N. E. Dixon, and P. A. Jennings. 2001. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA 98:11627-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duzen, J. M., G. C. Walker, and M. D. Sutton. 2004. Identification of specific amino acid residues in the E. coli beta processivity clamp involved in interactions with DNA polymerase III, UmuD and UmuD′. DNA Repair 3:301-312. [DOI] [PubMed] [Google Scholar]
- 10.Fijalkowska, I. J., P. Jonczyk, M. M. Tkaczyk, M. Bialoskorska, and R. M. Schaaper. 1998. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 95:10020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg, E. C., G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis, 2nd ed. ASM Press, Washington, DC.
- 12.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2:593-608. [DOI] [PubMed] [Google Scholar]
- 13.Gawel, D., M. Maliszewska-Tkaczyk, P. Jonczyk, R. M. Schaaper, and I. J. Fijalkowska. 2002. Lack of strand bias in UV-induced mutagenesis in Escherichia coli. J. Bacteriol. 184:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grompone, G., M. Seigneur, S. D. Ehrlich, and B. Michel. 2002. Replication fork reversal in DNA polymerase III mutants of Escherichia coli: a role for the beta clamp. Mol. Microbiol. 44:1331-1339. [DOI] [PubMed] [Google Scholar]
- 15.Ho, C., O. I. Kulaeva, A. S. Levine, and R. Woodgate. 1993. A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J. Bacteriol. 175:5411-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, R. C. 1978. Reduction of postreplication DNA repair in two Escherichia coli mutants with temperature-sensitive polymerase III activity: implications for the postreplication repair pathway. J. Bacteriol. 136:125-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce, C. M., and N. D. Grindley. 1983. Construction of a plasmid that overproduces the large proteolytic fragment (Klenow fragment) of DNA polymerase I of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyce, C. M., and N. D. Grindley. 1984. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J. Bacteriol. 158:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, D. R., and C. S. McHenry. 1996. Identification of the beta-binding domain of the alpha subunit of Escherichia coli polymerase III holoenzyme. J. Biol. Chem. 271:20699-20704. [DOI] [PubMed] [Google Scholar]
- 20.Kolodner, R., R. A. Fishel, and M. Howard. 1985. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J. Bacteriol. 163:1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornberg, A., and T. Baker. 1991. DNA replication, 2nd ed. W.H. Freeman, New York, NY.
- 22.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Saro, F. J., M. G. Marinus, P. Modrich, and M. O'Donnell. 2006. The beta sliding clamp binds to multiple sites within MutL and MutS. J. Biol. Chem. 281:14340-14349. [DOI] [PubMed] [Google Scholar]
- 24.Lopez de Saro, F. J., and M. O'Donnell. 2001. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl. Acad. Sci. USA 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahdi, A. A., and R. G. Lloyd. 1989. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol. Gen. Genet. 216:503-510. [DOI] [PubMed] [Google Scholar]
- 26.Maki, S., and A. Kornberg. 1988. DNA polymerase III holoenzyme of Escherichia coli. III. Distinctive processive polymerases reconstituted from purified subunits. J. Biol. Chem. 263:6561-6569. [PubMed] [Google Scholar]
- 27.Maliszewska-Tkaczyk, M., P. Jonczyk, M. Bialoskorska, R. M. Schaaper, and I. J. Fijalkowska. 2000. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proc. Natl. Acad. Sci. USA 97:12678-12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maor-Shoshani, A., and Z. Livneh. 2002. Analysis of the stimulation of DNA polymerase V of Escherichia coli by processivity proteins. Biochemistry 41:14438-14446. [DOI] [PubMed] [Google Scholar]
- 29.Maul, R. W., and M. D. Sutton. 2005. Roles of the Escherichia coli RecA protein and the global SOS response in effecting DNA polymerase selection in vivo. J. Bacteriol. 187:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 31.Okazaki, R., M. Arisawa, and A. Sugino. 1971. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc. Natl. Acad. Sci. USA 68:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ream, L. W., and A. J. Clark. 1983. Cloning and deletion mapping of the recF dnaN region of the Escherichia coli chromosome. Plasmid 10:101-110. [DOI] [PubMed] [Google Scholar]
- 33.Ream, L. W., L. Margossian, A. J. Clark, F. G. Hansen, and K. von Meyenburg. 1980. Genetic and physical mapping of recF in Escherichia coli K-12. Mol. Gen. Genet. 180:115-121. [DOI] [PubMed] [Google Scholar]
- 34.Richardson, C. C., R. B. Inman, and A. Kornberg. 1964. Enzymic synthesis of deoxyribonucleic acid. 18. The repair of partially single-stranded DNA templates by DNA polymerase. J. Mol. Biol. 116:46-69. [DOI] [PubMed] [Google Scholar]
- 35.Sakakibara, Y., and T. Mizukami. 1980. A temperature-sensitive Escherichia coli mutant defective in DNA replication: dnaN, a new gene adjacent to the dnaA gene. Mol. Gen. Genet. 178:541-553. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Sharma, R. C., and K. C. Smith. 1987. Role of DNA polymerase I in postreplication repair: a reexamination with Escherichia coli ΔpolA. J. Bacteriol. 169:4559-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton, M. D. 2004. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J. Bacteriol. 186:6738-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton, M. D., and J. M. Duzen. 2006. Specific amino acid residues in the beta sliding clamp establish a DNA polymerase usage hierarchy in Escherichia coli. DNA Repair 5:312-323. [DOI] [PubMed] [Google Scholar]
- 40.Sutton, M. D., J. M. Duzen, and R. W. Maul. 2005. Mutant forms of the Escherichia coli β sliding clamp that distinguish between its roles in replication and DNA polymerase V-dependent translesion DNA synthesis. Mol. Microbiol. 55:1751-1766. [DOI] [PubMed] [Google Scholar]
- 41.Sutton, M. D., M. F. Farrow, B. M. Burton, and G. C. Walker. 2001. Genetic interactions between the Escherichia coli umuDC gene products and the beta processivity clamp of the replicative DNA polymerase. J. Bacteriol. 183:2897-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 43.Wagner, J., S. Fujii, P. Gruz, T. Nohmi, and R. P. Fuchs. 2000. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]