Abstract
Orthologs of proteins SbcD (Mre11) and SbcC (Rad50) exist in all kingdoms of life and are involved in a wide variety of DNA repair and maintenance functions, including homologous recombination and nonhomologous end joining. Here, we have inactivated the sbcC and/or sbcD genes of Deinococcus radiodurans, a highly radioresistant bacterium able to mend hundreds of radiation-induced DNA double-strand breaks (DSB). Mutants devoid of the SbcC and/or SbcD proteins displayed reduced survival and presented a delay in kinetics of DSB repair and cell division following γ-irradiation. It has been recently reported that D. radiodurans DNA polymerase X (PolX) possesses a structure-modulated 3′-to-5′ exonuclease activity reminiscent of specific nuclease activities displayed by the SbcCD complex from Escherichia coli. We constructed a double mutant devoid of SbcCD and PolX proteins. The double-mutant ΔsbcCD ΔpolXDr (where Dr indicates D. radiodurans) bacteria are much more sensitive to γ-irradiation than the single mutants, suggesting that the deinococcal SbcCD and PolX proteins may play important complementary roles in processing damaged DNA ends. We propose that they are part of a backup repair system acting to rescue cells containing DNA lesions that are excessively numerous or difficult to repair.
Deinococcus radiodurans belongs to a family of bacteria characterized by an exceptional ability to withstand the lethal effects of DNA-damaging agents, including ionizing radiation, UV light, and desiccation (2, 31, 34). D. radiodurans R1 is able to reconstruct a functional genome from hundreds of radiation-induced chromosomal fragments, whereas the genomes of most organisms are irreversibly shattered under the same conditions (3). Reassembly of the shattered chromosomal fragments is accompanied by extensive DNA synthesis and has been proposed to involve an initial joining of fragments with overlapping homologies through an extended synthesis-dependent single-strand annealing (ESDSA) process to create long linear DNA intermediates, followed and completed by a classical homologous recombination (HR) process to generate circular chromosomes (44). In addition to these homology-driven processes, we cannot exclude the possibility that nonhomologous-end joining (NHEJ) of DNA fragments may also take place as a backup repair system in heavily irradiated cells. ESDSA, HR, and NHEJ could be favored by an unusual compactness of the D. radiodurans genome, restricting the diffusion of DNA fragments (27).
Recently, we have identified a D. radiodurans DNA polymerase that belongs to the X family (PolXDr) of DNA polymerases that comprises mainly eukaryotic enzymes such as Saccharomyces cerevisiae Pol4, human Pol β, Pol λ, and Pol μ, and terminal deoxyribonucleotidyl transferase (reviewed in reference 21). These polymerases have been proposed to play important roles in different DNA repair processes, including NHEJ (39). D. radiodurans cells devoid of PolXDr have been shown to display a delay in double-strand break (DSB) repair and an increased sensitivity to γ-irradiation (26). The PolXDr polymerase is peculiar in that it possesses a highly processive 3′-to-5′ exonuclease activity modulated by the structure of the DNA substrate, specifically recognizing and pausing when it encounters a stem-loop structure (4). The stem-loop-modulated exonuclease of PolXDr is required for efficient in vivo repair of DSBs, suggesting that it may play an important role in DNA repair by processing damaged DNA or repair intermediates, thus generating substrates for other repair proteins.
The activities of the deinococcal PolXDr nuclease are somewhat reminiscent of those displayed by the bacterial SbcC/SbcD complex and its eukaryotic homolog, the Rad50/Mre11 complex. The two complexes exhibit single-stranded endonuclease and 3′-to-5′ double-stranded exonuclease activities (7, 10, 17, 37, 41, 42). The 3′-to-5′ exonuclease activity of the Rad50/Mre11 complex is enhanced with substrates that have duplex DNA ends (40). This complex can also cleave ends sealed by hairpin structures (38, 40). In Escherichia coli, hairpin structures in single-stranded DNA formed during replication and protein-bound DNA ends are converted by the SbcCD nuclease to double-strand breaks that are repaired by recombination (7-9).
In Eukarya, a third protein, named Xrs2 in yeast and Nbs1 in humans, is associated with Rad50 and Mre11 in the formation of a stable complex, referred to as the RMX complex in yeast (or RMN in human). The RMX complex was previously seen, by atomic force microscopy or scanning force microscopy, to engage and juxtapose DNA ends to form DNA oligomers (6, 15), and the DNA end-bridging activity of the RMX complex is believed to be important for aligning the substrate molecules to prepare them for joining. In S. cerevisiae, the RMX complex is involved in meiotic recombination and in DNA double-strand break repair by NHEJ (36). In E. coli, the SbcCD complex stimulates repair of double-strand breaks generated by a restriction endonuclease (13), and in Bacillus subtilis, the SbcC protein appears to play a role in the repair of DNA interstrand cross-links (30).
Homologs of the Rad50/SbcC (DR1922) and the Mre11/SbcD (DR1921) proteins have been identified in the D. radiodurans genome (29, 43). Here, we investigate the involvement of the SbcC and SbcD proteins in DSB repair and radioresistance in D. radiodurans. We have shown that knockout cells devoid of SbcCD activity display increased sensitivity to γ rays, altered kinetics of DSB repair, and a delay in the resumption of cell division following γ-irradiation. Interestingly, these effects were more dramatic when the absence of SbcCD was associated with the absence of PolXDr, suggesting that SbcCD and PolXDr proteins may have complementary functions in DSB repair, acting on different substrates and/or in different repair pathways.
MATERIALS AND METHODS
Materials, media, and cultures.
All reagents, materials, and media were obtained from sources previously reported (5). When necessary, media were supplemented with the appropriate antibiotics used at the following final concentrations: kanamycin, 6 μg/ml for D. radiodurans; chloramphenicol, 25 μg/ml for E. coli and 3 μg/ml for D. radiodurans; and spectinomycin, 75 μg/ml for D. radiodurans and 40 μg/ml for E. coli.
Deletion of the sbcC, sbcD, or sbcCD gene from D. radiodurans.
Deletion of the sbcC, the sbcD, and the sbcCD gene was performed in two steps. First, alleles ΔsbcCΩkan, ΔsbcDΩkan, ΔsbCDΩkan, and ΔsbcCDΩcat were constructed in vitro by ligating a cam cassette expressing kanamycin or chloramphenicol resistance in D. radiodurans to the chromosomal sequences 500 bp upstream and downstream of the coding regions of the sbcC, sbcD, and sbcCD genes, respectively. Second, constructs were used to transform D. radiodurans R1, and the ΔsbcCDΩcam construct was used to transform GY12219 ΔpolXDrΩkan to replace the wild-type sbcCD allele by homologous recombination. The genetic structure of the Kanr or the Camr transformants was tested by PCR, and oligonucleotides used for the in vitro construction and diagnostic PCR are listed in Table S1 in the supplemental material.
Expression in trans of the sbcCD operon in a ΔsbcCD host.
A plasmid carrying the sbcCD operon under the control of its natural promoter was used to express SbcCD in the ΔsbcCD background. A SacI-BamHI PCR fragment containing the sbcCD operon, amplified by PCR from genomic DNA of strain D. radiodurans R1, using primers EB24 and EB26 (see Table S1 in the supplemental material), was cloned into the E. coli-D. radiodurans shuttle vector p11520, a derivative of pI8 containing a gene encoding resistance to spectinomycin, giving rise to plasmid p13002.
Expression in trans of the polXDr gene or the truncated polXc gene.
Plasmids p13008 and p13007 expressing polXDr and polXc (which encodes a mutant protein devoid of exonuclease activity for complementation), respectively, from a pSpac promoter are derivatives of p11549-polXDr and p11549-polXc (4) in which the 3,217-bp KpnI-BamH1 fragment containing a gene encoding resistance to chloramphenicol was replaced with the 3,121-bp KpnI-BamHI fragment containing a gene encoding resistance to spectinomycin of plasmid p11559 (33). These plasmids were used to transform GY12219 ΔpolXDrΩkan and GY12918 ΔpolXDrΩkan ΔsbcCDΩcat. Expression of polXDr or polXc was induced by adding 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the media.
Treatment of D. radiodurans with different DNA-damaging agents. (i) Gamma irradiation.
Bacteria were grown in 2× TGY (1% tryptone, 0.2% glucose, and 0.6% yeast extract) medium or in 2× TGY medium supplemented with spectinomycin when they contained plasmid p11520 or p13002-sbcCD or in 2× TGY medium with 10 mM IPTG and spectinomycin when bacteria contained plasmid p11559, p13008-polXDr, or p13007-polXc. At the exponential phase (A650 of 0.5 or 1.5) or the early stationary phase (A650 of 2), cultures were concentrated 10-fold in 2× TGY and irradiated on ice with a 137Cs irradiation system (Institut Curie, Orsay, France) at a dose rate of 56.6 Gy/min. Following irradiation, diluted samples were plated on TGY (0.5% tryptone, 0.1% glucose, and 0.3% yeast extract) plates or on TGY plates supplemented with 1 mM IPTG when bacteria contained plasmid p11559 or p13008-polXDr or p13007-polXc and then incubated for 3 to 4 days at 30°C before the colonies were counted.
(ii) UV irradiation.
Bacterial cultures at an A650 of 1.5 were centrifuged, washed in MgSO4 (10 mM), and centrifuged again before pellets were resuspended in MgSO4 to obtain an A650 of 1. Aliquots (100 μl) were transferred to 14-mm wells of Teflon-printed diagnostic slides prior to exposure to UV light at a dose rate of 3.5 J/m2/s. Appropriate dilutions were plated on TGY plates and incubated for 3 to 4 days at 30°C before the colonies were counted.
(iii) Mitomycin C treatment.
Bacterial cultures at an A650 of 1.5 were serially diluted in TGY broth and plated on TGY plates supplemented with increasing concentrations of mitomycin C. The colonies were counted after an incubation of 3 to 4 days at 30°C.
Pulsed-field gel electrophoresis.
Bacteria were grown in 2× TGY to an A650 of 1.2 and exposed to γ-irradiation (6,800 Gy). Irradiated cultures and unirradiated controls were diluted in 2× TGY to an A650 of 0.2 and incubated at 30°C. At different postirradiation incubation times, samples (5 ml) were taken to prepare DNA plugs, as described in the report by Mattimore and Battista (31). The DNA in the plugs was digested for 16 h at 37°C with 60 units of NotI restriction enzyme. After digestion, the plugs were subjected to pulsed-field gel electrophoresis for 28 h at 10°C using a CHEF MAPPER electrophoresis system (Bio-Rad) under the following conditions: 5.5 V/cm, linear pulse of 40 s, and a switching angle of 120° (−60° to +60°).
RESULTS
Cells devoid of SbcC and/or SbcD proteins showed an increased sensitivity to mitomycin C treatment and γ-irradiation.
To determine whether the SbcC (DR1922) and SbcD (DR1921) homologs encoded by the D. radiodurans genome are involved in DNA repair in this radioresistant organism, we constructed deletion mutants devoid of SbcC or SbcD or both proteins. The mutant alleles were first constructed in vitro by ligating the two regions flanking each gene to be inactivated to a cam (or a kan) cassette, as described in Materials and Methods. The tripartite ligation products were then introduced into D. radiodurans by transformation, selecting for chloramphenicol (or kanamycin) resistance to allow replacement of the wild-type alleles with their mutated counterparts via homologous recombination. Homogenotes of sbcC, sbcD, and sbcCD disruption mutants were easily obtained after just one cycle of purification on selective medium, and the purity of the strains was verified by PCR (see Fig. S1 in the supplemental material). Furthermore, disruption of the sbcCD genes did not show a significant effect on the growth of D. radiodurans cells in 2× TGY liquid medium, indicating that neither of the inactivated genes is essential for cell viability.
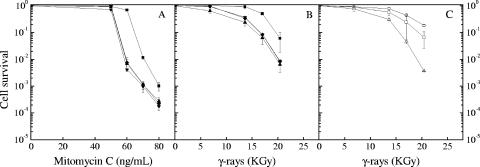
The ΔsbcC, ΔsbcD, and ΔsbcCD mutants showed a wild-type level of resistance to UV light (data not shown). In contrast, they were significantly more sensitive than the parental wild-type strain to mitomycin C treatment (Fig. 1A) and γ-irradiation (Fig. 1B). E. coli SbcC forms a complex with SbcD (7). The phenotypic similarities of the disruption mutants were consistent with the two proteins acting in a complex in D. radiodurans as they did in E. coli.
FIG. 1.
Mutant ΔsbcD, ΔsbcC, and ΔsbcCD strains of bacteria show an increased sensitivity to mitomycin C treatment and to γ-irradiation. Strain GY9613 (wild type, filled squares), GY12908 (ΔsbcD, filled inverted triangles), GY12909 (ΔsbcC, filled diamonds), and GY12910 (ΔsbcCD, filled triangles) bacteria grown to an A650 of 1.5 were exposed to mitomycin C (A) or γ-irradiation (B) at doses indicated on the abscissa. Strains GY12936 (wild type/p11520, squares), GY12923 (ΔsbcCD/p11520, triangle), and GY12912 (ΔsbcCD/p13002, sbcCD+, circles) were also exposed to γ-irradiation (C). Cell survival was measured as described in Materials and Methods.
To verify that the increased sensitivity to γ rays displayed by the ΔsbcCD mutant bacteria resulted from the absence of the SbcCD complex from these bacteria, the sbcCD operon, including its natural promoter, was cloned onto plasmid p11520 (a pI8-derived, low-copy-number shuttle vector in D. radiodurans [32]) and introduced into ΔsbcCD bacteria. The resulting ΔsbcCD/psbcCD+ bacteria expressing the sbcCD operon in trans recovered a wild-type level of γ-ray resistance (Fig. 1C), supporting an implication of the deinococcal SbcCD complex in DNA repair.
Additive effect of deletions of polXDr and sbcCD on cell survival after γ-irradiation.
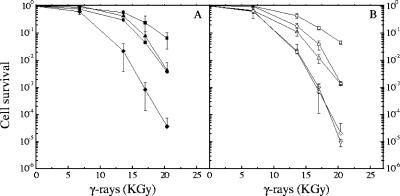
PolXDr possesses a stem-loop-modulated exonuclease activity that is reminiscent of the exo- and endonuclease activities associated with the E. coli SbcCD complex (4). To determine whether the deinococcal SbcCD complex might act in the same repair pathway as PolXDr, we constructed a double-mutant strain with deletions of sbcCD and polXDr and compared its γ-irradiation survival rate to those of single mutants sbcCD and polXDr and wild-type cells.
Disruption of polXDr alone resulted in an increase in γ-ray sensitivity that was of the same order of magnitude as that observed for the ΔsbcCD mutant. Indeed, in comparison to that of the wild type, both mutants showed a reduction in survival that ranged from 4-fold at 13.6 kGy to more than 100-fold at 20.4 kGy (Fig. 2A, filled circles, and B, circles). In contrast, the ΔsbcCD ΔpolXDr double-knockout cells were dramatically more sensitive to γ-irradiation than the single-deletion mutants and showed a decrease in survival of more than 2,000-fold after exposure to 20 kGy compared to that of the wild type (Fig. 2A, filled diamonds, and B, diamonds). When SbcCD (Fig. 2A, filled triangles) or PolXDr (Fig. 2B, triangles) was expressed in trans from the genes cloned into a plasmid in the double-deletion mutant, the cells regained the moderate γ-ray sensitivity characteristic of the single-mutant cells, indicating that their radiosensitive phenotype can be attributed to the combined absence of SbcCD and PolXDr. Since it was previously shown that the structure-modulated exonuclease activity of PolX plays an important role in DNA repair (4), we also tested PolXc, a PolXDr mutant protein devoid of this activity for complementation (4). In this case, expression in trans of the mutant PolXc protein failed to alleviate the radiation-sensitive phenotype of the double-knockout cells (Fig. 2B, inverted triangles).
FIG. 2.
ΔsbcCD and ΔpolX deletions are additive and confer increased sensitivity to γ-irradiation. (A) Bacterial strains GY12936 (wild type/p11520, filled squares), GY12923 (ΔsbcCD/p11520, filled circles), GY12924 (ΔsbcCD ΔpolXDr/p11520, filled diamonds), and GY12926 (ΔsbcCD ΔpolXDr/p13002 sbcCD+, filled triangles) and (B) GY12937 (wild type/p11559, squares), GY12931 (ΔpolXDr/p11559, circles), GY12925 (ΔsbcCD ΔpolXDr/p11559, diamonds), GY12930 (ΔsbcCD ΔpolXDr/p13008, polXDr, triangles), and GY12929 (ΔsbcCD ΔpolXDr/p13007 polXc, inverted triangles) grown to an A650 of 1.5 were exposed to γ-irradiation at doses indicated on the abscissa, and cell survival was measured as described in Materials and Methods.
It has been previously reported (4) that expression of PolXc in a ΔpolXDr host not only failed to restore γ-ray resistance but further decreased the γ-irradiation survival rate, leading to the hypothesis that the mutated protein was interfering with an alternate double-strand DNA repair pathway. We did not observe this negative effect on the survival of the ΔsbcCD ΔpolX mutant (Fig. 2B, inverted triangles). However, a closer examination of the ΔpolXDr/polXc strain used by Blasius et al. (4) indicated that it may contain an additional unidentified chromosomal mutation, decreasing its γ-ray survival rate, since cured cells, in which the polXc plasmid was segregated out, still retained their γ-ray-hypersensitive phenotype (data not shown).
Increased sensitivity of cells devoid of SbcCD or SbcCD and PolXDr proteins in early exponentially growing cultures.
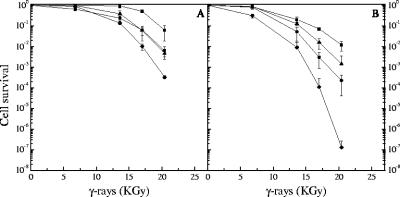
The effects of the absence of SbcCD or PolXDr on cell viability were observed only at doses in excess of 10 kGy, suggesting that these proteins function under specific conditions in vivo. To test whether these proteins have a role at lower doses, and taking into account the possibility that actively dividing cells could be more sensitive to γ rays than stationary growing cells, we compared the γ-ray survival rates of ΔsbcCD, ΔpolXDr, ΔsbcCD ΔpolXDr, and wild-type cells grown to an A650 of 0.5 (early exponential phase) to those of cells grown to an A650 of 2 (early stationary phase). Interestingly, whereas the wild-type and the ΔpolXDr cells were not more sensitive to γ rays at an A650 of 0.5 than at an A650 of 2, cells devoid of SbcCD were significantly more sensitive to γ rays at an A650 of 0.5 than at an A650 of 2, and the sensitivity of the ΔsbcCD ΔpolXDr double mutant increased dramatically at an A650 of 0.5 compared to those of the single ΔpolXDr mutants (Fig. 3). These results suggest a broader role of SbcCD in DNA repair than was suspected initially, based on observations at an A650 of 2.
FIG. 3.
Increased sensitivity of cells devoid of SbcCD or SbcCD and PolXDr proteins in early exponentially growing cultures. Bacterial strains GY9613 (wild type, squares), GY12910 (ΔsbcCD, circles), GY12918 (ΔsbcCD ΔpolXDr, diamonds), and GY12219 (ΔpolXDr, triangles) grown to an A650 of 2 (panel A) or an A650 of 0.5 (panel B) were exposed to γ-irradiation at doses indicated on the abscissa, and cell survival was measured as described in Materials and Methods.
Altered kinetics of reassembly of broken chromosomes and extended growth lag in γ-irradiated cells devoid of SbcCD and/or PolXDr proteins.
If cell survival implies that repair of DSBs goes to completion, the rate of DNA repair can be normal or reduced depending on the nature of the defect found in the DNA repair pathway involved in the completion of the intact genome. Moreover, decreased survival could be related to a defect independently of the completion of DNA repair, as shown previously with cells expressing limiting concentrations of RecA protein (22). Thus, we investigated whether the combined absence of SbcCD and PolXDr proteins results in an additive inhibitory effect on the kinetics of repair of radiation-induced DNA double-strand breaks. For this purpose, cells from the ΔsbcCD, ΔpolXDr, and ΔsbcCD ΔpolXDr mutants and the wild type were exposed to 6.8 kGy γ-irradiation, a dose that introduces approximately 200 DSBs per genome (2) without significantly affecting the survival of the mutant cells (Fig. 1 and 2), and the reassembly of the shattered chromosomes as a function of the postirradiation incubation time was monitored by pulsed-field gel electrophoresis. Recovery from DNA damage was monitored by the appearance of the complete pattern of the 11 resolvable fragments generated by NotI digestion of total genomic DNA (23). The absence of the 436-kb fragment in the digest from the restriction pattern of the ΔpolXDr ΔsbcCD mutant genome was noted, as it contains an additional NotI site located in the cam cassette used to inactivate sbcCD in this construct. An intact genome was reconstituted within 2.5 h of postirradiation incubation in wild-type cells (Fig. 3A). In contrast, the recovery process took longer to reach completion for all the mutants, requiring an additional 60 min for the ΔpolXDr strain (Fig. 3B), 90 min for the ΔsbcCD strain (Fig. 3C), and at least 150 min for the more radio-sensitive ΔsbcCD ΔpolXDr strain (Fig. 3D). Moreover, after the first step of extensive degradation of DNA, wild-type cells were able to reconstitute an intact genome in half an hour (Fig. 4A), whereas the increase of the size of the fragments was more progressive and seemed to require around 2 h for cells devoid of SbcCD (Fig. 4C). We observed the same influence of the absence of SbcCD on the kinetics of DNA DSB repair for ΔsbcCD ΔpolXDr bacteria (Fig. 4B and D).
FIG. 4.
Bacterial ΔsbcCD and ΔsbcCD ΔpolXDr strains show an increased delay in intact genomic DNA restoration and in cell division after γ-irradiation. Strains GY9613 (wild type, panels A and E, squares), GY12219 (ΔpolXDr, panels B and E, diamonds), GY12910 (ΔsbcCD, panels C and E, triangles), and GY12918 (ΔsbcCD ΔpolXDr, panels D and E, circles) grown to an A650 of 1.2 were exposed to γ-irradiation at a dose of 6,800 Gy (panel E, filled symbols) or not (panel E, open symbols), diluted in 2× TGY to an A650 of 0.2, and then incubated in 2× TGY. At different times after irradiation, aliquots were taken to prepare DNA agarose plugs, which were digested with NotI prior to analyses by pulsed-field gel electrophoresis (panels A, B, C, and D), and to measure the number of viable cells per ml of culture (panel E).
Since γ-irradiation induces growth lags whose duration depends on the times required for completion of DNA repair (2), we also measured, in the same experiment, the kinetics of recovery of exponential growth after irradiation. We found that all the mutants showed extended growth lags, consistent with their reduced capacity to perform rapid repair of DNA double-strand breaks. Indeed, when the cells were plated at different times during postirradiation incubation, the first doubling of the number of CFU was observed 5 h 30 min after irradiation for the wild-type strains (Fig. 3E, filled squares), whereas this doubling required 7 h for both the ΔsbcCD and the ΔpolXDr mutants (Fig. 3E, filled diamonds and filled triangles, respectively) and at least 8 h for the ΔsbcCD ΔpolXDr double mutant (Fig. 3E, filled circles). In contrast, all of the mutants exhibited the same exponential growth rate as the wild type in a control experiment in which the cells were mock irradiated (Fig. 3E, open symbols).
DISCUSSION
Orthologs of SbcC (Rad50) and SbcD (Mre11) exist in all kingdoms of life and are involved in a wide variety of DNA repair and maintenance functions, including homologous recombination and nonhomologous end joining (12, 14, 18, 36). These proteins appear to play a role in the processing of DNA ends (10, 37) and a structural role in tethering DNA ends (6, 11, 15, 19, 20). Here, we deleted the sbcC and/or the sbcD gene from the radioresistant bacterium D. radiodurans and showed that the knockout mutants had reduced cell survival after γ-irradiation and, to a lesser extent, after mitomycin C treatment. The mutated cells also showed an extended delay in the reconstitution of an intact genome, even at a γ-irradiation dose that had no detrimental effect on their survival. Our results suggest that the deinococcal SbcCD complex might be involved in at least one pathway of double-strand break repair.
Repair of double-strand breaks in D. radiodurans occurs mainly through accurate assembly of the broken chromosomal fragments, by the combined action of synthesis-dependent single-strand annealing and classical homologous recombination (44). Both processes require some processing of the ends of the DNA fragments to generate 3′ single-strand DNA ends. It is unlikely that the SbcCD complex plays a major role in this process, since the survival of cells devoid of the SbcCD complex was not dramatically affected by γ-radiation at doses of up to 10 kGy. Nevertheless, the SbcCD complex might be required for processing of a subset of DSBs blocked by hairpin structures, by covalently bound proteins, or by DNA interstrand cross-links to generate mature DNA ends that can be used as substrates in the ESDSA, the HR, or the NHEJ repair pathway. This can be inferred from the biochemical activities of the prototype E. coli SbcCD complex that possesses a structure-specific endonuclease that cleaves DNA hairpin loops and a 3′-to-5′ single-strand exonuclease that may function in rendering DNA ends flush (7). Although D. radiodurans lacks the RecBCD helicase/nuclease that processes double-strand breaks in E. coli (24), it encodes all the components of the alternate RecF pathway (29) that form an alternative system for initiation of recombination in E. coli (24). This pathway is inhibited by the SbcB nuclease (reference 24 and references therein), and D. radiodurans is naturally devoid of the SbcB protein. Moreover, it was shown that expression in trans of the SbcB protein from E. coli renders D. radiodurans cells radiation sensitive (35). Thus, a RecF-like pathway may operate in D. radiodurans to generate 3′ overhangs of single-stranded DNA.
PolXDr also possesses a structure-modulated 3′-to-5′ double-strand exonuclease activity involved in DSB repair (4). This activity is reminiscent of the hairpin-specific nuclease activity of the SbcCD complex in E. coli. This raises the question of whether PolXDr and SbcCD share redundant DNA repair activities. Inactivation of PolXDr or SbcCD alone resulted in a very similar radiation-sensitive phenotype. In contrast, double-knockout cells were conspicuously more γ-ray sensitive and showed an extended delay in DSB repair compared to that of single-mutant cells. Several hypotheses can be advanced to account for these findings. (i) They might act on different DNA substrates and/or be involved in different repair pathways, or (ii) SbcCD and PolXDr might have only partially redundant functions such that the activity of one enzyme can, in part, compensate for the loss of the other. For example, the DNA polymerase activity of PolXDr might be particularly important in the repair of DSBs with noncohesive and/or damaged termini (16, 28), whereas the SbcCD complex might be required to process DNA ends blocked by hairpin structures, by covalently bound proteins (11) or by DNA interstrand cross-links (30). In addition, the deinococcal SbcCD complex might also play a more general structural role and facilitate DNA repair by bridging the ends of contiguous DNA fragments like other complexes of the same family (11, 15, 20). An understanding of the role(s) of the SbcCD proteins in DBS repair in D. radiodurans awaits in vitro analysis of the activities of the purified deinococcal proteins.
In summary, our data indicate that the SbcCD complex is involved in DSB repair in D. radiodurans. The contribution of these proteins to deinococcal radioresistance is particularly important in cells devoid of the PolXDr DNA polymerase/structure-modulated exonuclease. Nonetheless, the double-mutant cells with deletions of sbcCD and polXDr showed enhanced sensitivity, especially at elevated γ-irradiation doses where the survival of the wild-type cells also started to decline. This suggests that these proteins might be part of a backup repair system acting to rescue cells containing excessively numerous (or particularly difficult to repair) DNA lesions.
Supplementary Material
TABLE 1.
Bacterial strains
| Strain | Revelant markersa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | hsdR17 recA1 endA1 lacZΔM15 | Invitrogen |
| SCS 110 | endA1 dam dcm (F′ lacIqlacZΔM15) | Stratagene |
| D. radiodurans | ||
| GY9613 | R1, ATCC 13939 | 1 |
| GY12219 | as R1 but ΔpolXDrΩkan | 26 |
| GY12908 | as R1 but ΔsbcDΩkan | This work |
| GY12909 | as R1 but ΔsbcCΩkan | This work |
| GY12910 | as R1 but ΔsbcCDΩkan | This work |
| GY12911 | as R1 but ΔsbcCDΩcat | This work |
| GY12918 | as GY12219 but ΔsbcCDΩcat | This work |
| GY12912 | GY12910 ΔsbcCDΩkan (p13002, sbcCD+) | This work |
| GY12923 | GY12910 ΔsbcCDΩkan (p11520) | This work |
| GY12924 | GY12918 ΔpolXDrΩkan Δ(sbcCD)Ωcat (p11520) | This work |
| GY12925 | GY12918 ΔpolXDrΩkan ΔsbcCDΩcat (p11559) | This work |
| GY12926 | GY12918 ΔpolXDrΩkan ΔsbcCDΩcat (p13002, sbcCD+) | This work |
| GY12929 | GY12918 ΔpolXDrΩkan ΔsbcCDΩcat (p13007, polXc) | This work |
| GY12930 | GY12918 ΔpolXDrΩkan ΔsbcCDΩcat (p13008, polXDr) | This work |
| GY12931 | GY12219 ΔpolXDrΩkan (p11559) | This work |
| GY12934 | GY12219 ΔpolXDrΩkan (p13007, polXc) | This work |
| GY12935 | GY12219 ΔpolXDrΩkan (p13008, polXDr) | This work |
| GY12936 | R1 (p11520) | This work |
| GY12937 | R1 (p11559) | This work |
Genes of D. radiodurans are named as their E. coli orthologs (29). Ω indicates an insertion.
TABLE 2.
Plasmids that replicate in E. coli and D. radiodurans
| Plasmid name | Relevant descriptiona | Source or reference |
|---|---|---|
| pI8 | A shuttle vector that replicates in E. coli and in D. radiodurans | 32 |
| p11520 | Derivative of pI8; Spcr in D. radiodurans | This work |
| p11549 | Expression vector; Pspac PtufA::lacI Camr in E. coli and in D. radiodurans | 25 |
| p11559 | Expression vector; Pspac PtufA::lacI Spcr in E. coli and in D. radiodurans | 33 |
| p11549-polXc | p11549 with a fragment encoding the PolXc domain of PolXDr; Pspac::polXc | 4 |
| p11549-polXDr | p11549 with a fragment encoding PolXDr; Pspac::polXDr | 26 |
| p13002 | p11520 with a SacI-BamHI PCR fragment encoding SbcCD | This work |
| p13007 | Replacement of the KpnI-BamHI fragment of p11549-polXc containing the chloramphenicol cassette by the KpnI-BamHI fragment of p11559 containing the spectinomycin cassette | This work |
| p13008 | Replacement of the KpnI-BamHI fragment of p11549-polXDr containing the chloramphenicol cassette by the KpnI-BamHI fragment of p11559 containing the spectinomycin cassette | This work |
Gene denominations and genetic symbols are as shown in Table 1.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the Commissariat à l'Energie Atomique (CEA LRC 42V), and Electricité de France.
We thank Adriana Bailone for valuable discussions and critical reading of the manuscript and Françoise Vannier for expert technical assistance. We also thank the Institut Curie for the use of the 137Cs irradiation system, V. Favaudon for help with γ-irradiation, and M. DuBow for help with English.
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, A. W., H. C. Nordon, R. F. Cain, G. Parrish, and G. Duggan. 1956. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 10:575-578. [Google Scholar]
- 2.Battista, J. R. 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51:203-224. [DOI] [PubMed] [Google Scholar]
- 3.Battista, J. R., A. M. Earl, and M. J. Park. 1999. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7:362-365. [DOI] [PubMed] [Google Scholar]
- 4.Blasius, M., I. Shevelev, E. Jolivet, S. Sommer, and U. Hubscher. 2006. DNA polymerase X from Deinococcus radiodurans possesses a structure-modulated 3′→5′ exonuclease activity involved in radioresistance. Mol. Microbiol. 60:165-176. [DOI] [PubMed] [Google Scholar]
- 5.Bonacossa de Almeida, C., G. Coste, S. Sommer, and A. Bailone. 2002. Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol. Genet. Genomics 268:28-41. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 7.Connelly, J. C., E. S. de Leau, and D. R. Leach. 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connelly, J. C., E. S. de Leau, and D. R. Leach. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amsterdam) 2:795-807. [DOI] [PubMed] [Google Scholar]
- 9.Connelly, J. C., L. A. Kirkham, and D. R. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connelly, J. C., and D. R. Leach. 1996. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells 1:285-291. [DOI] [PubMed] [Google Scholar]
- 11.Connelly, J. C., and D. R. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27:410-418. [DOI] [PubMed] [Google Scholar]
- 12.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 13.Cromie, G. A., and D. R. Leach. 2001. Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol. Microbiol. 41:873-883. [DOI] [PubMed] [Google Scholar]
- 14.D'Amours, D., and S. P. Jackson. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 15.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8:1129-1135. [DOI] [PubMed] [Google Scholar]
- 16.Fan, W., and X. Wu. 2004. DNA polymerase lambda can elongate on DNA substrates mimicking non-homologous end joining and interact with XRCC4-ligase IV complex. Biochem. Biophys. Res. Commun. 323:1328-1333. [DOI] [PubMed] [Google Scholar]
- 17.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hefferin, M. L., and A. E. Tomkinson. 2005. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amsterdam) 4:639-648. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney, and J. A. Tainer. 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105:473-485. [DOI] [PubMed] [Google Scholar]
- 20.Hopfner, K. P., C. D. Putnam, and J. A. Tainer. 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12:115-122. [DOI] [PubMed] [Google Scholar]
- 21.Hubscher, U., G. Maga, and S. Spadari. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71:133-163. [DOI] [PubMed] [Google Scholar]
- 22.Jolivet, E., F. Lecointe, G. Coste, K. Satoh, I. Narumi, A. Bailone, and S. Sommer. 2006. Limited concentration of RecA delays DNA double-strand break repair in Deinococcus radiodurans R1. Mol. Microbiol. 59:338-349. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi, M., I. Narumi, S. Kitayama, H. Watanabe, and K. Yamamoto. 1999. Genomic organization of the radioresistant bacterium Deinococcus radiodurans: physical map and evidence for multiple replicons. FEMS Microbiol. Lett. 174:151-157. [Google Scholar]
- 24.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lecointe, F., G. Coste, S. Sommer, and A. Bailone. 2004. Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans. Gene 336:25-35. [DOI] [PubMed] [Google Scholar]
- 26.Lecointe, F., I. V. Shevelev, A. Bailone, S. Sommer, and U. Hubscher. 2004. Involvement of an X family DNA polymerase in double-stranded break repair in the radioresistant organism Deinococcus radiodurans. Mol. Microbiol. 53:1721-1730. [DOI] [PubMed] [Google Scholar]
- 27.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minski. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance. Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 28.Ma, Y., H. Lu, B. Tippin, M. F. Goodman, N. Shimazaki, O. Koiwai, C. L. Hsieh, K. Schwarz, and M. R. Lieber. 2004. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 16:701-713. [DOI] [PubMed] [Google Scholar]
- 29.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mascarenhas, J., H. Sanchez, S. Tadesse, D. Kidane, M. Krisnamurthy, J. C. Alonso, and P. L. Graumann. 2006. Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol. Biol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meima, R., and M. E. Lidstrom. 2000. Characterization of the minimal replicon of a cryptic Deinococcus radiodurans SARK plasmid and development of versatile Escherichia coli-D. radiodurans shuttle vectors. Appl. Environ. Microbiol. 66:3856-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mennecier, S., G. Coste, P. Servant, A. Bailone, and S. Sommer. 2004. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Genet. Genomics 272:460-469. [DOI] [PubMed] [Google Scholar]
- 34.Minton, K. W. 1994. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol. Microbiol. 13:9-15. [DOI] [PubMed] [Google Scholar]
- 35.Misra, H. S., N. P. Khairnar, S. Kota, S. Shrivastava, V. P. Joshi, and S. K. Apte. 2006. An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol. Microbiol. 59:1308-1316. [DOI] [PubMed] [Google Scholar]
- 36.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paull, T. T., and M. Gellert. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1:969-979. [DOI] [PubMed] [Google Scholar]
- 38.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramadan, K., I. V. Shevelev, G. Maga, and U. Hubscher. 2004. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu and terminal deoxyribonucleotidyl transferase. J. Mol. Biol. 339:395-404. [DOI] [PubMed] [Google Scholar]
- 40.Trujillo, K. M., and P. Sung. 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 41.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 42.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 43.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahradka, K., D. Slade, A. Bailone, S. Sommer, D. Averbeck, M. Petranovic, A. B. Lindner, and M. Radman. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569-573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.