Abstract
Rapamycin is an important macrocyclic polyketide produced by Streptomyces hygroscopicus and showing immunosuppressive, antifungal, and antitumor activities as well as displaying anti-inflammatory and neuroregenerative properties. The immense pharmacological potential of rapamycin has led to the production of an array of analogues, including through genetic engineering of the rapamycin biosynthetic gene cluster. This cluster contains several putative regulatory genes. Based on DNA sequence analysis, the products of genes rapH and rapG showed high similarities with two different families of transcriptional activators, LAL and AraC, respectively. Overexpression of either gene resulted in a substantial increase in rapamycin biosynthesis, confirming their positive regulatory role, while deletion of both from the chromosome of S. hygroscopicus resulted in a complete loss of antibiotic production. Complementation studies indicated an essential role of the RapG regulator for rapamycin biosynthesis and a supportive role of RapH. A direct effect of rapH and rapG gene products on the promoter of the rapamycin polyketide synthase operon, rapA-rapB, was observed using the chalcone synthase gene rppA as a reporter system.
The polyketide rapamycin and its analogues are currently the most selective kinase inhibitors known and the only inhibitors of mTOR (mammalian target of rapamycin)-associated kinase activity (23). Rapamycins bind the cyclophilin FKBP12, and this complex binds mTOR and inhibits its function. mTOR, a serine/threonine kinase, appears to act as a central controller that senses the cellular environment and regulates translation initiation through the eukaryotic initiation factor 4E and ribosomal p70 S6 kinase pathways. Rapamycin (Sirolimus, Rapamune), which is licensed for use as an immunosuppressant after organ transplantation, also has potential therapeutic use in the treatment of cardiovascular, autoimmune, and neurodegenerative diseases (22). In addition, rapamycin and its derivatives represent exciting candidates for anticancer therapeutic development, with several presently in clinical trials as anticancer agents (44). Appreciation of the immense pharmacological potential of rapamycin analogues has led to a demand for the economical production of rapamycin and its biosynthetic analogues (19) at an industrial scale.
The study of polyketide synthase (PKS) systems has revealed a number of associated regulatory proteins (3), but much remains unknown about the regulation of the biosynthesis of some of the more complex polyketides (1, 12, 41), although the complexity of these regulatory networks is beginning to be appreciated. The expression of PKS gene cluster elements is often controlled by a number of different families of regulatory proteins that can have either a pathway-specific or a pleiotropic mode of action, affecting a broader range of morphological and physiological processes, including secondary metabolite production (3). Among the main families of PKS regulators are the SARP family (Streptomyces antibiotic regulatory proteins) (45), the LAL family (large ATP-binding regulators of the LuxR family) (14), γ-butyrolactone-binding regulatory proteins (26), and two-component regulators (8), which tend to act pleiotropically but are found in a number of PKS clusters.
Targeted genetic engineering offers an alternative or complementary approach to classical strain improvement and could potentially be applied to regulatory systems. Currently no known examples of industrial production strains in which regulatory genes have been manipulated in order to increase polyketide production have been reported (10).
The initial sequence analysis of the rapamycin biosynthetic gene cluster revealed a total of 27 open reading frames (ORFs) in a region of 107 kbp (38). Based on sequence similarity searches, the cluster contained several ORFs whose gene products may have a potential regulatory function. RapR and RapS were identified as a translationally coupled putative two-component system, RapY was found to contain a helix-turn-helix (HTH) motif similar to repressors of antibiotic export in the actinorhodin and tetracenomycin clusters (30), and RapK may be responsible for the availability of the PKS starter unit (17, 19). RapG showed sequence similarity to positive regulatory proteins such as the SoxS and the Rob proteins from Escherichia coli in a stretch of about 100 amino acids around the predicted HTH motif. RapH had a DNA-binding motif near the C terminus and an ATP-binding site at the N terminus (30). Both genes, rapG and rapH, also contain the rare Leu codon TTA that has been proposed to serve as part of a regulatory mechanism of secondary metabolism in Streptomyces (28).
As sequence analysis of rapH and rapG implicated their gene products as pathway-specific positive regulators of rapamycin biosynthesis, this study tested this hypothesis to evaluate their roles and to provide an initial understanding of elements of the regulatory pathways, with the long-term aim of exploiting this information to enhance the fermentation yields of rapamycin and its biosynthetic analogues.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Streptomyces hygroscopicus NRRL 5491 (referred to here as the wild type) and its derivative recombinant strains were propagated at 28°C on ISP3 agar plates (40). Cultivation of strains for rapamycin production was carried out as described previously (M. A. Gregory, S. Gaisser, H. Petkovic, and S. J. Moss, 22 January 2004, PCT International Application, WO 04/007709). Plasmid vectors were propagated in E. coli DH10B grown in 2TY medium. Nonmethylated DNA was prepared in E. coli ET12567, and conjugation of plasmids was carried out using E. coli ET12567 carrying the conjugation-facilitating plasmid pUZ8002 (32). Streptomyces strain manipulation was performed as described by Kieser et al. (25).
Recombinant DNA methods.
Standard techniques for DNA manipulation were performed as described by Sambrook and Russell (36). Primers for PCR amplification and cloning of the rapH and rapG genes were designed based on the original rapamycin cluster sequence (GI 987088), with NdeI and XbaI restriction sites incorporated at the start codon and after the stop codon, respectively. Primers for PCR amplification of rapH were H1 (5′ GGCATATGACCGGGCGGGCCAACGGC 3′) and H2 (5′ GGTCTAGAGGCTATTCCGCCTTGACGAGTTCGG 3′) (NdeI and XbaI sites are underlined). Primers for PCR amplification of rapG were G1 (5′ GGCATATGACCAACGGCGCTGGAGCGGAG 3′) and G2 (5′ GGTCTAGAGGTCAGCTGTCGGTCAGCCCGGTTG 3′). For subcloning of the complete rapH-rapG fragment, primers G2 and H2 were used. DNA fragments were cloned into the pSET152-derived φBT-based integrative expression vector (Gregory et al., PCT International Application, WO 04/007709) pGP9, provided by Gerard Peck, under the control of the ActII-ORF4/PactI activator/promoter system. For deletion of the rapH-rapG fragment, primers HG1 (5′ CGAATTCGGTCACGTCCTGGCGCTGGTG 3′) and HG2 (5′ GCTCTAGACGGCCGAACTCGTCAAGGCG 3′) were used to PCR amplify a 1.5-kb overlapping region on the left side of rapH, and primers HG3 (5′ GCTCTAGACTGACCGACAGCTGAACCCGG 3′) and HG4 (5′ CAAGCTTTCCAGCAACATGTTCGCCAACAAGGC 3′) were employed to PCR amplify a 2-kb overlapping region on the right side of rapG. Fragments were combined at the shared XbaI site and cloned into the temperature-sensitive vector pKC1139 (4), which was then used to introduce a genomic deletion by homologous recombination.
Expression of an additional copy of the rapH and rapG genes in the S. hygroscopicus wild-type strain.
An integrative φBT-based expression vector, pGP9, with the ActII-ORF4/PactI activator/promoter system was used to overexpress genes rapH (pEK28) and rapG (pEK30) in S. hygroscopicus NRRL 5491. The same pSET152-derived vector was used to introduce a second copy of both genes, rapH and rapG, together in that respective order, transcribed in the same orientation on a cassette under the control of ActII-ORF4/PactI (pEK48). The cloning technique employed to generate gene cassettes is based on the use of methylation-sensitive and -resistant versions of the XbaI restriction site. It was developed from a cloning system described previously (16). A more convenient, updated cloning version was used during this work, as described in detail in recent patent literature (S. Gaisser, P. F. Leadlay, S. F. Haydock, and H. A. McArthur, 16 June 2005, PCT International Application, WO 2005/054265; Gregory et al., PCT International Application, WO 04/007709).
To introduce and express an additional copy of both rapH and rapG together in their native form under the control of their native promoters, the ActII-ORF4/PactI activator/promoter system was removed from the pSET152-derived vector, and the rapH-rapG fragment, PCR amplified using G2 and H2 primers, was introduced in its place (pEK49). Plasmid transfer experiments were carried out using conjugation procedures as described by Kieser et al. (25).
Targeted gene deletion via homologous recombination.
The deletion plasmid pEK51 was created based on the temperature-sensitive vector pKC1139 and the appropriate 1.5-kb and 2-kb overlapping regions of the genes to be deleted. Conjugations were carried out as described above, and four clones containing the self-replicating vector pEK51 were isolated. These strains were grown in TSBMG (24) with 50 μg/ml apramycin antibiotic, initially at 28°C for the seed stage, followed by three rounds of liquid subculturing steps at 37°C before plating on agar plates to select for primary recombinants. Five colonies were chosen from each initial source and grown for seed in TSBMG without antibiotic at 28°C, followed by three rounds of subculturing again without antibiotic at 37°C before plating on agar plates to screen for secondary recombinants. Replica plating with velvet stamps was used to rapidly screen for loss of antibiotic resistance (secondary recombination). Apramycin-sensitive colonies were tested by Southern blot hybridization and PCR analysis to confirm the genomic deletion (data not shown).
Full and partial complementation of the S. hygroscopicus ΔHG strain with rapG and/or rapH.
The deleted genes were reintroduced (i) individually as a single copy, (ii) together in a cassette, or (iii) in their native form into the S. hygroscopicus ΔHG strain. Conjugation experiments were carried out using plasmids pEK28, pEK30, pEK48, and pEK49 according to procedures described previously (Gregory et al., PCT International Application, WO 04/007709).
Rapamycin production and analysis.
Engineered strains were tested for rapamycin production by growing the cultures in 50-ml tubes as described previously (Gregory et al., PCT International Application, WO 04/007709). Seed cultures in RapV7 were cultivated at 300 rpm for 2 days at 28°C. Production medium MD6 (Gregory et al., PCT International Application, WO 04/007709) was inoculated with seed culture at 10% (vol/vol) and shaken at 300 rpm for 6 days at 26°C. To obtain statistically significant results, each colony was represented by a triplicate sample set. The harvested cultures were extracted 1:1 with methanol, with shaking for 30 min at room temperature. The samples were pelleted by centrifugation, and the supernatants were analyzed by high-pressure liquid chromatography, which was carried out using a Phenomenex Hyperclone 3-μm BDS C18 column (4.6 mm by 150 mm), monitoring UV absorbance at 280 nm, and eluting at 1.0 ml/min with a gradient of 45% buffer A-55% buffer B to 5% buffer A-95% buffer B over 10 min. Buffer A consisted of 0.01 M ammonium acetate containing 10% (vol/vol) acetonitrile and 0.001% (vol/vol) trifluoroacetic acid. Buffer B consisted of 0.01 M ammonium acetate containing 90% (vol/vol) acetonitrile and 0.001% (vol/vol) trifluoroacetic acid.
Reporter plasmid for expression studies.
To study the activity of the PrapA promoter controlling the expression of the rapA-rapB PKS operon, rppA from the Saccharopolyspora erythraea chalcone synthase gene cluster was used as a reporter. The rppA gene was obtained from pBW219 (11) and introduced into a pIJ8660 vector backbone (42) to create pCHS (5). The rppA-based reporter plasmid pEK60 was made by taking the DNA fragment containing rppA and the strong terminator sequences tfd and to from pCHS and cloning them into pGP9 expression system from which ActII-ORF4/PactI had been removed. The PrapA promoter region (Fig. 1) was cloned in front of rppA in pEK60 to obtain pEK61. As control promoters, ActII-ORF4/PactI (pEK67) and ErmE* (pEK68) were also introduced into pEK60. All pEK60-based plasmids were introduced into both the S. hygroscopicus wild-type strain and the S. hygroscopicus ΔHG deletion strain by conjugation. Cultures were grown as described above for rapamycin production to study the expression of rppA. The resultant red pigment produced by the rppA gene product was analyzed by absorbance at 488 nm using a light spectrophotometer.
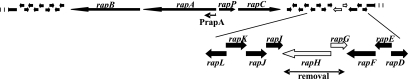
FIG. 1.
Rapamycin biosynthetic gene cluster, showing the location and the area of deletion of the rapH and rapG genes. Promoter PrapA, which controls the main PKS operon rapA-rapB, is also shown. To the right side of the rapA-rapB operon lie genes encoding RapP, a pipecolate-incorporating enzyme, and RapC, which together with RapA and RapB represent the rapamycin PKS.
RESULTS
To study the regulation of rapamycin biosynthesis, a standard gene deletion/complementation and overexpression approach was employed. The production of rapamycin in the wild-type strain was compared to that in various strains in which rapH and/or rapG was overexpressed or deleted from the chromosome (to produce S. hygroscopicus ΔHG strain) or was evaluated by partial or complete complementation of the ΔHG strain (Fig. 1; Table 1).
TABLE 1.
Strains used in this study
| Straina | Plasmid | Promoter | Gene(s) |
|---|---|---|---|
| wt | Native | ||
| wt pGP9 | pGP9 | ActII-ORF4/PactI | |
| wt rapH | pEK28 | ActII-ORF4/PactI | rapH |
| wt rapG | pEK30 | ActII-ORF4/PactI | rapG |
| wt rapHG-np | pEK49 | Native | rapH rapG |
| wt rapHG | pEK48 | ActII-ORF4/PactI | rapH rapG |
| ΔHG | ΔrapH ΔrapG | ||
| ΔHG rapH | pEK28 | ActII-ORF4/PactI | rapH |
| ΔHG rapG | pEK30 | ActII-ORF4/PactI | rapG |
| ΔHG rapHG-np | pEK49 | Native | rapH rapG |
| ΔHG rapHG | pEK48 | ActII-ORF4/PactI | rapH rapG |
| wt rppA-PrapA | pEK61 | PrapA | rppA |
| ΔHG rppA-PrapA | pEK61 | PrapA | rppA |
| wt rppA-PactI | pEK67 | ActII-ORF4/PactI | rppA |
| wt rppA-ErmE* | pEK68 | ErmE* | rppA |
wt, wild-type S. hygroscopicus NRRL 5491; ΔHG, deletion strain in which rapH and rapG were removed. np, gene under control of the native promoter.
The ActII-ORF4/PactI activator/promoter expression system was used in this study, as it is a robust activator/promoter expression system widely used for many actinomycetes (25). It has also been used as a reliable and well established activator/promoter expression system for S. hygroscopicus (19). The introduction of rapH and rapG into this artificial setting aimed to minimize the potential self-regulatory interference of RapH and/or RapG and other endogenous regulatory genes of the pathway, namely, rapY, rapR, rapS, and rapK. The reporter gene rppA of the chalcone synthase gene cluster, the product of which produces a red pigment, was used to evaluate the influence of rapH and rapG on the PrapA promoter activity to gain insight into the expression of the PKS genes rapA and rapB, which encode the major part of the rapamycin PKS (Fig. 1, Table 1).
Introduction of an additional copy of rapH and/or rapG genes under the control of the activator/promoter expression system ActII-ORF4/PactI.
To investigate the function of rapH and/or rapG, the constructs pEK28 (rapH), pEK30 (rapG), and pEK48 (rapG rapH), based on the integrative expression plasmid pGP9 derived from pSET152, were prepared as described in Materials and Methods. The integration function was φBT1 based (20), which was shown to be neutral both in previous studies (18) and in our control (Table 2). The expression of the genes of interest was controlled by the PactI promoter and its cognate activator ActII-ORF4 from Streptomyces coelicolor (4). The constructs were conjugated into S. hygroscopicus NRRL 5491, generating a second in trans copy of the rapH and/or rapG gene. The S. hygroscopicus NRRL 5491 wild-type strain with and without the integrated expression plasmid pGP9 was used as controls (Table 2).
TABLE 2.
Expression data obtained in the study
| Straina | Rapamycin expression
|
Red pigment production
|
||
|---|---|---|---|---|
| Avg % change (SD)b | No. of isolates/no. of expts | Avg rppA optical density at 488 nm (SD) | No. of isolates/no. of expts | |
| wt | 100 (2.3-8.1) | 21/5 | 0.013 (0.02) | 4/1 |
| wt pGP9 | 104 (18) | 5/1 | ||
| wt rapH | 127-155 (1.6-9.3) | 23/3 | ||
| wt rapG | 120-132 (2.4-9.3) | 12/2 | ||
| wt rapHG-np | 140 (9.3) | 3/1 | ||
| wt rapHG | 102 (8.1) | 4/1 | ||
| ΔHG | 0 (0) | 7/3 | ||
| ΔHG rapH | 0 (0) | 16/2 | ||
| ΔHG rapG | 6-13 (2.1-5.1) | 18/3 | ||
| ΔHG rapHG-np | 72-93 (2.7-5.7) | 18/3 | ||
| ΔHG rapHG | 3-16 (1.9-3.3) | 12/2 | ||
| wt rppA-PrapA | 1.244 (0.3) | 9/1 | ||
| ΔHG rppA-PrapA | 0.516 (0.1) | 9/1 | ||
| wt rppA-PactI | 0.246 (0.06) | 10/1 | ||
| wt rppA-ErmE* | 1.766 (0.26) | 6/1 | ||
wt, wild-type S. hygroscopicus NRRL 5491; ΔHG, deletion strain in which rapH and rapG were removed. np, gene under the control of the native promoter.
Each isolate was run in triplicate. Values were calculated with SAS/STAT, using means and the univariate procedure to test the normality of distribution. Using the GLM model, data were calculated as least mean square and are presented as an average change observed from all experiments when comparing least mean square values to the wild-type control least mean square value of each experiment.
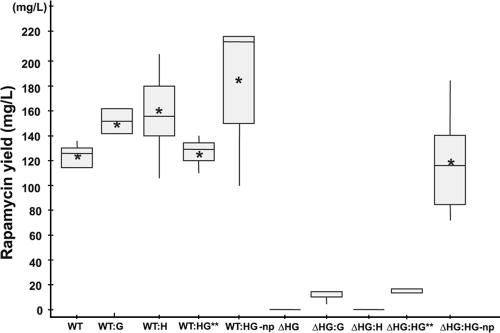
The additional copy of the rapH gene under the control of the ActII-ORF4/PactI promoter increased the production of rapamycin by between 27% and 55% (Table 2; Fig. 2 and 3). When rapG was expressed similarly, the production of rapamycin increased by between 20% and 32% (Table 2; Fig. 2). No difference in the production levels was detected between the S. hygroscopicus NRRL 5491 wild-type strain and the control transformed with pGP9, confirming the neutrality of the integration site (Table 2). These data also suggest that there is no detectable influence of the ActII-ORF4/PactI activator/promoter system alone on rapamycin production (Table 2).
FIG. 2.
Variability of rapamycin yields in S. hygroscopicus strains within a single fermentation run. **, data for WT:HG and ΔHG:HG were obtained from a separate fermentation run and were normalized against the wild-type control (WT). Bars encompass 95% of the sample population, with asterisks representing the mean values, horizontal line representing the median values, and perpendicular lines indicating outliers and extreme values. The data were analyzed using the SAS/STAT program. np, genes under control of the native promoters.
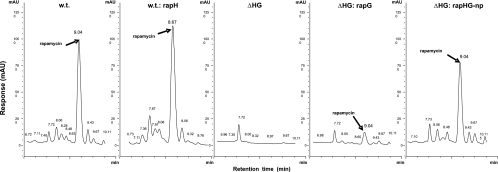
FIG. 3.
High-pressure liquid chromatography analysis of rapamycin (retention time, ca. 8.9 to 9.0 min). Wild-type (w.t.), rapH overexpression (w.t.:rapH), rapH rapG deletion (ΔHG), and partial (ΔHG:rapG) and full (ΔHG:rapHG-np) complementation strains are shown. All rapamycin peaks were verified against a rapamycin standard.
Introduction of an additional copy of rapH and rapG genes under control of the native promoter regions.
In the rapamycin cluster, rapH and rapG are linked by a bidirectional promoter sequence (Fig. 1). A PCR fragment containing rapH and rapG, including their native promoter sequence, was cloned into pGP9 from which the ActII-ORF4/PactI promoter was previously removed, as described in Materials and Methods. Both genes were therefore expressed under the control of their native promoters. The resulting plasmid, pEK49, was introduced by conjugation into the S. hygroscopicus NRRL 5491 wild-type strain, thus generating an additional in trans copy of the rapH and rapG genes expressed under the control of their natural promoter region. Exconjugants were tested, and an average increase in rapamycin production of 40% compared to that in the wild-type strain was observed (Table 2; Fig. 2).
Deletion of the rapG and rapH genes from the rapamycin biosynthetic gene cluster of S. hygroscopicus.
A deletion of rapH and rapG was introduced into the genome of S. hygroscopicus as described in Materials and Methods, and S. hygroscopicus ΔHG was isolated. The deletion comprising both genes in their entirety, including their native promoters (Fig. 1), was confirmed by PCR and Southern blot analysis. S. hygroscopicus ΔHG was tested, and no production of rapamycin was detected (Fig. 3), indicating that the expression of one or both genes is essential for rapamycin biosynthesis.
Complementation of S. hygroscopicus ΔHG by the pEK49 plasmid containing both rapH and rapG expressed from their native promoters.
Complementation of S. hygroscopicus ΔHG with plasmid pEK49 containing both genes under the control of their native promoters was assessed, and, as expected, restoration of rapamycin production to wild-type levels was detected (Table 2; Fig. 2 and 3).
Complementation of S. hygroscopicus ΔHG by rapH and rapG genes expressed under control of the ActII-ORF4/PactI activator/promoter system.
Complementation of deleted genes was carried out by reintroduction of each of the genes, rapH and rapG, separately and together using the integrative pSET152-based plasmids pEK28 (rapH), pEK30 (rapG), and pEK48 (rapH rapG) under the control of the promoter ActII-ORF4/PactI. Clones of S. hygroscopicus ΔHG conjugated with pEK28, pEK30, and pEK48 were tested for rapamycin production. No rapamycin could be detected in clones of S. hygroscopicus ΔHG complemented with rapH (Table 2; Fig. 2). A very small amount of rapamycin was detected in clones of S. hygroscopicus ΔHG complemented with rapG. Although the yield of rapamycin was only 6 to 13% compared to the yield of the wild-type strain (Table 2; Fig. 2 and 3), the compound made was fully processed rapamycin, which was confirmed by liquid chromatography-mass spectrometry. Complementation of S. hygroscopicus ΔHG by pEK48 (rapH rapG under the control of ActII-ORF4/PactI) gave titers equivalent to those after complementation with rapG alone (3 to 16%) (Table 2; Fig. 2). Analogously, introduction of an additional copy of rapH and rapG in trans, under the control of ActII-ORF4/PactI, into the wild-type strain gave no noticeable increase in titers (2% ± 8.1% [standard deviation]) (Table 2) compared to the experiment in which rapH and rapG were expressed in trans under control of the native promoters. This piece of data was unexpected, since the introduction of pEK30 (rapG under control of ActII-ORF4/PactI) into the wild-type strain gave improved rapamycin titers of 20 to 32% above the wild-type titer (Table 2). A complex interplay between RapG and RapH and co-/self-regulation involving the native bidirectional promoter sequence appear to be required to restore the full wild-type yield of rapamycin.
Influence of the rapH and rapG genes on expression of rapamycin PKS genes.
To evaluate the possible role of rapH and rapG in the expression of the polyketide synthase genes rapA and rapB, which are involved in the biosynthesis of the polyketide backbone of rapamycin (Fig. 1), a reporter system was employed that was based on the chalcone synthase (a type III PKS) gene rppA, which is responsible for production of the dark-red pigment flaviolin (5, 11). Plasmid pEK61 was constructed, in which the PrapA promoter from the PKS rapA-rapB operon was cloned in front of the rppA gene of the reporter plasmid pEK60. In addition, promoter control plasmids pEK67 and pEK68, containing the ActII-ORF4/PactI and ErmE* promoters, respectively, in front of the rppA gene, were also introduced into the S. hygroscopicus wild-type strain by conjugation. When inserted and expressed in the wild-type strain, good expression of rppA under the control of PrapA and production of red pigment were observed (Table 2; Fig. 4) and were only slightly lower than the values with the control ErmE* promoter, which is considered in the literature to be a strong Streptomyces promoter (46). When the same construct was inserted and expressed in the rapH rapG deletion strain, a reduction of ∼59% in red pigment production was observed compared to the wild-type strain (Table 2; Fig. 4). The deletion of rapH and rapG clearly results in a down regulation of the rapA-rapB operon, but this does not fully explain the total loss of rapamycin production in the deletion strain. This suggests that deletion of rapH and rapG may also be down regulating other essential components of the pathway or precursor supply.
FIG. 4.
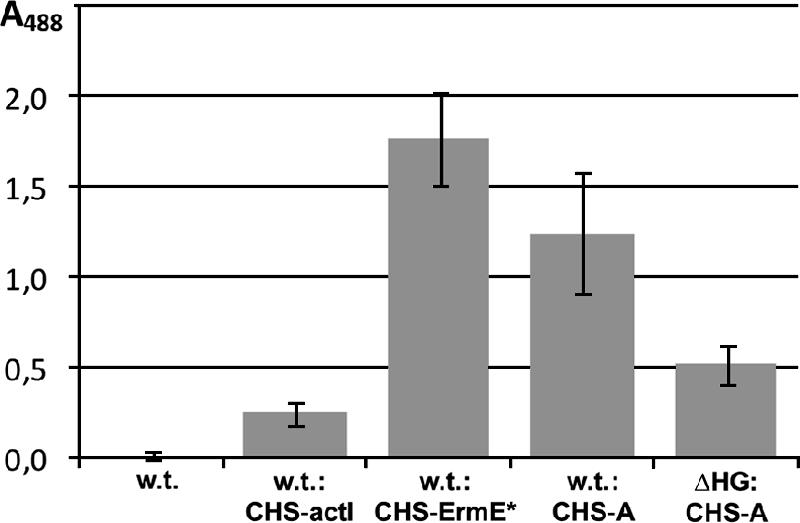
Chalcone synthase (CHS) reporter gene expression. The RppA-PrapA plasmid (A) contains the PrapA promoter in front of the rppA gene. Error bars indicate standard deviations.
DISCUSSION
The original sequence homology studies of the rapamycin gene cluster identified a number of ORFs with putative regulatory functions, suggesting that the biosynthesis of rapamycin is highly regulated (30). This is not surprising due to the complex biosynthesis of rapamycin, which involves an unusual shikimate-derived starter unit, three large PKS synthase proteins, incorporation of the amino acid l-pipecolate, and a number of post-PKS tailoring steps. It is reasonable to expect that a high degree of regulation is required to ensure the coordination of the PKS and post-PKS biosynthetic steps, in addition to the supply of the necessary building blocks from primary metabolism. Putative regulatory genes present in the cluster, such as rapR, rapS, rapY, and rapK, in addition to rapH and rapG, may have a direct or indirect influence on these aspects of biosynthesis. The complexity of regulatory systems would also suggest that one or more regulatory gene products may be dependent on and/or act in concert with another to affect a positive or negative regulatory role, e.g., as observed for tylosin (41) and nystatin (39) biosynthesis. The work presented here represents a first step in exploring the complexity of the regulation of rapamycin biosynthesis. Based on sequence similarity studies, rapH and rapG are likely candidates to have positive regulatory functions. We focused our initial study on these two genes, as their use might prove to be an important tool to increase the production yield of rapamycin and its analogues.
Overexpression studies with an additional copy of rapH and/or rapG expressed under the control of the ActII-ORF4/PactI activator/promoter expression system in wild-type S. hygroscopicus clearly demonstrated a strong positive effect of each of the genes on rapamycin production. We were also able to show that the genomic deletion of rapH and rapG in wild-type S. hygroscopicus eliminated rapamycin production, indicating an essential role of one or both genes in regulation of rapamycin biosynthesis. As expected, complementation with the fragment that had been deleted restored the production of rapamycin to wild-type levels (Table 2; Fig. 2).
Indeed, sequence analysis studies revealed that RapH (882 amino acids) contained an HTH DNA-binding motif near the C terminus and an ATP-binding site at the N terminus (30). Since the original assessment of the rapamycin gene cluster in 1996, a new family of transcriptional activators has been proposed. The LAL family (large ATP-binding regulators of the LuxR family) is characterized by a LuxR-type pattern of an HTH DNA-binding motif of about 65 amino acids at the C terminus (14) and a distinctive P loop at the N-terminal end that is specific for an ATP-binding site. The necessity for ATP binding and hydrolysis was demonstrated by work on PikD, a LAL regulator required for pikromycin biosynthesis. The ability of PikD to act as a transcriptional activator was dependent on the presence of a functional ATP-binding motif (47). RapH fits the profile of the LAL family and has a high sequence identity with a number of Streptomyces polyketide antibiotic biosynthetic cluster regulators of the LAL family, three of which have been identified in S. hygroscopicus strains (Table 3). The most extensively studied member of the LAL family of regulators is the Escherichia coli MalT protein, which is involved in the positive regulation of maltotriose uptake (ABC transporter) and catabolism (15). The transcription of this regulator is itself under direct catabolite repression control (9). MalT is activated by the binding of the inducer maltotriose, and this activation is modulated by interaction with other proteins of the maltose operon which prevent binding of the inducer (6). All regulators in this subclass have been found to be positive regulators.
TABLE 3.
Proteins similar to RapH and RapG
| Proteina | % Identity | % Similarity | System regulated | Organism | Reference |
|---|---|---|---|---|---|
| RapH homologues | |||||
| FkbN | 59 | 71 | FK520 PKS | S. hygroscopicus subsp. ascomyceticus | 48 |
| PikD | 33 | 45 | Pikromycin PKS | Streptomyces venezuelae | 47 |
| GdmRII | 36 | 49 | Geldanamycin PKS | S. hygroscopicus NRRL 3602 | 35 |
| HbmRII | 36 | 49 | Herbimycin PKS | S. hygroscopicus AM-3672 | 34 |
| NbmM | 33 | 45 | Desosamine biosynthesis | Streptomyces narbonensis | 7 |
| MonH | 34 | 44 | Monensin PKS | Streptomyces cinnamonensis | 31 |
| RapG homologues | |||||
| SCO7780 | 50 | 61 | Unknown | Streptomyces coelicolor A3(2) | 2 |
| NitR | 33 | 50 | Nitrilase | Rhodococcus rhodochrous | 27 |
| FasR | 34 | 58 | Leafy gall formation | Rhodococcus fascians | 43 |
| FeaR | 29 | 50 | 2-Phenylethylamine catabolism | Escherichia coli | 21 |
All of the identified proteins similar to RapH belong to the LAL family of Streptomyces regulators, with their characteristic LuxR-type pattern of an HTH DNA-binding motif at the C terminus and a P loop distinctive of an ATP-binding site at the N terminus, while all of the identified proteins similar to RapG contain the AraC-like HTH DNA-binding motif at the C-terminal region.
The sequence analysis of RapG (330 amino acids) also revealed an HTH DNA-binding motif with 38% sequence identity to the SoxS positive regulator of the superoxide response regulon from E. coli over a stretch of 100 amino acids with the predicted HTH motif (30). RapG was also found to be highly similar to a number of putative regulatory proteins which all share a signature pattern in the 100- to 150-amino-acid C-terminal region encompassing the HTH DNA-binding motif of the AraC family of transcriptional regulators (29). The N-terminal and central regions of members of the AraC family of proteins are believed to interact with effector molecules and potentially are involved in dimerization (37). However, RapG lacks any sequence similarity with other AraC family members in this region. Most AraC proteins have been found to contain both a C-terminal DNA-binding domain and an N-terminal dimerization and/or ligand-binding domain; however, several members of the AraC family have been shown to contain only the aforementioned DNA-binding domain (33), which may be the case in RapG. Some AraC proteins use DNA looping to repress transcription. Binding of the ligand to AraC protein dimers converts the AraC protein from a repressor to a transcriptional activator by changing the way DNA loops between AraC binding sites. Although not all AraC family members use DNA looping, the presence or absence of a bound ligand may still modify the function of the regulator (33). Not only have AraC family regulators been shown to bind small molecules as ligands, but some also interact directly with other proteins in order to become activated as transcriptional activators (13). RapG lacks any apparent ligand-binding domains; however, the data of this study suggest that it would be interesting to explore a possibility of DNA binding of either of the two proteins to the rapH-rapG promoter region and also to look into protein-protein interactions to investigate whether there may be some sort of interaction occurring between RapH and RapG.
The complex interplay between RapH and RapG in their regulatory role is observed through the complementation studies with S. hygroscopicus ΔHG. Complementation with rapH alone did not give rise to a restoration of rapamycin production (Table 2; Fig. 2). Complementation with rapG alone partially restored production of low levels of fully processed rapamycin, indicating that rapG may have a direct regulatory role in the initiation of rapamycin biosynthesis. Wild-type yields, however, were fully restored only when, rapG, rapH, and an intact native bidirectional promoter sequence were all present, suggesting that this interplay is mediated via promoter interactions of one or both gene products. In the absence of the native bidirectional promoter, but with ActII-ORF4/PactI instead, in either the wild-type or the deletion strains (S. hygroscopicus ΔHG with pEK48 and S. hygroscopicus NRRL 5491 with pEK48) (Tables 1 and 2), only a marginal increase in titer above that of their respective noncomplemented controls was observed. The presence of a rapamycin titer in S. hygroscopicus ΔHG with pEK48 that was of a similar level as in S. hygroscopicus ΔHG with pEK30 would indicate the proper activity of expression of rapH rapG in pEK48, as the rapG gene is the second gene in the cassette and is transcribed only after rapH. Titration by RapH and/or RapG of some other key factor of regulation may provide one explanation. The relative levels of free and promoter-bound RapH or RapG may also influence co-/self-activation of these proteins. Further studies will have to be carried out in order to explain the role of native promoters.
Additional evidence for a direct effect of these positive regulators on rapamycin biosynthesis was observed through the study of the PrapA promoter of the rapamycin PKS operon rapA-rapB by using a chalcone synthase reporter system. In the S. hygroscopicus ΔHG strain, a substantial drop, but not a complete loss, of the PrapA promoter activity was observed (Fig. 4). Deletion of rapH and rapG, however, eliminates rapamycin production entirely. RapH and RapG must be acting at one or more additional promoter sequences within the biosynthetic cluster, either directly or indirectly through additional regulatory components.
RapH and RapG clearly have a positive regulatory role in rapamycin biosynthesis. The data presented here also indicate a possible direct involvement of the rapG and/or rapH gene products in the regulation of rapamycin PKS gene expression. A genetically engineered producer strain which showed increased levels of rapamycin production after overexpression of the rapH and rapG genes indicates the potential of this approach for strain improvement. This work is the first step in deciphering the regulatory factors involved in the biosynthesis of rapamycin.
Acknowledgments
We thank the Government of Slovenia, Ministry of Science and Technology (Slovenian Research Agency, ARRS), for the award of studentship 3311-02-831487 to Enej Kuščer.
We thank Peter Raspor, Sabine Gassier, and Steve Kendrew for their guidance, helpful discussions, and critical reading of the manuscript. We are also grateful to Gerard Peck for the provision of pGP9.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Bate, N., G. Stratigopoulos, and E. Cundliffe. 2002. Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol. Microbiol. 43:449-458. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, M. J. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. [DOI] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. Nagaraja Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 5.Boakes, S., M. Oliynyk, J. Cortes, I. Bohm, B. A. Rudd, W. P. Revill, J. Staunton, and P. F. Leadlay. 2004. A new modular polyketide synthase in the erythromycin producer Saccharopolyspora erythraea. J. Mol. Microbiol. Biotechnol. 8:73-80. [DOI] [PubMed] [Google Scholar]
- 6.Bohm, A., J. Diez, K. Diederichs, W. Welte, and W. Boos. 2002. Structural model of MalK, the ABC subunit of the maltose transporter of Escherichia coli: implications for mal gene regulation, inducer exclusion, and subunit assembly. J. Biol. Chem. 277:3708-3717. [DOI] [PubMed] [Google Scholar]
- 7.Butler, A. R., N. Bate, D. E. Kiehl, H. A. Kirst, and E. Cundliffe. 2002. Genetic engineering of aminodeoxyhexose biosynthesis in Streptomyces fradiae. Nat. Biotechnol. 20:713-716. [DOI] [PubMed] [Google Scholar]
- 8.Champness, W., P. Riggle, T. Adamidis, and P. Vandervere. 1992. Identification of Streptomyces coelicolor genes involved in regulation of antibiotic synthesis. Gene 115:55-60. [DOI] [PubMed] [Google Scholar]
- 9.Chapon, C., and A. Kolb. 1983. Action of CAP on the malT promoter in vitro. J. Bacteriol. 156:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang, S. J. 2004. Strain improvement for fermentation and biocatalysis processes by genetic engineering technology. J. Ind. Microbiol. Biotechnol. 31:99-108. [DOI] [PubMed] [Google Scholar]
- 11.Cortés, J., J. Velasco, G. Foster, A. P. Blackaby, B. A. Rudd, and B. Wilkinson. 2002. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol. Microbiol. 44:1213-1224. [DOI] [PubMed] [Google Scholar]
- 12.Cundliffe, E. 1999. Organization and control of the tylosin-biosynthetic genes of Streptomyces fradiae. Actinomycetologica 13:68-75. [Google Scholar]
- 13.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Schrijver, A., and R. De Mot. 1999. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 145:1287-1288. [DOI] [PubMed] [Google Scholar]
- 15.Ehrmann, M., R. Ehrle, E. Hofmann, W. Boos, and A. Schlosser. 1998. The ABC maltose transporter. Mol. Microbiol. 29:685-694. [DOI] [PubMed] [Google Scholar]
- 16.Gaisser, S., R. Lill, J. Staunton, C. Méndez, J. Salas, and P. F. Leadlay. 2002. Parallel pathways for oxidation of 14-membered polyketide macrolactones in Saccharopolyspora erythraea. Mol. Microbiol. 44:771-781. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, M. A., S. Gaisser, R. E. Lill, H. Hong, R. M. Sheridan, B. Wilkinson, H. Petkovic, A. J. Weston, I. Carletti, H. L. Lee, J. Staunton, and P. F. Leadlay. 2004. Isolation and characterization of pre-rapamycin, the first macrocyclic intermediate in the biosynthesis of the immunosuppressant rapamycin by S. hygroscopicus. Angew. Chem. Int. 43:2551-2553. [DOI] [PubMed] [Google Scholar]
- 18.Gregory, M. A., H. Hong, R. E. Lill, S. Gaisser, H. Petkovic, L. Low, L. S. Sheehan, I. Carletti, S. J. Ready, M. J. Ward, A. L. Kaja, A. J. Weston, I. R. Challis, P. F. Leadlay, C. J. Martin, B. Wilkinson, and R. M. Sheridan. 2006. Rapamycin biosynthesis: elucidation of gene product function. Org. Biomol. Chem. 4:3565-3568. [DOI] [PubMed] [Google Scholar]
- 19.Gregory, M. A., H. Petkovic, R. E. Lill, S. J. Moss, B. Wilkinson, S. Gaisser, P. F. Leadlay, and R. M. Sheridan. 2005. Mutasynthesis of rapamycin analogues through the manipulation of a gene governing starter unit biosynthesis. Angew. Chem. Int. 44:4757-4760. [DOI] [PubMed] [Google Scholar]
- 20.Gregory, M. A., R. Till, and M. C. Smith. 2003. Integration site for Streptomyces phage φBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanlon, S. P., T. K. Hill, M. A. Flavell, J. M. Stringfellow, and R. A. Cooper. 1997. 2-Phenylethylamine catabolism by Escherichia coli K-12: gene organization and expression. Microbiology 143:513-518. [DOI] [PubMed] [Google Scholar]
- 22.Huang, S., M. A. Bjornsti, and P. J. Houghton. 2003. Rapamycins: mechanism of action and cellular resistance. Cancer Biol. Ther. 2:222-232. [DOI] [PubMed] [Google Scholar]
- 23.Huang, S., and P. J. Houghton. 2001. Mechanisms of resistance to rapamycins. Drug Resist. Update 4:378-391. [DOI] [PubMed] [Google Scholar]
- 24.Khaw, L. E., G. A. Böhm, S. Metcalfe, J. Staunton, and P. F. Leadlay. 1998. Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180:809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 26.Kinoshita, H., T. Tsuji, H. Ipposhi, T. Nihira, and Y. Yamada. 1999. Characterization of binding sequences for butyrolactone autoregulator receptors in streptomycetes. J. Bacteriol. 181:5075-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komeda, H., Y. Hori, M. Kobayashi, and S. Shimizu. 1996. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc. Natl. Acad. Sci. USA 93:10572-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leskiw, B. K., R. Mah, E. J. Lawlor, and K. F. Chater. 1993. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2). J. Bacteriol. 175:1995-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 30.Molnár, I., J. F. Aparicio, S. F. Haydock, L. E. Khaw, T. Schwecke, A. König, J. Staunton, and P. F. Leadlay. 1996. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Oliynyk, M., C. B. Stark, A. Bhatt, M. A. Jones, Z. A. Hughes-Thomas, C. Wilkinson, Z. Oliynyk, Y. Demydchuk, J. Staunton, and P. F. Leadlay. 2003. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol. Microbiol. 49:1179-1190. [DOI] [PubMed] [Google Scholar]
- 32.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plano, G. V. 2004. Modulation of AraC family member activity by protein ligands. Mol. Microbiol. 54:287-290. [DOI] [PubMed] [Google Scholar]
- 34.Rascher, A., Z. Hu, G. O. Buchanan, R. Reid, and C. R. Hutchinson. 2005. Insights into the biosynthesis of the benzoquinone ansamycins geldanamycin and herbimycin, obtained by gene sequencing and disruption. Appl. Environ. Microbiol. 71:4862-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rascher, A., Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis, and C. R. Hutchinson. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 218:223-230. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Saviola, B., R. Seabold, and R. F. Schleif. 1998. Arm-domain interactions in AraC. J. Mol. Biol. 278:539-548. [DOI] [PubMed] [Google Scholar]
- 38.Schwecke, T., J. F. Aparicio, I. Molnár, A. König, L. E. Khaw, S. F. Haydock, M. Oliynyk, P. Caffrey, J. Cortés, J. B. Lester, G. A. Böhm, J. Staunton, and P. F. Leadlay. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 92:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekurova, O. N., T. Brautaset, H. Sletta, S. E. Borgos, M. O. Jakobsen, T. E. Ellingsen, A. R. Strom, S. Valla, and S. B. Zotchev. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186:1345-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirling, E. B., and D. Gottlieb. 1966. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 13:313-340. [Google Scholar]
- 41.Stratigopoulos, G., N. Bate, and E. Cundliffe. 2004. Positive control of tylosin biosynthesis: pivotal role of TylR. Mol. Microbiol. 54:1326-1334. [DOI] [PubMed] [Google Scholar]
- 42.Sun, J. H., G. H. Kelemen, J. M. Fernández-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 43.Temmerman, W., D. Vereecke, R. Dreesen, M. Van Montagu, M. Holsters, and K. Goethals. 2000. Leafy gall formation is controlled by fasR, an AraC-type regulatory gene in Rhodococcus fascians. J. Bacteriol. 182:5832-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignot, S., S. Faivre, D. Aguirre, and E. Raymond. 2005. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann. Oncol. 16:525-537. [DOI] [PubMed] [Google Scholar]
- 45.Wietzorrek, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in Streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson, C. J., Z. A. Hughes-Thomas, C. J. Martin, I. Böhm, T. Mironenko, M. Deacon, M. Wheatcroft, G. Wirtz, J. Staunton, and P. F. Leadlay. 2002. Increasing the efficiency of heterologous promoters in actinomycetes. J. Mol. Microbiol. Biotechnol. 4:417-426. [PubMed] [Google Scholar]
- 47.Wilson, D. J., Y. Q. Xue, K. A. Reynolds, and D. H. Sherman. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183:3468-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, K., L. Chung, W. P. Revill, L. Katz, and C. D. Reeves. 2000. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251:81-90. [DOI] [PubMed] [Google Scholar]