Abstract
In aerobic and facultatively anaerobic bacteria, the degradation of para-cresol (p-cresol) involves the initial hydroxylation to p-hydroxybenzyl alcohol by water catalyzed by the soluble, periplasmatic flavocytochrome p-cresol methylhydroxylase (PCMH; α2β2 composition). In denitrifying bacteria the further metabolism proceeds via oxidation to p-hydroxybenzoate, the formation of p-hydroxybenzoyl-coenzyme A (CoA), and the subsequent dehydroxylation of the latter to benzoyl-CoA by reduction. In contrast, the strictly anaerobic Desulfobacterium cetonicum degrades p-cresol by addition to fumarate, yielding p-hydroxybenzylsuccinate. In this work, in vitro enzyme activity measurements revealed that the obligately anaerobic Geobacter metallireducens uses the p-cresol degradation pathway of denitrifying bacteria. Surprisingly, PCMH, which is supposed to catalyze both p-cresol hydroxylation and p-hydroxybenzyl alcohol oxidation to the corresponding aldehyde, was located in the membrane fraction. The α subunit of the enzyme was present in two isoforms, suggesting an αα′β2 composition. We propose that the unusual asymmetric architecture and the membrane association of PCMH might be important for alternative electron transfer routes to either cytochrome c (in the case of p-cresol oxidation) or to menaquinone (in the case of p-hydroxybenzyl alcohol oxidation). Unusual properties of further enzymes of p-cresol metabolism, p-hydroxybenzoate-CoA ligase, and p-hydroxybenzoyl-CoA reductase were identified and are discussed. A proteomic approach identified a gene cluster comprising most of the putative structural genes for enzymes involved in p-cresol metabolism (pcm genes). Reverse transcription-PCR studies revealed a different regulation of transcription of pcm genes and the corresponding enzyme activities, suggesting the presence of posttranscriptional regulatory elements.
Monomethylphenols (cresols) are produced in large amounts in the petrochemical industry as constituents of resins, solvents, disinfectants, and wood-preserving chemicals. Although para-cresol (p-cresol) can also be formed biologically from tyrosine via p-hydroxyphenylacetate (40), the major part of p-cresol-contaminated waste waters derives from industrial activities. The aromatic compound p-cresol (4-methylphenol) can be degraded by aerobic (19) and anaerobic bacteria (9, 24, 27, 32, 34, 36).
The metabolism of p-cresol was originally studied in the aerobic Pseudomonas putida. In this organism, p-cresol is hydroxylated by a periplasmatic p-cresol methylhydroxylase (PCMH) to p-hydroxybenzaldehyde with the transient formation of p-hydroxybenzyl alcohol (21). The enzyme consists of two subunits in an α2β2 composition: an active-site α subunit containing flavin adenine dinucleotide (FAD) covalently linked to tyrosine and a c-type cytochrome β subunit (29, 31). In Pseudomonas strains, electron equivalents generated by p-cresol oxidation are transferred to azurin (12). Meanwhile, many details about the structure, function, and mechanism of PCMH from Pseudomonas putida (for examples, see references 13, 14, and 22) and a denitrifying bacterium (20) have been elucidated. The product of PCMH, p-hydroxybenzaldehyde, becomes oxidized by a specific NAD+- or NADP+-dependent dehydrogenase to p-hydroxybenzoate.
The further metabolism of p-hydroxybenzoate differs in aerobic and denitrifying bacteria. In aerobic bacteria, p-hydroxybenzoate becomes hydroxylated by an oxygenase to protocatechuate (3,4-dihydroxybenzoate), which serves as substrate for dioxygenases (16). In contrast, denitrifying bacteria activate p-hydroxybenzoate first to p-hydroxybenzoyl-coenzyme A (CoA) by a specific ligase (4, 17), which becomes subsequently dehydroxylated by reduction to benzoyl-CoA and water. The latter reaction is catalyzed by the Mo-containing Fe/S-flavoprotein p-hydroxybenzoyl-CoA reductase (dehydroxylating). This enzyme has so far only been purified and characterized from the denitrifying bacterium Thauera aromatica (10). It consists of three subunits of 85, 35, and 17 kDa (α2β2γ2 composition) and belongs to the xanthine oxidase family of molybdenum cofactor-containing hydroxylases (for recent reviews see references 6 to 8). The resolution of the structure enabled insights into the unusual inverted electron transfer of this enzyme (38). The product of the reaction, benzoyl-CoA, represents a central intermediate of anaerobic aromatic metabolism and is dearomatized by an ATP-dependent benzoyl-CoA reductase in facultatively anaerobic bacteria (for recent reviews see references 5 and 6).
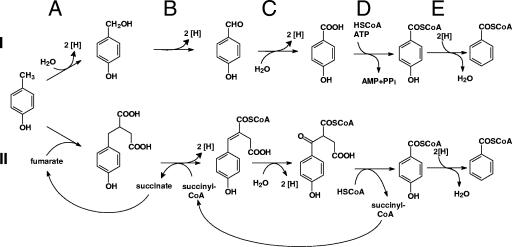
Much less is known about the metabolism of p-hydroxylated aromatic compounds in strictly anaerobic bacteria. In sulfate-reducing bacteria, two different pathways for p-cresol oxidation have been described (Fig. 1). In the gram-positive Desulfotomaculum sp. strain Groll, p-cresol appears to be oxidized by methylhydroxylation as in aerobic and facultatively anaerobic bacteria (24). This finding was not expected, as the redox potential of the electron-transferring heme group of the PCMH β subunit from facultative anaerobes appears to be too high (E°′ [standard redox potential] = +234 mV [15]) for electron transfer to the terminal electron acceptor adenosine-5′-phosphosulfate (E°′ = −60 mV) or sulfite (E°′ = −116 mV) of sulfate-reducing bacteria (37). In contrast, evidence for a different mechanism has been provided in the gram-negative Desulfobacterium cetonicum. In this organism, p-cresol is added to fumarate, yielding p-hydroxybenzylsuccinate, which is subsequently converted in a series of β-oxidation-like reactions to p-hydroxybenzoyl-CoA (32). The latter intermediate is then further metabolized by p-hydroxybenzoyl-CoA reductase to the central intermediate benzoyl-CoA (Fig. 1).
FIG. 1.
Pathways of p-cresol metabolism to benzoyl-CoA in anaerobic bacteria. I. Oxidation of p-cresol via methyl group hydroxylation. The reactions shown are catalyzed by (A and B) p-cresol methylhydroxylase, (C) p-hydroxybenzaldehyde dehydrogenase, (D) p-hydroxybenzoate-CoA ligase, and (E) p-hydroxybenzoyl-CoA reductase. II. Oxidation of p-cresol via addition to fumarate. The reactions shown are catalyzed by (A) p-hydroxybenzylsuccinate synthase, (B) succinyl-CoA:p-hydroxybenzylsuccinate-CoA transferase and p-hydroxybenzylsuccinyl-CoA dehydrogenase, (C) p-hydroxyphenylitaconyl-CoA hydratase and 3-hydroxyenoyl-CoA dehydrogenase, (D) benzoylsuccinyl-CoA thiolase, and (E) p-hydroxybenzoyl-CoA reductase.
To further elucidate whether p-cresol degradation in strict anaerobes is initiated by p-hydroxybenzylsuccinate formation or p-cresol methylhydroxylation, we carried out studies with the iron-reducing Geobacter metallireducens. This organism is able to grow on aromatic compounds such as benzoate, toluene, phenol, p-cresol, and p-hydroxybenzoate using Fe(III) as the terminal electron acceptor (25-28). The aim of this study was to elucidate whether p-cresol metabolism proceeds via methylhydroxylation (aerobic and facultatively anaerobic bacteria) or via p-hydroxybenzylsuccinate formation (at least some strictly anaerobic bacteria). The results obtained suggest that in the strictly anaerobic G. metallireducens, p-cresol metabolism is initiated by methylhydroxylation, as in aerobic and facultatively anaerobic bacteria; however, this activity was surprisingly found in the membrane fraction of cell extracts. Several further properties of the enzymes involved in p-cresol metabolism of G. metallireducens clearly differ from those in aerobic and facultatively anaerobic bacteria.
MATERIALS AND METHODS
Growth of bacterial cells and preparation of cell extracts.
G. metallireducens (Deutsche Sammlung von Mikoorgansimen; DSMZ no. 7210) was grown in a anaerobic batch culture in a mineral salt medium (28). Benzoate, p-hydroxybenzoate (3 mM each), acetate (30 mM), butyrate (10 mM), and p-cresol (1 mM), in addition to Fe(III)-citrate (60 mM), were used as carbon and energy sources. Larger cell amounts were obtained by culturing G. metallireducens in a 200-liter fermenter. The growth was monitored by cell counting by means of a Neubauer counting chamber. Cells were anaerobically harvested in the exponential growth phase by centrifugation (14,000 × g) and were stored in liquid nitrogen.
For the preparation of crude extracts, frozen cells were anaerobically suspended in 50 mM potassium phosphate buffer, pH 7.0 (1 g cells, wet mass, in 1.5 ml buffer), 0.1 mg of DNase I, and 10 mg dithioerythritol (40 mM) per liter. Cell extracts were obtained by passage through a French pressure cell at 137 MPa. After centrifugation at 100,000 × g (1 h, 4°C), the supernatant was immediately used for further studies (see below) or stored at −20°C. For the preparation of membrane fractions, cell lysates were obtained as described above. After passage through the French pressure cell and centrifugation at 6,000 × g (30 min, 4°C), the supernatant was centrifuged at 100,000 × g (1 h, 4°C). The pellet was washed twice by resuspension in the buffer described above and centrifugation at 100,000 × g (1 h, 4°C). The resulting pellet was resuspended in the same buffer and homogenized by pottering.
Enzyme assays.
Enzyme activity determinations were generally repeated at least three times in extracts from two different cell batches. The mean values and standard deviations are indicated.
p-Cresol methylhydroxylase/p-hydroxybenzyl alcohol-oxidizing activity.
Consumption of p-cresol and/or p-hydroxybenzyl alcohol and subsequent product formation by PCMH was monitored by high-performance liquid chromatography (HPLC). The 500-μl assay mixture (30°C) contained 50 mM Tris-HCl buffer, pH 7.6, 0.3 mM phenazine methosulfate (PMS), and 0.5 mM p-cresol or p-hydroxybenzyl alcohol. The reaction was started by addition of cell extract or membrane fractions (approximately 150 μg protein) of G. metallireducens cells grown on different substrates (p-cresol, p-hydroxybenzoate, benzoate, and acetate), and 100-μl samples were retrieved at several time points, after which the protein was precipitated with 10 μl 20% formic acid, followed by centrifugation at 10,000 × g for 10 min. The supernatant was applied to a Lichrospher 100 RP-C-18 HPLC column (125 by 4 mm; Merck, Darmstadt, Germany) at a flow rate of 1 ml min−1. HPLC separation of the reaction mixture was performed with a linear 10 to 60% methanol gradient formed from methanol and 40 mM aqueous formic acid buffer, pH 2.3. Detection of compounds was at 275 nm and 270 nm. p-Cresol, p-hydroxybenzyl alcohol, p-hydroxybenzaldehyde, and p-hydroxybenzoate eluted at 14.2 min, 5 min, 9 min, and 8.1 min, respectively.
p-Hydroxybenzaldehyde dehydrogenase.
The dehydrogenase activity forming p-hydroxybenzoate from p-hydroxybenzaldehyde was measured at 30°C in a discontinuous assay with subsequent HPLC analysis, as described above for PCMH activity. Alternatively, the same assay mixture was used in a continuous spectrophotometric assay following NADPH formation at 365 nm (ɛ365 = 3.4 mM−1 cm−1).
p-Hydroxybenzoate-CoA/benzoate-CoA ligase.
p-Hydroxybenzoate-CoA ligase activity could not be assayed by the typical spectrophotometric assay as described due to high levels of background reactions (4). Instead, enzyme activity was determined at 30°C in a discontinuous assay containing 150 mM morpholinepropanesulfonic acid buffer, pH 7.3, 10 mM MgCl2, 0.5 mM sodium p-hydroxybenzoate or sodium benzoate, 0.5 mM CoA, and 5 mM ATP in 125 μl. The reaction was started by adding 50 μl enzyme fraction (total volume, 175 μl). Fifty-microliter aliquots were taken at 0 and 10 min. The reaction was stopped with 7 μl 10% formic acid, vortexed, and centrifuged in an Eppendorf centrifuge for 15 min at 4°C. From the supernatant, 20 μl was diluted with distilled water to a final volume of 100 μl and applied to HPLC analysis. For this purpose a Waters 2695 chromatography system with a Waters 2996 photodiode array detector was used; separation was by a Eurospher RP C18 column (Knauer, Berlin) with gradients of acetonitrile in 50 mM aqueous potassium phosphate, pH 6.8, at a flow rate of 0.75 ml min−1. The gradient applied was the following: 0 to 7 min, 10% to 25% acetonitrile; 7 to 8 min, 25 to 40% acetonitrile; 8 to 10 min, 40% acetonitrile; 10 to 11 min, 40 to 10% acetonitrile; 11 to 15 min, 10% acetonitrile. Retention times were 7.2 min for p-hydroxybenzoyl-CoA and 8.8 min for benzoyl-CoA, respectively. Detection was carried out at 260 nm. The amount of CoA esters formed was calculated by comparison of the peak areas with standards of p-hydroxybenzoyl-CoA and benzoyl-CoA. The detection limit was 1 μM CoA ester. Determination of enzyme kinetics was carried out in the same test but was scaled up to a final volume of 350 μl. Time points for kinetic analysis were 0, 0.5, 1, 3, 6, and 12 min. Sample preparation and analysis was identical to that of the smaller assay.
p-Hydroxybenzoyl-CoA reductase.
p-Hydroxybenzoyl-CoA reductase from G. metallireducens could not be measured with the continuous spectrophotometric assay following the substrate-dependent oxidation of reduced benzyl viologen as described previously for the enzyme from Thauera aromatica (10). Instead, a direct assay following product formation was routinely performed under strictly anaerobic conditions at 30°C; all additions to the stoppered vials were made with gas-tight syringes. In this assay, the substrate p-hydroxybenzoyl-CoA was formed from p-hydroxybenzoate and CoA by enriched p-hydroxybenzoate-CoA ligase from T. aromatica in the presence of MgATP (volume activity, 13 μmol min−1 ml−1). Enrichment of the ligase included the first two purification steps (ammonium sulfate precipitation and DEAE anion exchange chromatography) of the purification protocol as described previously (4). No p-hydroxybenzoyl-CoA reductase activity was determined in the aerobically prepared enzyme fraction. Notably, p-hydroxybenzoyl-CoA reductase from T. aromatica is oxygen sensitive (10). The 500-μl assay mixture for p-hydroxybenzoyl-CoA reductase from G. metallireducens contained 50 mM potassium phosphate buffer, pH 7.0, 5 mM MgCl2, 5 mM ATP, 0.4 mM CoA, 25 μl enriched p-hydroxybenzoate-CoA ligase from T. aromatica (see above), 0.4 mM p-hydroxybenzoate, 5 mM titanium(III)-citrate, and 50 to 150 μl protein fraction. After preincubation for 10 min with enriched p-hydroxybenzoyl-CoA ligase from T. aromatica, the assay was started by addition of cell extract from G. metallireducens. Samples (50 μl) were retrieved at different time points. For analysis of CoA-thiol esters, protein was precipitated and pelleted by addition of 7 μl 10% formic acid and 10 min of centrifugation at 10,000 × g. The supernatant was applied to a Lichrospher 100 RP-C-18 HPLC column (125 by 4 mm; Merck, Darmstadt, Germany) at a flow rate of 1 ml min−1. HPLC separation of the reaction mixture was performed with a linear 5 to 40% acetonitrile gradient formed from acetonitrile and 50 mM potassium phosphate buffer, pH 6.8. Detection of CoA-thioesters was at 260 nm.
Partial purification of p-hydroxybenzoate-CoA ligase. (i) Preparation of cell lysates.
Enrichment of benzoate-CoA ligase was carried out under aerobic conditions and started from 10 g (wet mass) of cells. Frozen cells were suspended in 20 ml basal buffer (20 mM triethanolamine hydrochloride-KOH buffer, pH 7.8, 2.5 mM MgCl2, 10% glycerol) containing 1 mg of DNase I. Cell lysates were obtained by passage through a French press cell at 137 MPa. After centrifugation at 100,000 × g (1 h, 4°C), the supernatant was used for further studies.
(ii) Ammonium sulfate precipitation and dialysis.
The soluble protein fraction obtained after ultracentrifugation was precipitated with a saturated ammonium sulfate solution, pH 7.8, containing 1 mM Na2EDTA, to 33% saturation. After centrifugation (12,000 × g for 15 min), the supernatant was dialyzed overnight against 2 liters of basal buffer.
(iii) DEAE-Sepharose chromatography.
The dialyzed protein solution was applied at a flow rate of 1 ml min−1 to a DEAE-Sepharose column (diameter, 16 mm; volume, 15 ml; Fast Flow; GE Healthcare, Munich, Germany), which had been equilibrated with basal buffer. The column was washed with 30 ml 90 mM KCl in basal buffer. The ligase activity was eluted with a linear gradient of 90 to 200 mM KCl in basal buffer (100 ml). Fractions (5 ml) were collected and tested for p-hydroxybenzoate-CoA ligase activity.
(iv) Affinity chromatography.
The protein fraction containing p-hydroxybenzoate-CoA ligase activity obtained by DEAE-Sepharose chromatography was concentrated to a final volume of 3 ml using Vivaspin centrifugal concentrators (30-kDa molecular size cutoff; Sartorius AG, Göttingen, Germany), diluted with an equal volume of basal buffer, and applied at a flow rate of 1 ml min−1 to a Reactive Green cross-linked agarose column (diameter, 16 mm; volume, 8 ml; Reactive Green 19-agarose; Sigma-Aldrich), which had been equilibrated with 40 ml of basal buffer. After a washing step with 20 ml of 50 mM KCl in basal buffer, the ligase was eluted in a linear gradient of 50 to 250 mM KCl in basal buffer (50 ml); the p-hydroxybenzoate-CoA ligase activity eluted between 100 to 250 mM KCl.
(v) Gel filtration.
The protein fraction containing p-hydroxybenzoate-CoA ligase activity obtained by affinity chromatography was concentrated to a final volume of 1 ml using Vivaspin centrifugal concentrators (30-kDa molecular size cutoff; Sartorius AG, Göttingen, Germany) and applied to a gel filtration column (volume, 120 ml; Superdex, 200 pg; HiLoad 16/60; GE Healthcare, Munich, Germany), which was run with 20 mM triethanolamine, pH 7.8, 10% glycerol, and 150 mM KCl (1 ml min−1). Gel filtration was also used for determination of molecular mass analysis. Bovine serum albumin (BSA), ovalbumin, and lactate dehydrogenase served as molecular mass standards.
2-D gel electrophoresis.
Two-dimensional (2-D) gel electrophoresis was performed by isoelectric focusing (IEF) for the first dimension. Carrier ampholyte (pH 4 to 7) gels were prepared in glass tubes (2.3 mm diameter, 16 cm length) (1). Separation in the second dimension was by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12.5% gels; 20 cm by 20 cm) (23). Both separation steps were performed with a Protean II 2-D Electrophoresis Cell System (Bio-Rad). Prior to analysis, cell extracts were dialyzed against 20 mM Tris-HCl, pH 7.8 (4°C, 10 h), after which the proteins were precipitated overnight at −20°C by addition of a fivefold volume of ice-cold acetone in the presence of 1 mM dithiothreitol. Per separation, 500 μg protein was applied. Visual comparison of the protein patterns of cells grown on benzoate, p-hydroxybenzoate, or p-cresol after colloidal Coomassie blue staining (33) allowed the identification of p-hydroxybenzoate-induced proteins.
Mass spectrometry.
In-gel digestion of proteins was performed with an automated protein digestion system, MassPREP Station (Micromass, Manchester, United Kingdom), as described previously (39). Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) measurements was carried out with an Ultraflex TOF/TOF (Bruker Daltonik GmbH, Bremen, Germany), which was operated in the reflectron positive mode as previously described (18). Protein identification was made by peptide mass fingerprinting using the MASCOT program (Matrix Science, London, United Kingdom) and interrogation of different protein databases: the NCBI (National Center for Biotechnology) nonredundant protein database, SwissProt, and TrEMBL. Additionally, protein identification was made by nanoscale capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously (18). Briefly, analysis of the digested proteins was performed using a CapLC capillary LC system (Waters) coupled to a hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer, Q-TOF II (Waters). Collected mass data were submitted to the search software MASCOT (Matrix Science, London, United Kingdom) for protein identification using the NCBI nonredundant protein database. Searches were done with a tolerance on mass measurement of 0.25 Da in MS mode and 0.3 Da in MS/MS mode.
RT-PCR.
Total RNA from G. metallireducens grown on p-cresol, p-hydroxybenzoate, benzoate, butyrate, and acetate was used for reverse transcriptase PCR (RT-PCR). RNA was isolated from cells harvested in the exponential growth phase. An RNeasy Mini kit (QIAGEN, Hilden, Germany) for total RNA was used for RNA isolation and RNA clean-up after DNase treatment. Contaminating DNA was eliminated by on-column DNase digestion with an RNase-free DNase set (QIAGEN, Hilden, Germany) and treatment with DNase I (RNase free; 1 U per μg of total RNA; Fermentas, St. Leon-Rot, Germany) for 30 min at 37°C. Complete removal of DNA was verified by amplifying intergenic regions between two genes coding in different directions with cDNA as template. One microgram of total RNA was transcribed into cDNA by using a RevertAid H Minus First Strand cDNA Synthesis kit (Fermentas, St. Leon-Rot, Germany) with the provided random hexamer primer. Gene expression was studied by amplifying internal sequences of genes or intergenic regions between two genes from undiluted, 10-fold-diluted, and 100-fold-diluted cDNA, using genomic DNA of G. metallireducens as a positive control. PCR products (approximately 600 to 700 bp) were visualized by agarose gel electrophoresis. Standard protocols were used for DNA isolation and amplification (2, 35).
Further determinations.
SDS-PAGE (12.5% polyacrylamide) was performed as described by Laemmli (23). For the one-dimensional electrophoresis of membrane proteins, 5 μl of the suspended membrane fraction (25 mg ml−1) was mixed with fourfold sample buffer containing 12% SDS, 6% mercaptoethanol, 30% glycerol, and 0.05% Serva Blue G. The solution was incubated at 60°C for 30 min prior to electrophoresis. Proteins were visualized using Coomassie blue staining (41). Protein was routinely determined by the method of Bradford (11) using BSA as a standard.
RESULTS AND DISCUSSION
In vitro enzyme activities involved in p-cresol metabolism. (i) p-Cresol methylhydroxylase and p-hydroxybenzyl alcohol-oxidizing activity.
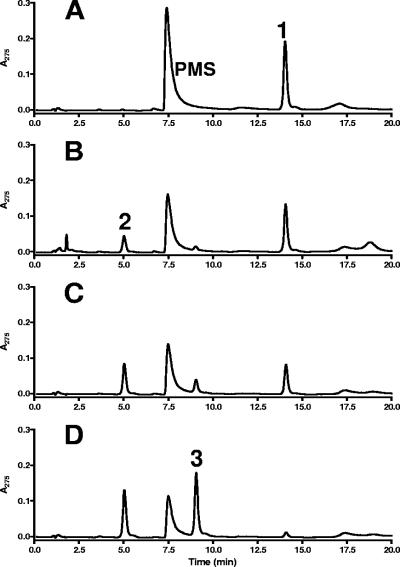
Extracts of G. metallireducens grown on p-cresol were tested for p-cresol methylhydroxylase activity by HPLC analysis following substrate consumption and product formation in the presence of PMS as an electron acceptor. In the soluble protein fraction (100,000 × g supernatant after ultracentrifugation of cell extracts), no p-cresol-oxidizing activity was detected. Instead, a p-cresol-hydroxylating activity was found in the washed membrane fraction (25 nmol min−1 mg−1; Table 1). HPLC analysis revealed that p-cresol was first converted to p-hydroxybenzyl alcohol using PMS as an electron acceptor (Fig. 2). This intermediate was then further converted at a slower rate to p-hydroxybenzaldehyde with the same electron acceptor (Table 1; Fig. 2). No alcohol dehydrogenase activity acting on p-hydroxybenzyl alcohol was detected in the soluble cell extract fraction of G. metallireducens using NAD+, NADP+, ferricenium, or PMS as electron acceptor. These results suggest that both p-cresol and p-hydroxybenzyl alcohol are oxidized by membrane-bound PCMH. In contrast, all other so-far characterized PCMHs are periplasmatic, soluble enzymes.
TABLE 1.
Activities measured in cell extracts (soluble protein or membrane fraction) of G. metallireducens grown on different growth substratesa
| Substrate | sp act (nmol min−1 mg−1) of cell extract
|
||||
|---|---|---|---|---|---|
|
p-Cresol methylhydroxylase with:
|
p-Hydroxy-benzaldehyde dehydrogenase | p-Hydroxy-benzoate- CoA ligase | p-Hydroxybenzoyl- CoA reductase | ||
| p-Cresol | p-Hydroxybenzyl alcohol | ||||
| p-Cresol | 25 ± 3 | 10 ± 1.5 | 10 ± 2 | 12 ± 1.5 | ND |
| p-Hydroxybenzoate | 1 ± 0.4 | 0.25 ± 0.05 | 1.25 ± 0.5 | 1.4 ± 0.3 | 16.6 ± 1.4 |
| Benzoate | <0.1 | <0.1 | 1 ± 0.2 | 0.2 ± 0.05 | <0.1 |
| Acetate | <0.1 | <0.1 | 0.25 ± 0.05 | <0.1 | <0.1 |
The mean deviations are indicated. ND, not determined.
FIG. 2.
p-Cresol methylhydroxylase activity. Time-dependent oxidation of p-cresol (0.5 mM; compound 1) via p-hydroxybenzyl alcohol (2) to p-hydroxybenzaldehyde (3). HPLC analysis of samples taken at 0 min (A), 15 min (B), 30 min (C), and 60 min (D) are shown. Detection of all compounds was carried out with a UV monitor at 275 nm.
Only a minor PCMH activity (<5% of the activity in p-cresol-grown cells) was measured in the washed membrane fraction of cells grown on p-hydroxybenzoate, whereas no PCMH activity was detected in fractions from cells grown on benzoate or acetate (Table 1). This result indicates a strong regulation of the enzyme activity by p-cresol.
(ii) p-Hydroxybenzaldehyde dehydrogenase.
The enzymatic oxidation of p-hydroxybenzaldehyde to p-hydroxybenzoate was tested in a spectrophotometric assay using NAD+, NADP+, or ferricenium as the electron acceptor. Only with NADP+ was p-hydroxybenzaldehyde oxidized to p-hydroxybenzoate by cell extracts of G. metallireducens grown on p-cresol, with a rate of approximately 10 nmol min−1 mg−1 (Table 1). All activity was found in the soluble protein fraction. p-Hydroxybenzaldehyde dehydrogenase activity was considerably lower in extracts of cells grown on p-hydroxybenzoate, benzoate, or acetate than in extracts of cells grown on p-cresol (Table 1).
(iii) p-Hydroxybenzoate-CoA ligase.
The presence of the putative next step in p-cresol metabolism, catalyzed by p-hydroxybenzoate-CoA ligase, was tested in the soluble protein fraction after ultracentrifugation and in protein fractions obtained after ammonium sulfate precipitation. Both a complete continuous spectrophotometric assay coupling CoA-ester formation to the reduction of NADH and a direct discontinuous assay following substrate consumption/product formation by HPLC analysis were applied. Only with the latter assay was a p-hydroxybenzoate-CoA ligase activity determined in extracts of cells grown in p-cresol (12 nmol min−1 mg−1; Table 1). Surprisingly, the activity was 10-fold lower in cells grown on p-hydroxybenzoate and was only poorly detectable in cells grown on benzoate and acetate. As p-hydroxybenzoate-CoA ligase is required during growth on both p-cresol and p-hydroxybenzoyl-CoA, the reason for the lower expression in p-hydroxybenzoate-grown cells remains unknown. Notably, p-hydroxybenzoate-CoA ligase from T. aromatica is also hardly measurable in extracts of cells grown on p-hydroxybenzoate (4).
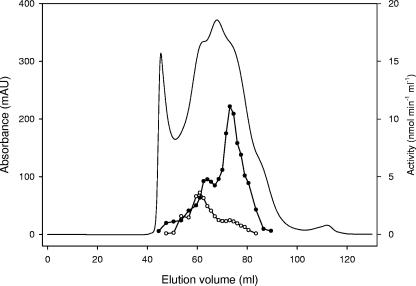
In extracts of G. metallireducens grown on p-hydroxybenzoate or p-cresol and p-hydroxybenzoate, a benzoate-CoA ligase activity was determined (0.7 and 0.1 nmol min−1 mg−1, respectively). To unambiguously test whether benzoate and p-hydroxybenzoate were converted by the same or different CoA ligases, the p-hydroxybenzoate-CoA ligase activity was partially purified from cells grown on p-cresol to a sixfold enrichment using ammonium sulfate precipitation, followed by chromatography on DEAE-Sepharose and Reactive Green (specific activity with p-hydroxybenzoate after this enrichment was 25 nmol min−1 mg−1). As this protein preparation still contained benzoate-CoA ligase activity (12.7 nmol min−1 mg−1), an additional gel filtration step was carried out in order to separate both activities. The major part of p-hydroxybenzoyl-CoA-forming activity eluted in a peak corresponding to a molecular mass of approximately 65 ± 15 kDa (Fig. 3). Most importantly, the p-hydroxybenzoate-CoA ligase activity could be clearly separated from the benzoate-CoA ligase activity, which eluted at 178 ± 20 kDa. Thus, G. metallireducens expressed two different aromatic carboxylic acid-CoA ligases: one acting on p-hydroxybenzoate, and the other acting on benzoate. Both differ in their substrate preference and their molecular properties (Fig. 3). This result is in accordance with a previous study, where it was shown that benzoate-CoA ligase from G. metallireducens does not accept p-hydroxybenzoate as substrate (39).
FIG. 3.
Separation of benzoate-CoA and p-hydroxybenzoate-CoA ligase by gel filtration. A partially purified protein fraction from G. metallireducens grown on p-cresol containing both p-hydroxybenzoate-CoA and benzoate-CoA ligase activity was applied. Line with no symbols, protein; line with the filled circles, p-hydroxybenzoate-CoA ligase activity; line with the open circles, benzoate-CoA ligase activity. Absorbance was measured at 280 nm.
(iv) p-Hydroxybenzoyl-CoA reductase.
Using a discontinuous assay coupled to HPLC analysis of substrate consumption and product formation, a p-hydroxybenzoyl-CoA reductase activity was determined in G. metallireducens grown on p-hydroxybenzoate or p-cresol. In contrast, virtually no such activity was determined in cells grown on benzoate or acetate (Table 1). These results suggest that the dehydroxylating activity was strongly regulated by the growth substrates.
Proteomic studies. (i) Membrane fraction.
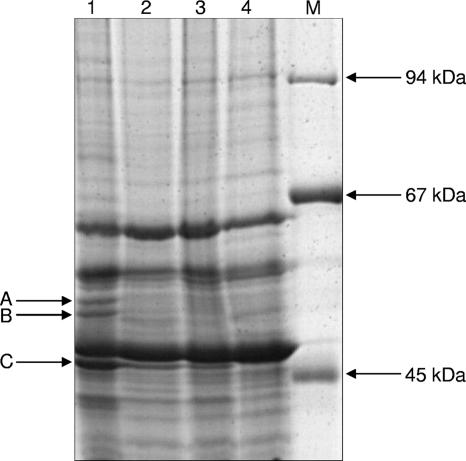
The results obtained so far indicated that p-cresol-induced PCMH is membrane bound. To test this further, the membrane fractions of cells grown on p-cresol, p-hydroxybenzoate, benzoate, and acetate were separated by one-dimensional gel electrophoresis, and the protein patterns were compared (Fig. 4). In cells grown on p-cresol, two proteins bands at 60 and 57 kDa were produced in a significantly higher amount, suggesting that they play a specific role in p-cresol metabolism. The two p-cresol-induced spots were digested by trypsin and analyzed by peptide mass fingerprinting using matrix-assisted laser desorption ionization-time of flight mass spectrometry. In addition, a liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis step was carried out. The results from peptide mass analysis obtained clearly identified the genes coding for the two proteins in the genome of G. metallireducens (accession no. NC_007515; gi7822332 and gi7822333), which were both annotated as FAD-linked oxidase-like proteins with deduced molecular masses of 59.3 and 57 kDa, respectively. The deduced amino acid sequences of both proteins were highly similar to each other (62% identity). In addition, a BLAST search revealed high amino acid sequence similarities (45%) to the large FAD-containing α subunit of PCMH from several organisms. Both proteins were produced in a 1:1 ratio, suggesting that PCMH has an αα′β2 composition, with α and α′ representing two isoforms with 59.3 and 57 kDa.
FIG. 4.
SDS-PAGE of the membrane fraction of G. metallireducens grown on different substrates. Extracts from cells grown on p-cresol (lane 1), p-hydroxybenzoate (lane 2), benzoate (lane 3), and acetate (lane 4) are shown. Spots A to C were analyzed by mass spectrometry and identified as two isoforms of the FAD-containing α subunit of p-cresol methylhydroxylase (A and B) and as a hypothetical protein, Gmet_2148 (C). Lane M, molecular size marker.
A third protein band (48 kDa) also appeared to be present at a higher amount in the membrane fraction of p-cresol-grown cells (band C in Fig. 4); however, this band was also induced in cells grown on other aromatic compounds, albeit to a lesser extent. Mass spectrometric analysis gave the best result with the BamT gene product (gi 78223356; coverage, 60%), which is part of a benzoate-induced gene cluster described previously (39). The deduced protein was annotated in the genome as Gmet_2148, and the amino acid sequence showed no similarities to other known proteins.
(ii) Soluble fraction.
The soluble protein fractions of G. metallireducens grown on p-cresol, p-hydroxybenzoate, benzoate, and acetate were subjected to two-dimensional SDS-polyacrylamide gel electrophoresis and were screened for putative soluble proteins involved in the metabolism of para-hydroxylated compounds. As previously shown, the benzoate-induced bam gene products were identified in extracts of all cells grown on an aromatic substrate but not in extracts of cells grown on butyrate (39) (the representative BamA, BamB, and BamD gene products are marked in Fig. 5). Moreover, two novel bands were identified which, after mass spectrometric analysis, were assigned to two open reading frames located close to the two genes coding for isoforms of the large subunit of PCMH. The amino acid sequence of the deduced gene products showed highest similarities to molybdenum cofactor binding (gi7822343; 52% amino acid sequence identity; 84.5 kDa) and FAD binding (gi7822341; 39% sequence identity; 30.8 kDa) subunits of p-hydroxybenzoyl-CoA reductases from facultative anaerobes. Surprisingly, the two proteins were present in cells grown on p-cresol, p-hydroxybenzoate, and benzoate, although no p-hydroxybenzoyl-CoA reductase activity was determined in cells grown on benzoate (Table 1).
FIG. 5.
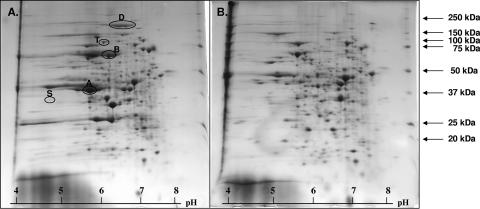
Representative 2-D gel electrophoresis (IEF/SDS-PAGE) of soluble extracts from G. metallireducens. Shown are extracts from cells grown on benzoate (A) and butyrate (B). Selected spots of gene products of the bam gene cluster are the following: BamA (A), BamB (B), and BamD (D). p-Hydroxybenzoate-induced spots of the pcm gene cluster are PcmT (T) and PcmS (S). The synthesis of PcmS and PcmT in cells grown on benzoate was not expected. The pH gradient applied was from pH 4 to 8; the polyacrylamide gradient was from 8% to 14%. The numbered protein spots were analyzed by MALDI-TOF MS and LC-MS/MS, and the corresponding genes were identified.
The bam and pcm gene clusters.
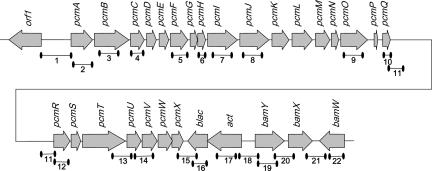
In a recent study, two gene clusters involved in benzoate metabolism of G. metallireducens were identified, including the bamA-bamY genes (for “benzoic acid metabolism” [39]). The bamY gene, coding for a benzoate-CoA ligase, marked the margin of bam cluster II; it is flanked by two genes in opposite reading directions coding for a putative N-acetyltransferase (act; gi78223349) and a β-lactamase-like protein (blac; gi78223348) (Table 2). The detection of genes putatively coding for p-hydroxybenzoyl-CoA reductase and PCMH nearby the bam genes suggested the presence of a genomic island containing genes for the catabolism of several aromatic compounds. Indeed, gi78223323 to gi78223347 are located on the same strand (Fig. 6). Among these 24 open reading frames, many putatively represent genes coding for proteins with amino acid sequence similarities to enzymes involved in the degradation of para-hydroxylated compounds. As they were all induced during growth on p-cresol (see below), they are therefore termed pcm genes (for “p-cresol metabolism”) pcmA to pcmX.
TABLE 2.
Gene clusters involved in p-cresol and p-hydroxybenzoate metabolism of G. metallireducensa
| GI no. | Name | Annotation | Molecular mass (Da) | Induction of mRNA by cells grown on:
|
||||
|---|---|---|---|---|---|---|---|---|
| p-Cresol | 4-OH-Benzoate | Benzoate | Butyrate | Acetate | ||||
| 78223324 | pcmA | Hypothetical membrane protein | 41,805 | ++ | ++ | − | − | − |
| 78223325 | pcmB | FAD-dependent oxidoreductase | 65,958 | ++ | ++ | − | − | − |
| 78223326 | pcmC | Cytochrome bc-like, N terminal | 27,911 | ++ | ++ | − | − | − |
| 78223327 | pcmD | Cytochrome bc-like, C terminal | 19,230 | ND | ND | ND | ND | ND |
| 78223328 | pcmE | Rieske protein, [2Fe-2S] binding | 18,459 | ND | ND | ND | ND | ND |
| 78223329 | pcmF | Cytochrome c, class I | 35,417 | ++ | ++ | − | − | − |
| 78223330 | pcmG | Cytochrome c with twin arginine translocation motif | 7,563 | ND | ND | ND | ND | ND |
| 78223331 | pcmH | Hypothetical protein | 14,828 | ++ | +++ | − | − | − |
| 78223332 | pcmI | p-Cresol methylhydroxylase, FAD containing | 59,372 | ++ | ++ | − | − | − |
| 78223333 | pcmJ | p-Cresol methylhydroxylase, FAD containing | 57,029 | ++ | + | − | − | − |
| 78223334 | pcmK | Phenol degradation-like protein | 27,258 | ND | ND | ND | ND | ND |
| 78223335 | pcmL | Acetate and butyrate kinase | 38,949 | ND | ND | ND | ND | ND |
| 78223336 | pcmM | Short-chain dehydrogenase | 26,687 | ND | ND | ND | ND | ND |
| 78223337 | pcmN | Pyridoxamine 5′-phosphate oxidase-related, FMN binding protein | 15,260 | ND | ND | ND | ND | ND |
| 78223338 | pcmO | Aldehyde dehydrogenase | 51,623 | ++ | ++ | − | − | − |
| 78223339 | pcmP | Twin-arginine translocation protein TatA/E | 7,153 | ND | ND | ND | ND | ND |
| 78223340 | pcmQ | Transcriptional regulator, MarR family | 15,388 | ++ | +++ | ++ | (+) | (+) |
| 78223341 | pcmR | Molybdopterin dehydrogenase, FAD binding | 30,815 | +++ | +++ | +++ | (+) | (+) |
| 78223342 | pcmS | Molybdenum dehydrogenase, Ferredoxin [2Fe-2S] binding protein | 17,852 | ND | ND | ND | ND | ND |
| 78223343 | pcmT | Aldehyde oxidase and xanthine dehydrogenase, molybdopterin binding | 84,541 | ++ | ++ | ++ | (+) | (+) |
| 78223344 | pcmU | XdhC-like, xanthine and CO dehydrogenase maturation factor | 28,018 | + | + | + | − | − |
| 78223345 | pcmV | XdhC-like, xanthine and CO dehydrogenase maturation factor | 28,265 | + | + | + | − | − |
| 78223346 | pcmW | MobA-related protein | 27,544 | ND | ND | ND | ND | ND |
| 78223347 | pcmX | Conserved hypothetical protein, possibly involved in molybdenum cofactor biosynthesis | 21,905 | ND | ND | ND | ND | ND |
| 78223348 | blac | β-Lactamase like | 39,903 | +/− | +/− | +/− | +/− | +/− |
| 78223349 | act | GCN5-related N-acetyltransferase, acetyl-CoA hydrolase/transferase | 69,020 | +/− | +/− | +/− | +/− | +/− |
| 78223350 | bamY | Benzoate-CoA ligase family | 58,412 | +++ | ++ | +++ | − | − |
| 78223351 | bamX | Hypothetical protein | 46,310 | + | + | + | − | − |
| 78223352 | bamW | Two-component, sigma 54-specific, transcriptional regulator, Fis family | 49,891 | + | + | + | − | − |
The gene products identified by proteome analysis are shown in boldface. The annotation is taken from the NCBI gene bank (NZ_AAAS00000000). ND, not determined.
FIG. 6.
Gene cluster in the genome of G. metallireducens containing p-cresol- and p-hydroxybenzoate-induced open reading frames. The abbreviations of genes and their annotations in the gene bank are listed in Table 2. The numbered short lines below the genes symbolize DNA fragments that were tested for amplification in RT-PCR experiments (see Table S1 in the supplemental material for sequences of the primers).
Reverse transcription-PCR studies of pcm gene expression.
The expression of the 24 pcm genes was investigated in cells grown on p-cresol, p-hydroxybenzoate, benzoate, butyrate, and acetate. As a control, expression of the flanking bamW-Y genes (genes involved in benzoic acid metabolism) were also analyzed (39). For this purpose, total RNA was isolated from cells harvested in the exponential growth phase, and the induction of the individual genes was tested by RT-PCR. A set of 22 DNA oligonucleotide pairs as primers for PCRs were constructed for the analysis of gene expression (Fig. 7; for sequences, see Table S1 in the supplemental material). For positive controls, genomic DNA instead of cDNA was used, which gave positive results in all cases. In an additional control experiment, a primer pair for the amplification of an internal DNA fragment of the gene coding for the β subunit of RNA polymerase was tested. The results indicated that a gene of this housekeeping enzyme was transcribed in all cells independent of the carbon source utilized.
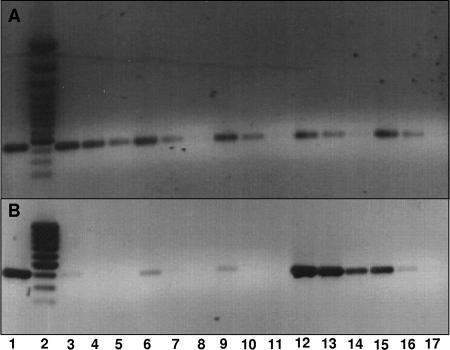
FIG. 7.
Representative reverse transcriptase PCR studies of a characteristic gene involved in p-cresol metabolism. Ethidium bromide/agarose gel of PCR products formed with different primers using cDNA obtained from mRNA of G. metallireducens cells as template. Induction of (A) RNA-polymerase β-subunit (gi45593176) and (B) pcmI (gi78223332) coding for an FAD-linked oxidase-like protein with similarities to the α subunit of p-cresol methylhydroxylases. The cDNA was produced from mRNA isolated from G. metallireducens cells grown on acetate (lanes 3 to 5), butyrate (lanes 6 to 8), benzoate (lanes 9 to 11), p-hydroxybenzoate (lanes 12 to 14), or p-cresol (lanes 15 to 17). In lane 1, genomic DNA of G. metallireducens was used as a positive control. The cDNA templates used in these reactions were undiluted (lanes 3, 6, 9, 12, and 15) or diluted 10-fold (lanes 4, 7, 10, 13, and 16) and 100-fold (lanes 5, 8, 11, 14, and 17). Lane 2 shows a DNA ladder.
A representative result of an RT-PCR experiment is shown in Fig. 7, where the pcmI gene, assigned to one of the isoforms of the FAD-binding α subunit of PCMH (Table 2), is shown. With cDNA from cells grown on p-cresol and p-hydroxybenzoate, a DNA fragment of the expected size was amplified, whereas no DNA amplification was observed with cDNA from cells grown on benzoate or acetate. Similar results were obtained with all primer pairs amplifying internal or intergenic DNA fragments of pcmA-pcmP (using primer pairs 1 to 8). In contrast, using primer pairs for amplifying internal or intergenic DNA fragments of pcmQ-pcmX, positive results were obtained using cDNA from cells grown on p-cresol, p-hydroxybenzoate, and benzoate but not with cDNA from cells grown on butyrate or acetate. Expression analysis of the bamW-Y genes indicated that these genes were expressed in cells grown on p-cresol, p-hydroxybenzoate, and benzoate but not on acetate. No amplificates were obtained using primer pairs 1 and 15 to 18, which were used to amplify the blac and act genes (Table 2) in opposite directions, and an intergenic fragment between pcmA and orf1 (Fig. 6).
The data obtained from enzyme activity measurements, proteome analysis, and RT-PCR experiments allowed us to compare the regulation of benzoate, p-hydroxybenzoate, and p-cresol metabolism on the transcriptional, translational, and posttranslational levels (Fig. 4; Table 3). The pcmA-P genes are expressed during growth on p-cresol and p-hydroxybenzoate, although proteins encoded by these genes were only produced in cells grown on p-cresol (e.g., PCMH α subunits; Table 3). In accordance with this, PCMH and p-hydroxybenzaldehyde dehydrogenase activities were only found in p-cresol-grown cells. These results suggest that in p-hydroxybenzoate-grown cells posttranscriptional regulatory elements inhibit the synthesis of PcmA-D. The bam and pcmQ-X genes are expressed in cells grown on benzoate, p-hydroxybenzoate, and p-cresol, although the pcmQ-X genes putatively coding for p-hydroxybenzoyl-CoA reductase and proteins involved in its maturation (see below) are not required for benzoate metabolism. Moreover, p-hydroxybenzoyl-CoA reductase was produced in cells grown on benzoate (Fig. 5), although the enzyme could not be measured in these extracts (Table 1). This finding can only be explained by the presence of posttranslational regulation of the enzyme activities (Table 3).
TABLE 3.
Regulation of transcription, translation, and enzyme activities of the pcm gene cluster in G. metallireducens cells grown on p-cresol, p-hydroxybenzoate, or benzoatea
| Activity | Growth substrate | pcmA-P | pcmQ-X | bamY-W |
|---|---|---|---|---|
| Transcription | p-Cresol | + | + | + |
| p-Hydroxybenzoate | + | + | + | |
| Benzoate | − | + | + | |
| Protein synthesis | p-Cresol | + | + | + |
| p-Hydroxybenzoate | − | + | + | |
| Benzoate | − | + | + | |
| Enzyme activity | p-Cresol | + | + | + |
| p-Hydroxybenzoate | − | + | + | |
| Benzoate | − | − | + |
Transcription was tested by RT-PCR analysis and protein synthesis was tested by proteome analysis (for details, see the text). The representative gene products used for enzyme assays in the pcmA-P, pcmQ-X, and bamY-W regions were PCMH (PcmGIJ), p-hydroxybenzoyl-CoA reductase (PcmRST), and benzoate-CoA ligase (BamY).
Assignment of individual pcm genes to function.
The results from RT-PCR studies suggest that the pcm genes are organized in a gene cluster flanked by orf1 and by two open reading frames in the opposite reading direction (Fig. 6). The function of a number of genes of the pcm cluster may be involved in regulation (e.g., PcmQ with similarities of a MarR type of transcriptional regulator) or remains unknown at this initial state (Table 2). However, many pcm gene products can be assigned to enzymes involved in p-cresol and p-hydroxybenzoate metabolism, which are discussed below.
(i) Genes involved in p-hydroxybenzoate metabolism.
The gene products of pcmR and pcmT were unambiguously identified on two-dimensional polyacrylamide gel electrophoresis and are assigned to two of the three subunits of a p-hydroxybenzoyl-CoA reductase (Fig. 5; Table 2). The third [2Fe-2S] cluster containing a subunit of the putative p-hydroxybenzoyl-CoA reductase, putatively encoded by pcmS, could not be determined on 2-D gels, probably due to its small size (17.8 kDa as deduced from the gene sequence). Notably, PcmR, assigned to the FAD-containing subunit of p-hydroxybenzoyl-CoA reductases, does not contain an approximately 40 amino acids containing a [4Fe-4S] cluster binding loop. This loop is considered to play an important role in the reverse electron transport from the external donor-reduced ferredoxin to the substrate (38). Thus, p-hydroxybenzoyl-CoA reductase from G. metallireducens appears to accomplish electron transfer to the substrate in a different mode than common p-hydroxybenzoyl-CoA reductases (6). The deduced pcmU-X gene products show similarities to proteins involved in maturation of molybdenum cofactor proteins and are proposed to play a role in Mo cofactor assembly of p-hydroxybenzoyl-CoA reductase.
In a previous work, the benzoate-induced bamY gene (gi78223350) was characterized as a structural gene of a highly specific benzoate-CoA ligase, which virtually did not convert p-hydroxybenzoate to the corresponding thiol ester (39). In accordance, the separation of two distinct benzoate-CoA and p-hydroxybenzoate-CoA ligase activities by gel filtration indicated the presence of a specific p-hydroxybenzoate-CoA ligase. In spite of the rather low activity of enriched p-hydroxybenzoate-CoA ligase, activity measurements excluded the presence of a CoA-transferase using benzoyl-CoA, succinyl-CoA, crotonyl-CoA, or acetyl-CoA as the CoA donor and p-hydroxybenzoate as the CoA acceptor (data not shown). In general, the amino acid sequences of aromatic carboxylic acid-CoA ligases are much more similar to each other than to nonaromatic carboxylic acid-CoA ligases. However, a BLAST search in the genome of G. metallireducens indicated that bamY appears to be the only gene coding for a typical aromatic carboxylic acid-CoA ligase. Thus, the gene coding for p-hydroxybenzoate-CoA ligase in G. metallireducens remains unknown.
(ii) Genes involved in p-cresol metabolism.
PcmO is assigned to an aldehyde dehydrogenase with high similarities to known or deduced p-hydroxybenzaldehyde or p-hydroxyphenylacetaldehyde dehydrogenases (up to 75% amino acid sequence identity). Thus, it is likely that PcmO catalyzes the observed NADP+-dependent oxidation of p-hydroxybenzaldehyde to p-hydroxybenzoate.
The two isoforms of the α subunits of PCMH could unambiguously be assigned to pcmI and pcmJ gene products. However, only the deduced pcmJ gene product contains the typical amino acid sequence and Tyr384, both of which are important for the typical covalent FAD attachment of PCMH (30). Thus, PcmJ appears to represent a functional α subunit of PCMH, whereas the function of PcmI in the suggested αα′β2 complex is not known. The deduced pcmG product shows clear amino acid sequence similarities with cytochrome c-like β subunits of PCMH from other organisms, including the conserved ligands involved in heme ligation.
A hydropathy plot of the deduced pcmG-J gene products clearly indicated that they all represent soluble proteins, most probably located in the periplasm (using the transmembrane helices prediction tool at http://www.cbs.dtu.dk/services/TMHMM-2.0/). Thus, the location of the PCMH activity is considered to result from a strong interaction with a membrane anchor. Further upstream of the genes coding for putative PCMH in G. metallireducens there are four p-cresol-induced open reading frames (pcmC-F). The deduced products share amino acid sequence similarities (30 to 66% identity) with the subunits of membrane-bound bacterial ubiquinol:cytochrome b oxidoreductases from some acidobacteria, deltaproteobacteria, or green sulfur bacteria (bc1 complex) (Fig. 6; Table 2). These enzymes are usually composed of a cytochrome c subunit, a [2Fe-2S] cluster binding, soluble Rieske protein, and a membrane-bound cytochrome b subunit (3). The presence of p-cresol-induced genes putatively coding for a membrane-bound bc1-like complex explains the membrane location of PCMH. A PCMH/bc1 complex may result in an alternative electron transfer route from PCMH to the menaquinone pool (E°′ = −80 mV). In this case the bc1 complex would operate in the reverse direction (menaquinol oxidation). However, the two-electron reduction potential of the active-site p-cresol/p-hydroxybenzyl alcohol couple is −5 to −10 mV, while the one for the p-hydroxybenzyl alcohol/aldehyde is approximately −200 mV (37). Thus, the transfer of electrons from p-cresol hydroxylation to menaquinone is thermodynamically rather unlikely, while the transfer of electrons from p-hydroxybenzyl alcohol oxidation would be possible. An electron transfer from p-hydroxybenzyl alcohol to the menaquinone pool could theoretically result in a higher energy yield compared with conventional soluble PCMH using cytochrome c or azurin as electron acceptor. Thus, the unusual asymmetric architecture of PCMH with two differing active-site α subunits might play a role for channeling electrons to different electron acceptors.
Supplementary Material
Acknowledgments
We thank MPhil Nasser Gad'on and Simon Wischgoll for help with the cultivation of cells.
This work was funded by the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 20 April 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abromeit, M., P. Askman, E. Sarnighausen, and K. Dörfling. 1992. Accumulation of high-molecular-weight proteins in response to cold hardening and abscisic acid treatment in two winter wheat varieties with different frost tolerance. J. Plant Physiol. 140:617-622. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Berry, E. A., M. Guergova-Kuras, and A. R. Crofts. 2000. Structure and function of cytchrome bc complexes. Annu. Rev. Biochem. 69:1005-1075. [DOI] [PubMed] [Google Scholar]
- 4.Biegert, T., U. Altenschmidt, C. Eckerskorn, and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur. J. Biochem. 213:555-561. [DOI] [PubMed] [Google Scholar]
- 5.Boll, M. 2005. Dearomatizing benzene ring reductases. J. Mol. Microbiol. Biotechnol. 10:132-142. [DOI] [PubMed] [Google Scholar]
- 6.Boll, M. 2005. Key enzymes in the anaerobic aromatic metabolism catalysing Birch-like reductions. Biochim. Biophys. Acta 1707:34-50. [DOI] [PubMed] [Google Scholar]
- 7.Boll, M., and G. Fuchs. 2005. Unusual reactions involved in anaerobic metabolism of phenolic compounds. Biol Chem. 386:989-997. [DOI] [PubMed] [Google Scholar]
- 8.Boll, M., B. Schink, A. Messerschmidt, and P. M. Kroneck. 2005. Novel bacterial molybdenum and tungsten enzymes: three-dimensional structure, spectroscopy, and reaction mechanism. Biol. Chem. 386:999-1006. [DOI] [PubMed] [Google Scholar]
- 9.Bossert, I. D., and L. Y. Young. 1986. Anaerobic oxidation of p-cresol by a denitrifying bacterium. Appl. Environ. Microbiol. 52:1117-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brackmann, R., and G. Fuchs. 1993. Enzymes of anaerobic metabolism of phenolic compounds. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from a denitrifying Pseudomonas species. Eur. J. Biochem. 213:563-571. [DOI] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Causer, M. J., D. J. Hopper, W. McIntire, and T. P. Singer. 1984. Azurin from Pseudomonas putida: an electron acceptor for p-cresol methylhydroxylase. Biochem. Soc. Trans. 12:1131-1132. [Google Scholar]
- 13.Cunane, L. M., Z. W. Chen, W. S. McIntire, and F. S. Mathews. 2005. p-Cresol methylhydroxylase: alteration of the structure of the flavoprotein subunit upon its binding to the cytochrome subunit. Biochemistry 44:2963-2973. [DOI] [PubMed] [Google Scholar]
- 14.Cunane, L. M., Z. W. Chen, N. Shamala, F. S. Mathews, C. N. Cronin, and W. S. McIntire. 2000. Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism. J. Mol. Biol. 295:357-374. [DOI] [PubMed] [Google Scholar]
- 15.Efimov, I., C. N. Cronin, and W. S. McIntire. 2001. Effects of noncovalent and covalent FAD binding on the redox and catalytic properties of p-cresol methylhydroxylase. Biochemistry 40:2155-2166. [DOI] [PubMed] [Google Scholar]
- 16.Entsch, B., and W. J. van Berkel. 1995. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J. 9:476-483. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, J., M. Dispensa, G. C. Fogg, D. T. Evans, and C. S. Harwood. 1994. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J. Bacteriol. 176:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintz, D., V. Wurtz, A. A. High, A. Van Dorsselaer, R. Reski, and E. Sarnighausen. 2004. An efficient protocol for the identification of protein phosphorylation in a seedless plant, sensitive enough to detect members of signalling cascades. Electrophoresis 25:1149-1159. [DOI] [PubMed] [Google Scholar]
- 19.Hopper, D. J. 1976. The hydroxylation of P-cresol and its conversion to p-hydroxybenzaldehyde in Pseudomonas putida. Biochem. Biophys. Res. Commun. 69:462-468. [DOI] [PubMed] [Google Scholar]
- 20.Hopper, D. J., I. D. Bossert, and M. E. Rhodes-Roberts. 1991. p-Cresol methylhydroxylase from a denitrifying bacterium involved in anaerobic degradation of p-cresol. J. Bacteriol. 173:1298-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keat, M. J., and D. J. Hopper. 1978. p-Cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem. J. 175:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., J. H. Fuller, V. Kuusk, L. Cunane, Z. W. Chen, F. S. Mathews, and W. S. McIntire. 1995. The cytochrome subunit is necessary for covalent FAD attachment to the flavoprotein subunit of p-cresol methylhydroxylase. J. Biol. Chem. 270:31202-31209. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Londry, K. L., J. M. Suflita, and R. S. Tanner. 1999. Cresol metabolism by the sulfate-reducing bacterium Desulfotomaculum sp. strain Groll. Can. J. Microbiol. 45:458-463. [PubMed] [Google Scholar]
- 25.Lovley, D. R., M. J. Baedecker, D. J. Lonergan, I. M. Cozzarelli, E. J. Phillips, and D. I. Siegel. 1989. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature 339:297-299. [Google Scholar]
- 26.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 27.Lovley, D. R., and D. J. Lonergan. 1990. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism GS-15. Appl. Environ. Microbiol. 56:1858-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovley, D. R., and E. J. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIntire, W., D. E. Edmondson, D. J. Hopper, and T. P. Singer. 1981. 8 Alpha-(O-tyrosyl)flavin adenine dinucleotide, the prosthetic group of bacterial p-cresol methylhydroxylase. Biochemistry 20:3068-3075. [DOI] [PubMed] [Google Scholar]
- 30.McIntire, W., D. E. Edmondson, T. P. Singer, and D. J. Hopper. 1980. 8 Alpha-O-Tyrosyl-FAD: a new form of covalently bound flavin from p-cresol methylhydroxylase. J. Biol. Chem. 255:6553-6555. [PubMed] [Google Scholar]
- 31.McIntire, W., D. J. Hopper, and T. P. Singer. 1985. p-Cresol methylhydroxylase. Assay and general properties. Biochem. J. 228:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller, J. A., A. S. Galushko, A. Kappler, and B. Schink. 2001. Initiation of anaerobic degradation of p-cresol by formation of 4-hydroxybenzylsuccinate in Desulfobacterium cetonicum. J. Bacteriol. 183:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuhoff, V., R. Stamm, I. Pardowitz, N. Arold, W. Ehrhardt, and D. Taube. 1990. Essential problems in quantification of proteins following colloidal staining with coomassie brilliant blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11:101-117. [DOI] [PubMed] [Google Scholar]
- 34.Rudolphi, A., A. Tschech, and G. Fuchs. 1991. Anaerobic degradation of cresols by denitrifying bacteria. Arch. Microbiol. 155:238-248. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Smolenski, W. J., and J. M. Suflita. 1987. Biodegradation of cresol isomers in anoxic aquifers. Appl. Environ. Microbiol. 53:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unciuleac, M., E. Warkentin, C. C. Page, M. Boll, and U. Ermler. 2004. Structure of a xanthine oxidase-related 4-hydroxybenzoyl-CoA reductase with an additional [4Fe-4S] cluster and an inverted electron flow. Structure 12:2249-2256. [DOI] [PubMed] [Google Scholar]
- 39.Wischgoll, S., D. Heintz, F. Peters, A. Erxleben, E. Sarnighausen, R. Reski, A. Van Dorsselaer, and M. Boll. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58:1238-1252. [DOI] [PubMed] [Google Scholar]
- 40.Yu, L., M. Blaser, P. I. Andrei, A. J. Pierik, and T. Selmer. 2006. 4-Hydroxyphenylacetate decarboxylases: properties of a novel subclass of glycyl radical enzyme systems. Biochemistry 45:9584-9592. [DOI] [PubMed] [Google Scholar]
- 41.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low background coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.