Abstract
Myxococcus xanthus is a soil bacterium that undergoes a unique life cycle among the prokaryotes upon starvation, which includes the formation of macroscopic structures, the fruiting bodies, and the differentiation of vegetative rods into coccoid myxospores. This peculiarity offers the opportunity to study the copper response in this bacterium in two different stages. In fact, M. xanthus vegetative rods exhibit 15-fold-greater resistance against copper than developing cells. However, cells preadapted to this metal reach the same levels of resistance during both stages. Analysis of the M. xanthus genome reveals that many of the genes involved in copper resistance are redundant, three of which encode proteins of the multicopper oxidase family (MCO). Each MCO gene exhibits a different expression profile in response to external copper addition. Promoters of cuoA and cuoB respond to Cu(II) ions during growth and development; however, they show a 10-fold-increased copper sensitivity during development. The promoter of cuoC shows copper-independent induction upon starvation, but it is copper up-regulated during growth. Phenotypic analyses of deletion mutants reveal that CuoB is involved in the primary copper-adaptive response; CuoA and CuoC are necessary for the maintenance of copper tolerance; and CuoC is required for normal development. These roles seem to be carried out through cuprous oxidase activity.
Like other myxobacteria, Myxococcus xanthus bacteria glide on the soil either as single cells or more typically as groups feeding cooperatively. However, under conditions of nutrient depletion, this bacterium undergoes a developmental program, unique among the prokaryotes, in which cells come together in a coordinated way to produce multicellular fruiting bodies within which they sporulate, converting the vegetative cells into myxospores. During this differentiation process, long vegetative rods reshape and become coccoids. This aggregation and differentiation of bacteria requires a complex interaction of signals controlling gene expression in time and space (22). This ability has converted this deltaproteobacterium in a model for studying prokaryotic morphogenesis and differentiation.
M. xanthus is common in soils rich in organic materials and rotting wood. In these niches, this bacterium has to frequently cope with toxic concentrations of copper ions, since levels of this metal in noncontaminated soils fluctuate between 2 and 100 mg/kg. This means that the M. xanthus life cycle under normal environmental circumstances is accomplished in the presence of relatively high concentrations of this transition metal. Copper is an essential trace element, but an excess is toxic. The importance of maintaining copper homeostasis is further reinforced by the presence in the cells of several families of proteins which are known to control the intracellular concentration of metal ions or that help confine them to vital roles (for reviews, see references 10, 29, 33, and 37). Moreover, in higher-order organisms, such as humans, disturbed copper homeostasis has been implicated in diseases such as Menkes and Wilson syndrome, Alzheimer's disease, Parkinson′s disease, and cystic fibrosis (5).
The copper response in M. xanthus seems to be more complex than in other bacteria. It has been reported that copper induces the accumulation of carotenoids in dark-grown cultures of M. xanthus, activating the transcription of the structural genes for carotenoid synthesis (27). This work has also shown that copper induces other unknown cellular mechanisms that confer tolerance to this metal during vegetative growth. It remains unknown which other mechanisms are responsible for this Cu-regulated adaptive response. The M. xanthus genome was recently sequenced (12), and analysis of this genome by our group has revealed a plethora of gene products with sequence similarities to proteins known to be involved in copper handling and trafficking in other organisms, most of which are redundant. This finding is indicative that copper homeostasis mechanisms in this bacterium could be different during growth and development. Among others, we have identified genes that encode three copper-translocating P-type ATPases, three Cus systems, and three multicopper oxidases (MCOs).
First, we concentrated our attention on three paralogous genes that encode MCOs, which have been named cuoA, cuoB, and cuoC (for cuprous oxidases). The MCO family is defined by the presence of three spectroscopically different copper centers. Enzymes that belong to this family include laccases, ascorbate oxidases, and ceruloplasmin in eukaryotes. Even though several bacterial MCOs have also been described, the biological roles of MCOs of prokaryotic origins are diverse and apparently unrelated (9, 11, 15, 17, 19, 20, 35, 36). Some of them have been implicated in copper tolerance (3, 6, 23, 31). Nevertheless, in those bacteria where two MCOs have been involved in copper homeostasis, only one gene is chromosomally encoded, while the other is plasmid borne (10, 33, 38). The coexistence in the M. xanthus genome of three paralogs for MCOs raises intriguing questions about their physiological roles in copper homeostasis along the life cycle.
To clarify possible alternatives, we first studied how growth and development alter the Cu-adaptive response in M. xanthus. Second, after characterization of the three MCO genes, we considered the effect of copper and the life cycle on the expression of each promoter. The correlation between the copper phenotypes of the three mutants during each developmental stage with differential expression of the MCO genes indicates that M. xanthus needs the coordinate action of the three proteins to complete a life cycle in the presence of elevated levels of copper ions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. xanthus and Escherichia coli strains used in this study are summarized in Table 1, together with their genotypes and origins. M. xanthus strains were grown with shaking at 30°C in liquid-rich Casitone-Tris (CTT) medium (18); CTT agar plates contained 1.5% Bacto agar (Difco). When necessary, kanamycin (40 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (100 μg/ml), or galactose (10 mg/ml) was added. E. coli strains were grown at 37°C in Luria-Bertani medium, which was supplemented with ampicillin (50 μg/ml), kanamycin (25 μg/ml), or X-Gal (25 μg/ml) when needed. Specific growth conditions are described in the corresponding Results section.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 40 |
| M. xanthus | ||
| DZF1 | pilQ1 | 28 |
| JM51AIF | ΔcuoA Galr Kms | This study |
| JM51AZY | cuoA-lacZ Kmr | This study |
| JM51ADWZY | cuoA-lacZ Kmr | This study |
| JM51BIF | ΔcuoB Galr Kms | This study |
| JM51BZY | cuoB-lacZ, Kmr | This study |
| JM51CIF | ΔcuoC Galr Kms | This study |
| JM51CDWZY | cuoC-lacZ Kmr | This study |
| Plasmids | ||
| pBJ113 | KmrgalK | 21 |
| pKY481 | lacZ Kmr | 7 |
| pKY481-CuoA | cuoA-lacZ Kmr | This study |
| pKY481-FICuoA | cuoA-lacZ Kmr | This study |
| pKY481-CuoB | cuoB-lacZ Kmr | This study |
| pKY481-CuoCDW | cuoC-lacZ Kmr | This study |
| pAMDcuoA | ΔcuoA Kmr | This study |
| pAMDcuoB | ΔcuoB Kmr | This study |
| pAMDcuoC | ΔcuoC Kmr | This study |
Galr means that the strain is able to grow in the presence of galactose.
In metal tolerance experiments, the ranges of metals assayed (in mM) were as follows: nickel, 0.1, 0.5, 1, 2, 3, 4, 5, and 6; zinc, 0.05, 0.1, 0.25, 0.5, and 1; cobalt, 0.05, 0.25, 0.5, 1, 2, 3, 4, and 5; cadmium, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.5, and 0.6; silver, 0.02, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, and 0.5; ferrous ions, 0.5, 1, 2, 3, 4, 5, 6, 7, and 8; and ferric ions, 0.1, 0.5, 1, 2, and 3. Copper was tested as Cu2+ by adding 0.02, 0.04, 0.06, 0.08, 0.1, 0.25, 0.5, 1, or 2 mM of copper sulfate. To test Cu+, an excess of ascorbate (1 mM) was added to the medium plus the concentrations of Cu2+ indicated above.
Developmental conditions, spore quantification, and germination.
Clone fruiting (CF) medium (16) was used to induce development. Developmental conditions used were the same as those previously reported (25). When required, metals where added at the concentrations indicated in the figures. Specific developmental conditions are described in Results. For counting spores, 10 drops of 20 μl each were spotted on a 10-cm petri dish containing CF medium with various copper sulfate concentrations. At different times, the fruiting bodies of one plate were harvested and resuspended in 200 μl of TM buffer (10 mM Tris-HCl [pH 7.6], 1 mM MgSO4). Fruiting bodies were dispersed and rod-shaped cells disrupted by sonication. Myxospores were counted in a Petroff-Hausser chamber. For germination, sonicated samples obtained at 72 h of starvation were heated at 50°C for 2 h and then diluted and inoculated on CTT agar plates for 6 to 7 days to obtain colonies.
Construction of in-frame deletion strains.
Plasmids pAMΔcuoA, pAMΔcuoB, and pAMΔcuoC were constructed for in-frame deletion of the cuoA, cuoB, and cuoC genes, respectively. Two fragments, upstream and downstream of each gene, were PCR amplified using the appropriate pairs of primers specified in Table 2. The amplified products were then digested and cloned into pBJ113, also digested with compatible enzymes. The resulting plasmids carrying the corresponding gene deletion were introduced into wild-type (WT) M. xanthus by electroporation. Several randomly chosen Kmr merodiploids (cuo+) were analyzed by Southern blot hybridization for the proper recombination event. One positive strain was then grown in the absence of kanamycin and plated on CTT agar plates containing galactose. Southern blot analysis was again used to analyze positives and select a galactose-resistant (Galr) Kms strain with in-frame deletion for each gene. The strains thus obtained have been designated JM51AIF, JM51BIF, and JM51CIF, containing deletions in cuoA, cuoB, and cuoC, respectively.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Purpose | Sequence (5′→3′)a |
|---|---|---|
| lcsABamA | Amplification of upstream of cuoA (pKY481-CuoA) | TTGGGATCCATGAAGCCTCTTCACGAATG |
| lcsAKpn | Amplification of upstream of cuoA (pKY481-CuoA) | CAGGGTACCGGCCATGAACGGCACTTCAC |
| lcsBam2 | Amplification of upstream of cuoA (pAMDcuoA) | CAGGGATCCCGATGCATGAAGCCTCTTCACG |
| lacEco | Amplification of upstream of cuoA (pAMDcuoA) | GTGGAATTCATGAACGGCACTTCACGGCTC |
| lacPst | Amplification of downstream of cuoA (pAMDcuoA) | CGGCTGCAGGGCCTCCAGCAGTGAAACCG |
| lacBam2 | Amplification of downstream of cuoA (pAMDcuoA) | TGGGGATCCATGCGGAGGGTTCCCAG |
| lcsAXhoF | Amplification of upstream of cuoA (pKY481-FICuoA) | GCCTCGAGCCGAGGTCACGCGCCTCGGG |
| lcsABamR | Amplification of upstream of cuoA (pKY481-FICuoA) | CGGGATCCATCCGCAGTCGCACCCGCTC |
| lcsBBamA | Amplification of upstream of cuoB (pKY481-CuoB) | CTGGGATCCATGGTGGTGTGTCCTCTCA |
| lcsBKpn | Amplification of upstream of cuoB (pKY481-CuoB) | CGCGGTACCCCGTGGCCGAAGCCGAGAAC |
| lcsBBam2 | Amplification of upstream of cuoB (pAMDcuoB) | CTGGGATCCATGGTGGTGTGTCCTCTC |
| lcsBXba | Amplification of upstream of cuoB (pAMDcuoB) | GGTCTAGAGCACGGCGGTGGAGCTGGAG |
| lcsBEco | Amplification of downstream of cuoB (pAMDcuoB) | CCCGAATTCACTCGGGCTCCTGCCGGCTTG |
| lcsBam | Amplification of downstream of cuoB (pAMDcuoB) | GAGGATCCCAATCCGGACGTGCGCTTTG |
| lcsCB2 | Amplification of upstream of cuoC (pAMDcuoC); amplification of upstream of cuoA (pKY481-CuoCDW) | TGGGATCCATGCTCCGCCGCGTCAGCG |
| lcsCK | Amplification of upstream of cuoC (pAMDcuoC); amplification of upstream of cuoA (pKY481-CuoCDW) | CTGGTACCTTCGGGCGCGTGGACGTGG |
| lcsCB | Amplification of downstream of cuoC (pAMDcuoC) | GTGGATCCCACCTCGGACGAGCTGCGC |
| lcsCE | Amplification of downstream of cuoC (pAMDcuoC) | CTGAATTCTGCTCCCCCTTCCCCTGGC |
Restriction sites added to the sequences are underlined.
Construction of strains harboring translational lacZ fusions and gene reporter studies.
Appropriate oligonucleotide pairs (Table 2) were used to amplify the fragments encompassing the upstream regions of the corresponding genes. A BamHI site in each primer was introduced either at the start codon or inside the genes and in-frame with the BamHI site existing in the lacZ gene of plasmid pKY481. PCR products were digested and ligated to pKY481, which was previously linearized by digestion with compatible enzymes for each PCR product. The resulting plasmids were introduced into M. xanthus WT by electroporation. Kanamycin-resistant colonies were analyzed as previously reported (4). All of the strains thus constructed harbor a WT copy of the three MCO genes. Strains containing fusions were incubated at 30°C on CTT and CF agar plates containing different copper concentrations. Cell extracts were obtained at various times by sonication and analyzed for β-galactosidase as previously reported (26). The protein content was determined by using the Bio-Rad protein assay. Specific activities are expressed as nmol of o-nitrophenol produced per minute and mg of protein.
Transmission and scanning electron microscopy.
For transmission electron microscopy, cells were plated on CTT or CF agar plates to an optical density at 600 nm (OD600) of 15. Cells were treated as previously described (26). Photographs were taken in a Zeiss TEM902 transmission electron microscope (TEM) at 80 kV. For scanning electron microscopy, cells and fruiting bodies were fixed with glutaraldehyde vapors for 24 h at room temperature. They were then postfixed with osmium tetroxide under the same conditions described above. Dehydration was accomplished by a graded series of ethanol. Samples were then critical-point dried and sputter coated with gold. Photographs were taken in a Zeiss DSM950 scanning electron microscope (SEM).
MCO activity determinations.
Cell extracts were obtained by sonication as described above for β-galactosidase activity determinations. Cells were harvested at 24 h of incubation in the absence or presence of 300 or 600 μM copper. Assays were performed utilizing the following aromatic compounds as substrates under the conditions reported: 3,3-dimetoxybenzidine (30); 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) and 4-hydroxy-3,5-dimethoxybenzaldehyde (24); and 2,6-dimethoxyphenol (14).
Cuprous oxidase activities were measured in terms of rates of oxygen consumption by using an oxygraph instrument (Hansatech) at 37°C as described by Singh et al. (35). Initial rates were calculated from the recorded curves of oxygen consumption versus time. The substrate, Cu(I), was added as a complex, [Cu(I)(MeCN)4]PF6, which releases free Cu(I) in solution. Stock solutions of Cu(I) were freshly prepared in degassed and argon-purged acetonitrile and subsequently diluted anaerobically by using gas-tight syringes. Reactions were initiated by addition of the substrate (0.5 mM) to an air-saturated mixture containing cell extract, 100 mM sodium acetate buffer, pH 5, and 5% acetonitrile. Control assays lacking the cell extracts or the substrate were also carried out. Specific activity is expressed as nmol of Cu(I) oxidized per minute per mg of protein, and activities are mean values from six assays with crude extracts prepared from cells derived from two independent cultures.
Nucleotide sequence accession numbers.
The gene identifier for cuoA is MXAN_3420, accession number ABF91184; for cuoB, MXAN_3425, accession number ABF86571; and for cuoC, MXAN_3432, accession number ABF89931.
RESULTS
Effect of copper on M. xanthus and Cu-regulated adaptive response during growth and development.
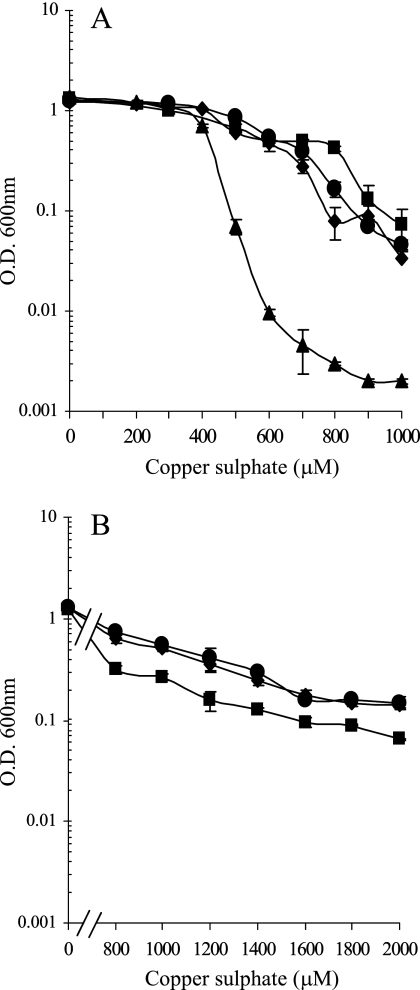
We have studied M. xanthus copper tolerance during growth and development under two different conditions. In the first set of experiments, cells were grown in the absence of external copper. For vegetative growth studies, these cells were diluted into fresh CTT liquid media supplemented with different copper concentrations and incubated for 24 h at 30°C. As can be observed in Fig. 1A, WT cells grew with copper concentrations up to 400 μM at rates similar to those observed in the absence of metal. At higher concentrations, growth yields were lower, and no growth was observed at copper concentrations of 1,000 μM. The difference in growth was the result of an increase in the generation time but not in the length of the lag phase (data not shown).
FIG. 1.
Effect of copper on M. xanthus growth. The WT strain is represented by circles, the ΔcuoA mutant by squares, the ΔcuoB mutant by triangles, and the ΔcuoC mutant by diamonds. (A) Copper tolerance of nonpreadapted cells to copper. Cells grown in the absence of the metal were diluted to an OD600 of 0.05 in fresh CTT liquid media containing the indicated copper sulfate concentrations. The OD600 indicated in the figure was monitored after 24 h of incubation. (B) Copper tolerance of Cu-preadapted cells. The WT strain and ΔcuoA and ΔcuoC mutants were grown in the presence of 600 μM copper sulfate prior to dilution into fresh CTT liquid media containing different copper sulfate concentrations. As above, the OD600 was determined at 24 h of incubation. Error bars indicate standard deviations.
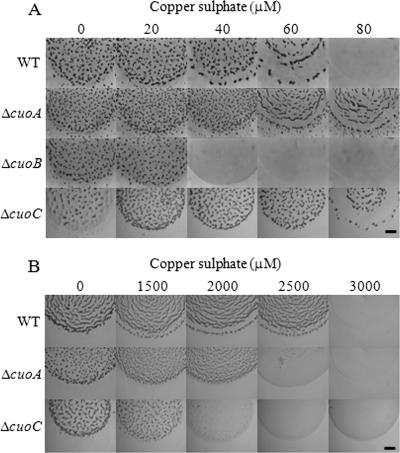
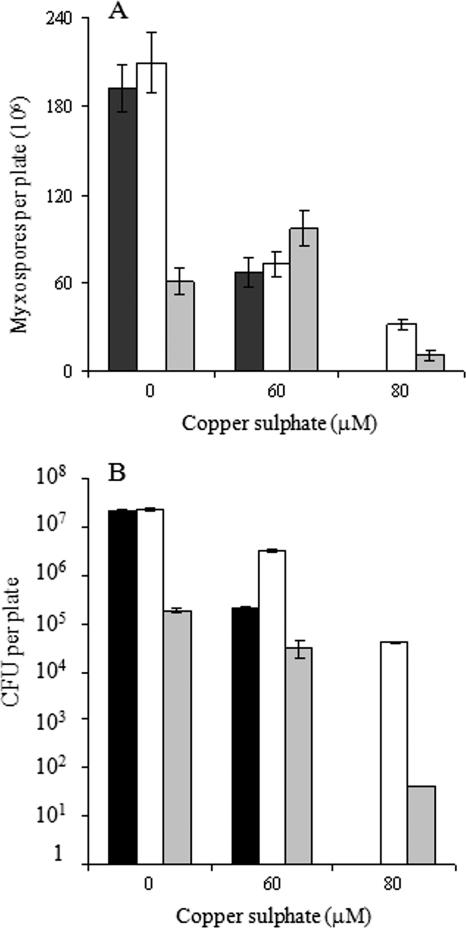
To study M. xanthus copper sensitivity during development, cells grown in the absence of copper were centrifuged and concentrated in TM buffer, and 10-μl drops were then spotted on starvation CF agar (where cells develop) containing different copper concentrations. As shown in Fig. 2A, WT cells aggregated only with copper concentrations up to 60 μM. At higher concentrations, cells died. We have also analyzed the sporulation and germination efficiencies of this bacterium in the presence of copper, and we have found that the myxospore yield was decreasing as the copper concentration was increasing (Fig. 3A, black bars). Copper addition also reduced the percentage of myxospores that were able to germinate (Fig. 3B, black bars).
FIG. 2.
Effect of copper on fruiting body formation. (A) Cells incubated in the absence of copper were concentrated at an OD600 of 15 and spotted on CF medium with the copper sulfate concentrations indicated in each photograph. (B) Cells grown in the presence of 600 μM copper sulfate were spotted on CF agar plates with the metal concentrations illustrated. Photographs were taken at 72 h of incubation. Bar, 1 mm.
FIG. 3.
Quantification of spore yield (A) and germination of the myxospores (B) for the WT strain (black bars) or the ΔcuoA (white bars) or ΔcuoC (gray bars) mutant. Error bars, standard deviations.
The copper tolerance ratio (tolerance of growing cells versus tolerance of developing cells) was around 17. This metal tolerance ratio was much higher for copper than for other metals, such as nickel, zinc, cobalt, cadmium, silver, ferrous, and ferric ions (Table 3). The addition of an excess of ascorbate to the medium containing Cu2+, to promote the reduction of the metal to Cu+, increased the copper sensitivity only of developing cells (Table 3).
TABLE 3.
M. xanthus tolerance of different metals during growth and developmenta
| Metal | Maximum concn of metal (mM) allowing viability in medium
|
TR (CTT/CF)b | |
|---|---|---|---|
| CTT | CF | ||
| Cu2+ | 1.0 | 0.06 | 17.0 |
| Cu+c | 1.0 | 0.04 | 25.0 |
| Ni2+ | 6.0 | 4.0 | 1.5 |
| Zn2+ | 0.5 | 0.5 | 1.0 |
| Co2+ | 1.0 | 4.0 | 0.25 |
| Cd2+ | 0.3 | 0.1 | 3.0 |
| Ag+ | 0.25 | 0.05 | 5.0 |
| Fe2+ | 7.0 | 7.0 | 1.0 |
| Fe3+ | 2.0 | 1.0 | 2.0 |
Cells grown in the absence of copper were concentrated to an OD600 of 15, and 10 μl was spotted on CTT (for growth) or CF (for development) agar plates with different metal concentrations. Values shown represent the highest metal concentrations tested where cell viability was detected. Viability was determined by taking cells from the media with metals and streaking them on CTT agar plates with no copper. These plates were incubated for 1 week at 30°C to get colonies.
The tolerance ratio (TR) was calculated as the quotient between the maximal metal concentrations where viability was observed.
Cu+ was obtained by the addition of Cu2+ and 1 mM ascorbate. The concentrations indicated correspond to those of Cu2+ added to the medium.
Since we recently demonstrated that M. xanthus copper resistance mechanisms are inducible (27), in a second set of experiments we tested the tolerance of cells during growth and development when they were preincubated in the presence of a copper concentration high enough to induce all of the mechanisms involved in resistance to this metal. In this case, we performed the same type of experiments reported above but with cells previously grown in CTT medium containing 600 μM copper sulfate. Preadapted cells were able to grow with up to 2,000 μM copper, but generation times lengthened considerably as copper concentrations were increased (Fig. 1B). However, it is noteworthy that during development, preadapted cells aggregated even with copper concentrations as high as 2,500 μM (Fig. 2B).
This elevated copper tolerance of preadapted cells during development versus that during growth could be attributed to the fact that experiments were carried out in different experimental settings. In the case of vegetative growth, cells were diluted to an OD600 of 0.05 and incubated in liquid medium with shaking, whereas they were concentrated to an OD600 of 15 before being plated on CF agar plates to study development. It is known that M. xanthus is a social bacterium that can face unfavorable environmental changes more efficiently when it is forming a community than when it is isolated. Therefore, we decided to test copper sensitivities of copper-preincubated and nonpreincubated cultures on CTT agar plates after concentrating cells to an OD600 of 15. In this case, we observed that cultures exhibited the same copper sensitivity on CTT agar plates as that reported above for liquid medium (data not shown).
These results clearly indicate that the copper preincubation step amplified the M. xanthus copper response only 2.5-fold in growing cells but as much as 41.7-fold in developing cells, indicating that development is the critical step for copper toxicity.
MCOs in M. xanthus genome.
To study the M. xanthus copper response at the genetic level, we searched the genome for genes that have been involved in copper resistance and homeostasis in other bacteria, and we found that most of them are redundant. We concentrated our attention on M. xanthus MCOs, named CuoA, CuoB, and CuoC. We defined as the start codon for cuoA the first ATG found downstream of a termination codon in the same reading frame. This start codon exhibits a putative ribosome-binding site (AGAGG) 5 bp upstream. The coding sequence starting at this codon encodes a protein of 481 amino acids, where 80% of the triplets use either C or G as the third base, a codon usage typical of M. xanthus genes. The protein encoded by this gene exhibits a signal sequence at the N-terminal end, which indicates that it must be secreted to the periplasmic space by the Sec system.
In the case of cuoB, an ATG triplet with a putative ribosome-binding site (AGAGG) 10 bp upstream has been chosen as the start codon. The protein encoded by this gene contains 440 amino acids, with a codon usage where 88% of the triplets contain G or C as the third base. CuoB shows a signature in the N-terminal portion (RRQFI followed by a hydrophobic region) typical of proteins that are secreted by the twin-arginine translocation (Tat) pathway (1).
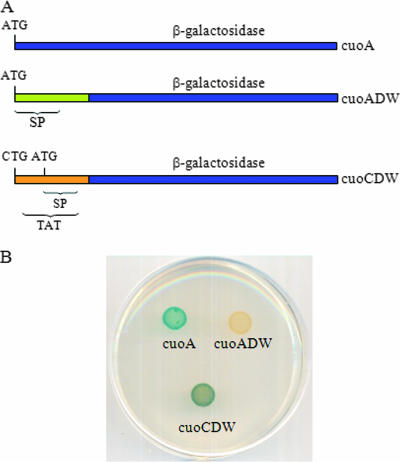
In case of cuoC, two different start codons could be chosen. One of them was a CTG codon with a putative ribosome-binding site (GAGGG) 4 bp upstream; the other was an ATG codon with no appropriate candidate ribosome-binding site. The distance between both codons was only 12 bp, with CTG being located upstream of ATG. However, the predicted secretion system used to export the protein would be different depending on the start codon used: the protein would be secreted by the Tat system if CTG were the start codon, but it would be secreted by the Sec system in the case of its being ATG. In order to test the secretion system used by CuoC, a translational fusion between cuoC and lacZ from E. coli was constructed (strain JM51CDWZY). The fusion was performed at a codon located after both signal secretion signatures (Fig. 4A), so the chimeric protein would be secreted to the periplasmic space. It is known that the Tat system translocates proteins that are completely folded, while the Sec system secretes unfolded protein (2). It is also known that β-galactosidase has to be properly folded in a tetrameric form in order to be active (13). Therefore, the chimeric protein would be active if secreted by the Tat system, but it would be inactive if secreted by the Sec system. When this experiment was performed, it was observed that the chimeric protein was active (see Fig. 4B). As controls, we constructed two translational fusions between cuoA and lacZ, one at the predicted start codon (strain JM51AZY) and the other at a codon after the signal peptide sequence (strain JM51ADWZY), as depicted in Fig. 4A. In this case, the fusion at the start codon was active, while the chimeric protein was inactive (Fig. 4B), indicating that CuoA is indeed secreted by the Sec system. We therefore concluded that CuoC is secreted by the Tat system and that CTG functions as the start codon for cuoC, with the twin-arginine motif being RRSML. This gene encodes a protein with 461 residues.
FIG. 4.
Determination of the start codon of cuoC and the secretion systems used by CuoC and CuoA. (A) Relevant features of the three fusions constructed between cuoA or cuoC and lacZ. The construct cuoA is harbored in strain JM51AZY, cuoADW in JM51ADWZY, and cuoCDW in JM51CDWZY (see Table 1 for details). (B) Analysis of β-galactosidase activity in the bacteria harboring the three fusions mentioned above. These bacteria were incubated for 24 h on CTT agar plates containing 600 μM copper sulfate and 100 μg/ml X-Gal.
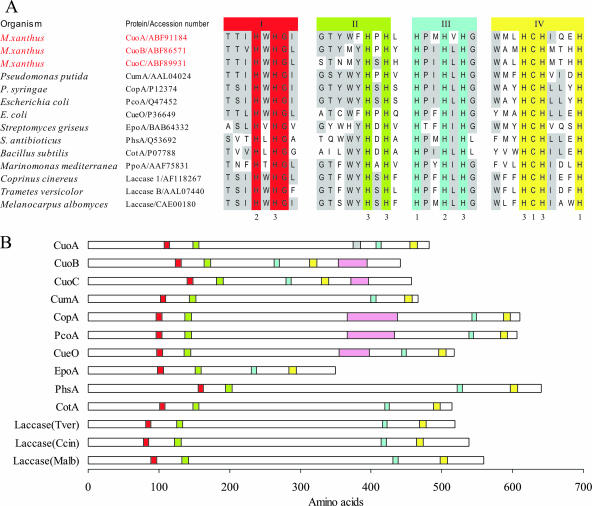
The three MCO genes are located nearby in the chromosome in a 26.5-kb region that codes for 20 proteins, including at least 9 with conserved domains known to be involved in copper handling and trafficking. Figure 5A shows the amino acid sequence alignments of the four predicted copper-binding domains from M. xanthus MCOs, characterized bacterial MCOs, and crystallized fungal laccases. The three M. xanthus proteins contain perfect matches with the sequence signature motifs for MCOs. However, CuoA is quite different from CuoB and CuoC, which are at the same time very similar to one another. Thus, the distance between motifs II and III is shorter in CuoB and CuoC than in CuoA (Fig. 5B). CuoA exhibits only 24.45 and 22.91% identity with CuoB and CuoC, respectively, while this parameter rises to 56.18% when CuoB and CuoC are compared. Another difference is that only CuoA exhibits a histidine-rich region between domains II and III (Fig. 5B), a peculiarity that has not been observed in the other MCOs. CuoB and CuoC, on the contrary, exhibit methionine-rich motifs in their C-terminal domains.
FIG. 5.
Domain organization of M. xanthus MCOs. (A) Alignments of the four copper domains of CuoA, CuoB, and CuoC of M. xanthus with those of other bacterial and fungal MCOs. Histidine and cysteine residues involved in the coordination of copper atoms are numbered according to the copper type. Invariant residues in all the proteins are shown on a red, green, blue, or yellow background for motif I, II, III, or IV, respectively, whereas conserved residues in at lest six proteins are shaded in gray. (B) Schematic representation of the motifs in M. xanthus, fungal, and bacterial MCOs. The four copper domains are represented with the same color as in panel A. The gray rectangle in CuoA represents a histidine-rich motif. Methionine-rich motifs in several MCOs are represented by pink rectangles.
Expression profiles of cuoA, cuoB, and cuoC during growth.
To get insights into the expression of the three MCO genes, three different M. xanthus strains harboring a fusion between cuoA, cuoB, or cuoC and lacZ from E. coli were constructed.
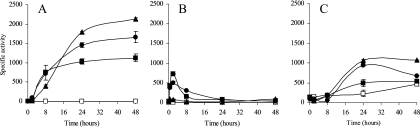
In the absence of copper, the expression of cuoA and cuoB remained undetectable (Fig. 6A and B). On the contrary, basal expression levels for cuoC were around 200 units (Fig. 6C).
FIG. 6.
Copper up-regulation of cuoA (A), cuoB (B), or cuoC (C) promoter during growth. Strains JM51AZY, JM51BZY, and JM51CDWZY (see Table 1) were incubated on CTT agar plates containing 0 (□), 300 (▪), 600 (•), or 800 μM (▴) copper sulfate. β-Galactosidase specific activity in cell extracts was determined as described in Materials and Methods. Error bars indicate standard deviations.
The three promoters were highly dependent on the addition of external copper, although the amount of metal and the incubation times required for maximum induction were clearly different for each gene (Fig. 6). In the case of cuoB, the maximal expression level was higher with 300 μM copper than with 600 μM, indicating that the cuoB promoter responds very efficiently to lower copper concentrations (Fig. 6B). The cuoA promoter was induced in a stepwise fashion as the external Cu(II) ions were increased, reaching the maximum levels at subinhibitory copper concentrations (Fig. 6A). The promoter of cuoC, on the contrary, was induced only at high copper concentrations (Fig. 6C). As a summary, the cuoB and cuoC promoters seemed to respond differentially to low and high copper concentrations, respectively, while cuoA responded to a broad range of copper concentrations, exhibiting a linear dependency on the external copper.
The maximum expression time for each gene was also clearly different. The early induction of cuoB, reaching a peak 2 h after the metal addition, suggests that this MCO forms part of the primary copper response (Fig. 6B). With longer incubation times, expression decreased nearly to prestress levels after 24 h. In the case of cuoA and cuoC, the expression increased with time, reaching a plateau between 24 and 48 h of incubation with copper (Fig. 6A and C).
Expression of cuoA, cuoB, and cuoC during development.
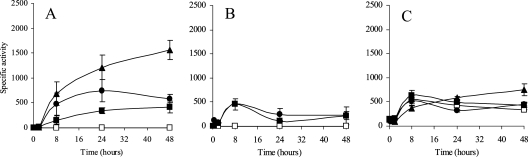
To analyze the MCO promoter responses to copper during development, a 10-fold-lower concentration of the metal had to be used in order to prevent cells from dying (Fig. 2A). Very similar to the case with growing cells, cuoA and cuoB were not expressed in the absence of copper (Fig. 7A and B). However, cuoC expression profiles changed under developing conditions, and this promoter exhibited a copper-independent induction by starvation (Fig. 7C).
FIG. 7.
Copper up-regulation of cuoA (panel A), cuoB (panel B), or cuoC (panel C) promoter during development. Strains JM51AZY, JM51BZY, and JM51CDWZY (see Table 1) were incubated on CF agar plates containing 0 (□), 20 (▪), 40 (•) or 60 μM (▴) copper sulfate. β-Galactosidase specific activity in cell extracts was determined as described in Materials and Methods. Error bars indicate standard deviations.
The copper-responsive expression profile for cuoA was similar to that shown during growth, although with only 1/10 of the metal in the medium. That means that cuoA exhibited a linear induction dependent on the external copper concentration (Fig. 7A). On the contrary, several differences could be observed in the expression of cuoB during development. First, the induction was not so immediate, and the time of maximal induction changed from 2 h during growth to 8 h (Fig. 7B). Second, all the concentrations tested exhibited similar expression levels. As in the case of cuoA, a 10-fold-lower amount of copper was required to achieve maximum expression during development. In contrast, the cuoC promoter did not respond to the addition of metal during development, even with copper concentrations as high as 60 μM (Fig. 7C).
Role of MCOs in copper resistance.
To analyze whether the three MCOs are involved in copper tolerance, three in-frame deletion mutants were constructed (Table 1). The three mutants and the WT strain were incubated in liquid CTT medium containing several copper concentrations. As shown in Fig. 1A, the copper tolerances for ΔcuoA and ΔcuoC mutants were very similar to that of the WT strain. On the contrary, the ΔcuoB mutant was markedly more sensitive to copper than the other three strains, as no growth was observed with copper concentrations of 500 μΜ.
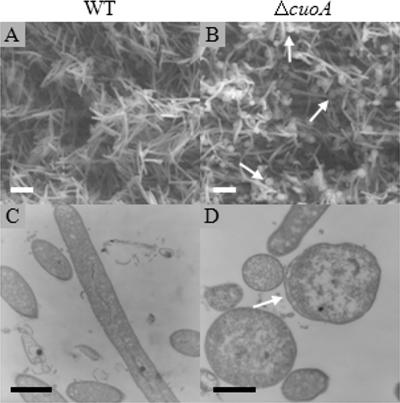
Although the ΔcuoA mutant and the WT strain exhibited similar growth rates with 700 μΜ copper, optical, transmission, and scanning microscopy analyses revealed that the mutant cells exhibited altered morphologies, since many of them (approximately one-third) became spherical instead of remaining typical long rods (Fig. 8). The number of spherical forms increased with higher copper concentrations for the mutant. Some of these forms could also be observed for the WT strain, but only with 900 μΜ copper. We were also able to detect spherical forms for the ΔcuoC mutant grown with 700 μM copper but at a much lower ratio than for the ΔcuoA strain.
FIG. 8.
Morphology and ultrastructure of vegetative cells of the WT (A and C) or ΔcuoA (B and D) strain grown on CTT medium containing 700 μM copper sulfate. Pictures were taken with a SEM (A and B) or TEM (C and D) after 72 h of incubation. Bars represent 3 μm in panels A and B or 0.5 μm in panels C and D. Arrows point to spherical forms (B) or a region where the distance between the outer and inner membranes is unusual (D).
We obtained two contradictory observations with the ΔcuoA and ΔcuoC strains: mutant cells exhibited alterations in morphology, but they could tolerate the same metal concentration as the WT strain or even higher. Although other alternatives cannot be ruled out, one possible explanation for these paradoxical observations could be that the lack of CuoA or CuoC might prevent a proper detoxification of the periplasmic space, provoking the disruption of the peptidoglycan. Simultaneously, this lack of detoxification might induce other mechanisms involved in copper resistance, enough to maintain or even increase viability of cells but not to detoxify the periplasmic space. As a result, the mutant cells would survive in the presence of high copper concentrations, like the WT cells, but not as long rods; rather, as spherical forms. If this premise is correct, mutant cells that have been grown in the presence of copper to induce all the mechanisms involved in resistance should be more sensitive to the metal than the WT cells. To test this hypothesis, ΔcuoA and ΔcuoC mutants and the WT strain were grown in the presence of 600 μM copper sulfate (to induce all of the copper resistance mechanisms) and then diluted into fresh medium containing higher copper concentrations. As shown in Fig. 1B, the ΔcuoA mutant was more sensitive to copper than the WT strain under these conditions, which is consistent with our initial hypothesis. On the other hand, we could not observe a clear decrease in sensitivity in the case of the ΔcuoC mutant.
Role of MCOs in development.
When no copper was added to the starvation CF medium, all mutants exhibited a delay in aggregation compared to the WT strain (data not shown). However, after 72 h of incubation, the mutant fruiting bodies were very similar to those of the WT strain. Only the ΔcuoC fruiting bodies exhibited some abnormalities in morphology (Fig. 2A). The number and ability to germinate of the myxospores obtained in the absence of metal were similar in the ΔcuoA and ΔcuoB mutants compared to the WT strain (data for WT and ΔcuoA mutant are shown in Fig. 3A and B). On the contrary, the yield of myxospores of the ΔcuoC mutant decreased to one-third of that of the WT strain (Fig. 3A). In addition, the ability of these mutant spores to germinate was even more reduced. As shown in Fig. 3B, only 0.39% of the ΔcuoC myxospores were able to germinate, versus 11% obtained in the case of the WT strain (Fig. 3B).
In the presence of copper, the ΔcuoB mutant was much more sensitive than the WT strain, as was observed during growth. As shown in Fig. 2A, this mutant could make fruiting bodies only with copper concentrations up to 20 μM. With greater concentrations, cells died. The capacity of spore formation and germination remained unaltered in this mutant compared to the WT in the presence of 20 μM of copper (data not shown). The ΔcuoA and ΔcuoC mutants not only aggregated at higher copper concentrations than the WT strain but also were able to reshape and originate myxospores. The yield of myxospores in the ΔcuoA mutant and the WT strain was decreasing at the same rate as the copper concentration was increasing (Fig. 3A). However, the ΔcuoA mutant exhibited a greater germination efficiency, which is consistent with the fact that these mutant cells are more resistant to copper. As shown in Fig. 3B, only 0.3% of the WT myxospores obtained with 60 μM copper were able to germinate, versus 4.4% in the case of the ΔcuoA mutant. The ΔcuoC strain yielded even more myxospores with 60 μM copper than in the absence of metal (Fig. 3A). But this mutant was clearly affected in germination, and only 0.032% of the spores obtained with that copper concentration were able to originate colonies in rich medium (Fig. 3B). Moreover, the ΔcuoA and ΔcuoC mutants yielded myxospores with the ability to germinate with 80 μM copper (Fig. 3).
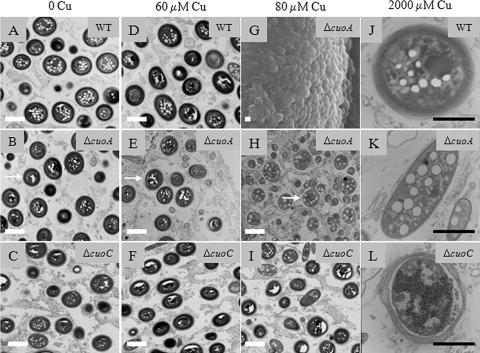
When cells inside the aggregates of the three mutants were compared to those of the WT strain under a TEM, it was observed that ΔcuoB, ΔcuoC, and WT myxospores were identical at all the copper concentrations that could be tested (Fig. 9A, C, D, and F). The ΔcuoC cells maintained an unaltered spore coat even at 80 μM copper (Fig. 9I). In the case of the ΔcuoA mutant, the external coat gradually became fainter as the copper concentration increased, completely disappearing with 80 μM copper (compare panels B, E, and H in Fig. 9). These myxospore-like cells obtained with 80 μM copper were different from the spherical forms observed during vegetative growth for this mutant. First, myxospore-like cells were observed only in the aggregates and not in the peripheral rod population surrounding the fruiting bodies. Second, the same type of inclusion bodies as those observed in the typical myxospores appeared inside these spherical cells (Fig. 9). Third, some of these cells were able to germinate.
FIG. 9.
Effect of copper on the spore coat. Fruiting bodies from WT, ΔcuoA, and ΔcuoC strains were harvested from CF agar containing the indicated copper sulfate concentrations and treated as described in Materials and Methods. All the pictures were taken with a TEM, with the only exception being panel G. Picture G, taken with a SEM, shows a fruiting body filled with myxospores formed by the ΔcuoA mutant at 80 μM of copper sulfate. Myxospores formed by copper-preadapted cells on CF containing 2,000 μM copper sulfate are shown in pictures J (WT strain) and K (ΔcuoA mutant). In the case of the ΔcuoC mutant (L), a copper concentration of 1,500 μM was used, because with greater concentrations this mutant does not make fruiting bodies. White bars, 1 μm; black bars, 0.5 μm. Arrows in panels B, E, and H point to the outer coat of the myxospores.
In concordance with the phenotype observed during growth, we assayed development of ΔcuoA and ΔcuoC mutants when cells were previously grown with 600 μM copper. In these experiments, the ΔcuoA mutant also turned out to be more sensitive to copper than the WT strain. Under these circumstances, the ΔcuoA mutant formed fruiting bodies only up to 2,000 μM, versus the WT strain, which was able to develop with a 2,500 μM concentration (Fig. 2B). Mutant cells died at higher copper concentrations. In addition, the spore ultrastructure was absolutely different in this mutant. As shown in Fig. 9K, the coat of cells inside the aggregates disappeared. Furthermore, the morphology of the mutant cells was also affected, and more than 90% failed to completely reshape, remaining short rods. Contrary to the case with the mutant, most of the WT spores exhibited an unaltered cell envelope (Fig. 9J).
In the case of the ΔcuoC mutant, preincubated cells exhibited the same viability as the WT strain (swarming was observed, and cells grew when transferred to rich medium). However, they were unable to aggregate and sporulate with copper concentrations higher than 1,500 μM (Fig. 2B). Spores obtained with 1,500 μM copper were covered with a deteriorated spore coat (Fig. 9L).
Cuprous oxidase activity in M. xanthus cell extracts.
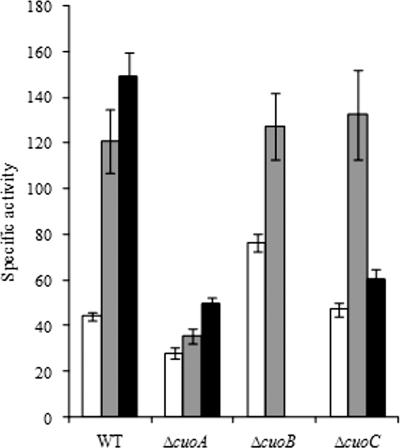
MCOs have been reported to oxidize a wide variety of aromatic compounds (phenolics and nonphenolics) and also metals, such as cuprous ions. To investigate how M. xanthus MCOs exert their functions, we searched for several enzymatic activities in WT cell extracts obtained from cells grown in the absence or presence of copper. With use of cell crude extracts, no oxidation activity was detected towards the oxidation of any the aromatic compounds tested, either in cells grown in the absence of copper or in those incubated with this metal. Instead, we were able to measure cuprous oxidase activity in crude extracts of WT and mutant strains from cells harvested after 24 h of growth in the absence of copper. Furthermore, significant changes were found in cell extracts grown in the presence of increasing concentrations of copper (Fig. 10). WT cells grown in the presence of 300 or 600 μM copper exhibited a 2.7- or 3.4-fold increase in the enzymatic rate, respectively, compared with cells grown in the absence of copper. In addition, cuprous activity in cell extracts from the ΔcuoA mutant at all of the copper concentrations tested and the ΔcuoC mutant at 600 μM was clearly reduced. This reduction was not observed with the ΔcuoB strain, although it should be noted that the expression of the cuoB promoter in the WT strain at this growth stage was negligible (Fig. 6). These results are in accordance with the observed copper up-regulation response of the MCO genes.
FIG. 10.
Cuprous oxidase activity in crude extracts of WT and mutant strains harvested after 24 h of growth in absence (white bars) or presence of 300 (gray bars) or 600 μM (black bars) copper sulfate.
DISCUSSION
The copper response has been deeply studied so far with two bacteria, E. coli and Enterococcus hirae (33, 37, 39), but the elucidation of the copper systems in M. xanthus will provide a missing piece in the complicated puzzle of bacterial adaptation to this metal under natural environmental conditions: how do cells handle different copper concentrations during a complete life cycle?
In this report we have first studied the effect of copper on developing cells, and we have found that they are approximately 15-fold more sensitive to this metal than growing cells. It should be noted that developing cells have to undergo many changes in the cell envelope in order to reshape and become myxospores. These alterations could make developing cells extremely sensitive to copper stress. Additionally, these cells are physiologically different. As they are starving, their energy levels are considerably lower than those of vegetative cells. It has been reported for E. coli that anaerobic cells are also more sensitive to copper than those that are grown in the presence of oxygen (30). These authors have proved that growth in the absence of oxygen clearly alters the copper physiology and that two different systems involved in copper tolerance are differently regulated during aerobic and anaerobic growth. The data obtained for E. coli and those reported in the present study on M. xanthus during development and growth indicate that the physiological stage of cells is an important factor in copper tolerance.
However, M. xanthus copper-preadapted cells are as resistant to this metal during development as during growth. Elucidation of this huge increase in resistance during development probably will reveal a very complex and intricate process, where many different proteins will be involved. Our data indicate that once the copper adaptation mechanisms are turned on, cells are able to tolerate higher copper concentrations. In fact, we have demonstrated that the induction of cuoA and cuoC (and, therefore, the resistance conferred by CuoA and CuoC) persists during long periods of time.
Due to the large variety of functions where MCOs seem to be involved, we have investigated the role that the three MCOs from M. xanthus play during growth and development in an attempt to find the reasons for such a high gene redundancy in the chromosome. From the sequence data analyses and translocation systems used by the three MCOs, we can distinguish two groups: CuoA and CuoB/CuoC. CuoB and CuoC possess the methionine-rich region proximal to the copper centers found in all of the MCOs with a function in copper resistance. This domain has been suggested to function as a regulator that binds copper to activate the enzymes or as a mediator in protein-protein interaction (34). In addition, they are secreted by the Tat pathway, which transports fully folded proteins with their cofactors bound before translocation into the periplasm (2). These two features indicate that these two MCOs will eliminate copper from the cytoplasm during translocation. On the contrary, CuoA lacks this methionine-rich region, and it is translocated by the Sec system, what means that this protein has to incorporate copper as a cofactor into the periplasm. These differences reinforce our interest in the compared study of regulation and physiological role of these proteins.
One of the reasons for MCO redundancy must be the differences in the expression profiles exhibited by each gene during growth and development. A first difference observed is that cuoC is the only MCO expressed during growth and development in the absence of copper. Although the three promoters are induced by this metal during growth, the timing and copper levels required for maximum expression differ for each gene. These data, along with the phenotypic analyses of deletion mutants, indicate that each gene is specialized in a different function. CuoB seems to be involved in the primary adaptive response to copper; hence, its promoter responds rapidly after copper addition, and its expression decreases as the expression of the other two genes increases. As a result, ΔcuoB mutant cells are extremely sensitive to this metal. On the contrary, CuoA and CuoC seem to be responsible for the maintenance of the response. They are induced more slowly than CuoB, and their expression levels reach a plateau. These results clearly demonstrate that along with differential induction with increased copper concentrations, differences in timing of expression to adjust the cells to the Cu-regulated adaptive response justify the redundancy of the three MCOs in M. xanthus.
One interesting observation is that the induction of cuoA and cuoB during development requires a 10-fold-lower amount of copper than during growth to reach similar levels of expression. This difference in promoter sensitivity has not been observed either with cuoC or with genes involved in carotenoid biosynthesis (unpublished results). On the contrary, cuoC is induced by starvation. An explanation for these differences could be that the three genes are regulated by two different mechanisms, one functioning during development and the other during growth. A multiple regulation has been previously described for several proteins involved in the copper response. Hence, the Cu(I)-translocating P-type ATPase CopA, which is the central component in copper homeostasis in E. coli, is regulated by the metal-responsive regulator CueR, but its expression is also influenced by the cell envelope stress response regulator CpxR (31). In addition, the oxygen sensor FNR also induces copA expression under microaerobic conditions (32). Only the identification of the factors involved in the regulation of the three M. xanthus MCOs will answer these intriguing questions.
Phenotypic analysis of the ΔcuoA deletion mutant revealed that the morphology of one-third of the mutant cells dramatically changes during growth with copper, and the long rods become spherical forms (Fig. 8). This change in shape suggests a loss of peptidoglycan, a phenotype that has not been reported for any other bacteria where the copper response has been studied, such as E. coli. The different behavior of the M. xanthus ΔcuoA mutant cells can be attributed to the peculiarities of peptidoglycan in this myxobacterium. This peptidoglycan consists of patches that are connected by bridges that are sensitive to treatments such as sodium dodecyl sulfate and proteases (8). These bridges must also be sensitive to the oxidative stress originated by copper, causing the loss of the morphology of the mutant cells in the presence of this metal. Since this phenotype is not observed so drastically with the cuoB and cuoC mutants, probably CuoA is the main MCO responsible for periplasmic detoxification. This hypothesis is further reinforced by the lack of a spore coat in the ΔcuoA myxospores obtained with 80 and 2,000 μM copper. The spore coat has to be secreted from the cytoplasm to the exterior surface, and during this traffic, materials have to go across the periplasm. They are there subjected to copper deleterious side reactions in the cuoA mutant, preventing proper assembly and originating myxospores with no coat.
The MCO gene cotA from B. subtilis encodes a structural component of the outer coat but is not copper inducible (24). CuoC could be a good candidate to exert a function similar to that of CotA in the myxospore coat because of the regulation of the gene upon starvation in the absence of extracellular copper. However, several evidences point in another direction. Thus, this gene is expressed at quite high basal levels and is induced by copper during growth. Therefore, if CuoC were located in the spore coat as is CotA, this protein should exert another function during growth in the presence of high copper concentrations. Nevertheless, CuoC seems to have an important role during development, because severe defects are observed in the mutant strain, with or without copper. We believe that all of these defects most likely can be attributed to a lack of detoxification in the mutant cells.
The role of CueO from E. coli in copper homeostasis is difficult to discern due to the fact that this protein is able to oxidize a broad range of substrates, such as cuprous compounds, catechols, and other aromatic compounds or iron-chelating siderophores (15, 35). From this perspective, it could also be discussed that the coexistence and expression of three MCOs in M. xanthus are due to different enzymatic activities exerted by these proteins. This question will be clarified only by biochemical characterization of the three purified enzymes, an investigation that is currently under way in our laboratories. However, the results obtained from oxidase assays with M. xanthus WT and mutant cell extracts have revealed that most likely they will exert their functions through cuprous oxidase activity. The cuoA promoter exhibits not only the highest levels of expression but also the strongest dependency on the copper concentration in the medium. Accordingly, the deletion of this gene in the ΔcuoA mutant results in the lowest enzymatic activities measured together with the lowest activity dependency on external copper addition. Therefore, the kinetic analysis supports an essential role for this enzyme in the full copper tolerance of M. xanthus during vegetative growth, presumably through Cu(I) detoxification in the periplasmic space (Fig. 1B). Furthermore, the levels of enzymatic activities in the ΔcuoB and ΔcuoC mutants confirm the weak involvement of CuoB in copper responses at late stages of growth and a major contribution of CuoC during vegetative growth at higher copper concentrations.
As a summary, we conclude that the three paralogous genes that encode MCOs in M. xanthus are differentially regulated; that CuoA is mainly involved in periplasm detoxification and is necessary for full copper tolerance during growth and development; that CuoB is essential in the primary adaptive response to copper oxidative stress; and that CuoC plays an important role in development, especially in the sporulation process. These different functions might explain why M. xanthus requires three MCO paralogs.
Acknowledgments
This work was supported by Ministerio de Educación y Ciencia (grants BMC2003-02038 and BFU2006-00972/BMC, 70% funded by FEDER, and the integrated action HP1005-0034). M.C.S.-S. and N.G.-S. are predoctoral fellows from Plan Propio, Universidad de Granada. A.M.-M. was granted a postdoctoral fellowship from Universidad de Granada.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Bendtsen, J. D., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks, B. C., T. Palmer, and F. Sargent. 2005. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8:174-181. [DOI] [PubMed] [Google Scholar]
- 3.Brown, N. L., S. R. Barrett, J. Camakaris, B. T. O. Lee, and D. A. Rouch. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17:1153-1166. [DOI] [PubMed] [Google Scholar]
- 4.Carrero-Lérida, J., A. Moraleda-Muñoz, R. García-Hernández, J. Pérez, and J. Muñoz-Dorado. 2005. PhoR1-PhoP1, a third two-component system of the family PhoRP from Myxococcus xanthus: role in development. J. Bacteriol. 187:4976-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerpa, W., L. Valera-Nallar, A. E. Reyes, A. N. Minniti, and N. C. Inestrosa. 2005. Is there a role for copper in neurodegenerative diseases? Mol. Aspects Med. 26:405-420. [DOI] [PubMed] [Google Scholar]
- 6.Cha, J., and D. A. Cooksey. 1991. Copper resistance in Pseudomonas syringae by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 88:8915-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, K., and D. R. Zusman. 1999. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34:268-281. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin, M. 1993. Cell surfaces and appendages, p. 63-83. In M. Dworkin and D. Kaiser. (ed.), Myxobacteria II. ASM Press, Washington, DC.
- 9.Endo, K., Y. Hayashi, T. Hibi, K. Hosono, T. Beppu, and K. Ueda. 2003. Enzymological characterization of EpoA, a laccase-like phenol oxidase produced by Streptomyces griseus. J. Biochem. 133:671-677. [DOI] [PubMed] [Google Scholar]
- 10.Finney, L. A., and T. V. O'Halloran. 2003. Metals: impact on health and the environment. Science 300:931-936. [DOI] [PubMed] [Google Scholar]
- 11.Francis, C. A., and B. M. Tebo. 2001. Multicopper oxidase genes from diverse Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains. Appl. Environ. Microbiol. 67:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gött, P., and W. Boos. 1988. The transmembrane topology of the sn-glycerol 3-phosphate permease of Escherichia coli analysed by phoA and lacZ protein fusions. Mol. Microbiol. 2:655-663. [DOI] [PubMed] [Google Scholar]
- 14.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grass, G., K. Thakali, P. E. Klebba, D. Thieme, A. Müller, G. F. Wildner, and C. Rensing. 2004. Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J. Bacteriol. 186:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagen, D. C., A. P. Betscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 17.Hernández-Romero, D., F. Solano, and A. Sanchez-Amat. 2005. Polyphenol oxidase activity expression in Ralstonia solanacearum. Appl. Environ. Microbiol. 71:6808-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 19.Hullo, M. F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huston, W. M., M. P. Jennings, and A. G. McEwan. 2002. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45:1741-1750. [DOI] [PubMed] [Google Scholar]
- 21.Julien, B., D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 23.Lee, Y., M. Hendson, N. J. Panopaulos, and M. N. Schroth. 1994. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Pseudomonas campestris pv. juglandis: homology with small proteins and multicopper oxidases. J. Bacteriol. 176:173-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins, L. O., C. M. Soares, M. M. Pereira, M. Teixeira, T. Costa, G. H. Jones, and A. O. Henriques. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277:18849-18859. [DOI] [PubMed] [Google Scholar]
- 25.Moraleda-Muñoz, A., J. Carrero-Lérida, A. L. Extremera, J. M. Arias, and J. Muñoz-Dorado. 2001. Glycerol 3-phosphate inhibits swarming and aggregation of Myxococcus xanthus. J. Bacteriol. 183:6135-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moraleda-Muñoz, A., J. Carrero-Lérida, J. Pérez, and J. Muñoz-Dorado. 2003. Role of two novel two-component regulatory systems in development and phosphatase expression in Myxococcus xanthus. J. Bacteriol. 185:1376-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moraleda-Muñoz, A., J. Pérez, M. Fontes, F. J. Murillo, and J. Muñoz-Dorado. 2005. Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus. Mol. Microbiol. 56:1159-1168. [DOI] [PubMed] [Google Scholar]
- 28.Morrison, C. E., and D. R. Zusman. 1979. Myxococcus xanthus mutants with temperature-sensitive, stage-specific defects: evidence for independent pathways in development. J. Bacteriol. 140:1036-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 30.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 31.Outten, F. W., C. E. Outten, J. A. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275:31024-31029. [DOI] [PubMed] [Google Scholar]
- 32.Partridge, J. D., G. Sanguinetti, D. Dibden, R. E. Roberts, R. K. Poole, and J. Green. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 282:11230-11237. [DOI] [PubMed] [Google Scholar]
- 33.Rensing, C., and G. Grass. 2003. Escherichia coli mechanism of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 34.Roberts, S. A., G. F. Wildner, G. Grass, A. Weichsel, A. Ambrus, C. Rensing, and W. R. Montfort. 2003. A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J. Biol. Chem. 278:31958-31963. [DOI] [PubMed] [Google Scholar]
- 35.Singh, S. K., G. Grass, C. Rensing, and W. R. Montfort. 2004. Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 186:7815-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solano, F., E. Lucas-Elio, E. Fernández, and A. Sánchez-Amat. 2000. Marinomonas mediterranea MMB-1 transposon mutagenesis: isolation of a multipotent polyphenol oxidase mutant. J. Bacteriol. 182:3754-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solioz, M., and J. V. Stoyanov. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27:183-195. [DOI] [PubMed] [Google Scholar]
- 38.Voloudakis, A. V., T. M. Reignier, and D. A. Cooksey. 2005. Regulation of resistance to copper in Xanthomonas axonopodis pv. vesicatoria. Appl. Environ. Microbiol. 71:782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, K., and A. Ishihama. 2005. Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56:215-227. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]