Abstract
The ybxF gene is a member of the streptomycin operon in a wide range of gram-positive bacteria. In Bacillus subtilis, it codes for a small basic protein (82 amino acids, pI 9.51) of unknown function. We demonstrate that, in B. subtilis, YbxF localizes to the ribosome, primarily to the 50S subunit, with dependence on growth phase. Based on three-dimensional structures of YbxF generated by homology modeling, we identified helix 2 as important for the interaction with the ribosome. Subsequent mutational analysis of helix 2 revealed Lys24 as crucial for the interaction. Neither the B. subtilis ybxF gene nor its paralogue, the ymxC gene, is essential, as shown by probing ΔybxF, ΔymxC, or ΔybxF ΔymxC double deletion strains in several functional assays.
The streptomycin (str) operon belongs to the most conserved operons in prokaryotic evolution (14, 16). The str operon of gram-negative Escherichia coli, from which most of our knowledge about this operon is derived, is composed of four genes: rpsL, coding for ribosomal protein S12; rpsG, coding for ribosomal protein S7; fus, coding for elongation factor G; and tufA, coding for elongation factor Tu. These proteins are essential components of the protein synthesis machinery of the cell.
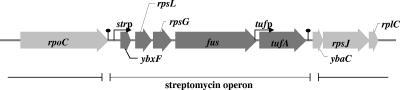
In contrast to E. coli, we found out in our previous studies that the str operon of two gram-positive organisms, Bacillus stearothermophilus and Bacillus subtilis, is a transcriptional unit composed of five genes (Fig. 1). An additional gene, preceding the rpsL gene, and designated ybxF, was found to extend the 5′ end of the operon in both organisms. The ybxF gene is transcribed in the form of two transcripts: (i) as a part of the polycistronic str mRNA carrying messages for the production of all five proteins encoded by the operon genes and (ii) as a separate ybxF mRNA carrying a message for the production of YbxF protein only. Both mRNAs start from the main promoter, strp, situated upstream of the ybxF (17).
FIG. 1.
Streptomycin operon of B. subtilis. Genes belonging to the operon are colored dark gray. Black arrows indicate promoters; transcription terminators are shown as stem-loop structures.
The ybxF gene of B. subtilis codes for a protein of 82 amino acid residues with a calculated molecular mass of 8.325 Da and a basic pI of 9.51. It shares about 57% amino acid identity with an analogous open reading frame of B. stearothermophilus. A database search revealed that more than 20 other microorganisms, including e.g., Bacillus halodurans (27), Bacillus anthracis, Clostridium acetobutylicum, and Staphylococcus aureus (8), also harbor the ybxF gene or an unnamed gene (open reading frame) with a high degree of sequence similarity to ybxF. All of these bacterial proteins have been tentatively classified as putative L7ae/L30e-like ribosomal proteins.
In contrast to the archeal/eukaryotic L7ae/L30e proteins, neither the cellular localization nor any function of YbxF has been reported to date. The presence of ybxF on the same transcript with genes for other ribosomal proteins suggests that it may be a ribosome-associated protein as well. However, the deduced amino acid sequence of the YbxF protein matches none of the amino acid sequences of the known ribosomal proteins of B. stearothermophilus or B. subtilis (2, 11, 23). Also, a previously published factorial correspondence analysis of the str operon genes argues against ybxF being a ribosomal protein (17).
In this report, we show that a fusion of green fluorescent protein (GFP) to YbxF localizes predominantly to ribosomes in log-phase B. subtilis cells. The specific localization to ribosomes appears to be a dynamic process because ribosomes isolated from stationary-phase cells displayed no fluorescence. Three-dimensional (3D) in silico modeling further confirms YbxF as a eubacterial L7ae/L30e homologue. Based on mutational analysis, we demonstrate that Lys24 is crucial for the ribosomal localization of YbxF. Finally, gene deletion experiments show that YbxF, unlike L30e, is not an essential protein.
MATERIALS AND METHODS
Bacterial strains.
All plasmid cloning was carried out in Escherichia coli DH5α. The B. subtilis strains used in this study are listed in Table 1.
TABLE 1.
B. subtilis strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| 168 | Wild type, prototroph | Laboratory stock |
| 168trp | trpC2 | Alison Hunt |
| ALMT | purB6 leuA8 metB5 thr-5 | 6 |
| ALMTstr | purB6 leuA8 metB5 thr-6 strA1 | 6 |
| 168 ΔybxF cat | ΔybxF Cmr | This study |
| 168 ΔybxF cat str | ΔybxF Cmr Strr | This study |
| ALMT ΔybxF cat | purB6 leuA8 metB5 thr-5 ΔybxF Cmr | This study |
| ALMT ΔybxF cat str | purB6 leuA8 metB5 thr-5 ΔybxF Cmr Strr | This study |
| Co1 | amyE::cat Cmr | This study |
| Co2 | purB6 leuA8 metB5 thr-5 amyE::cat Cmr | This study |
| Co3 | purB6 leuA8 metB5 thr-5 strA1 amyE::cat Cmr Strr | This study |
| 168 ΔymxC spc | ΔymxC Spcr | This study |
| 168 ΔybxF cat ΔymxC spc | ΔybxF Cmr ΔymxC Spcr | This study |
| I (ybxF+ybxF-gfp) | trpC2 ybxF+amyE::ybxF-gfp Spcr | This study |
| II (ΔybxF ybxF-gfp) | trpC2 ΔybxF CmramyE::ybxF-gfp Spcr | This study |
| III (ybxF+gfp) | trpC2 ybxF+amyE::gfp Spcr | This study |
| IV (ΔybxF gfp) | trpC2 ΔybxF CmramyE::gfp Spcr | This study |
| V (ΔybxF ybxF K17A-gfp) | trpC2 ΔybxF CmramyE::ybxF K17A-gfp Spcr | This study |
| VI (ΔybxF ybxF K21A-gfp) | trpC2 ΔybxF CmramyE::ybxF K21A-gfp Spcr | This study |
| VII (ΔybxF ybxF K24A-gfp) | trpC2 ΔybxF CmramyE::ybxF K24A-gfp Spcr | This study |
| Plasmids | ||
| pUC18 | Original vector used for knockout construct preparation (2,686 bp) | Commercial source |
| pDG1728 | Original vector used for spc gene PCR amplification (10,776 bp) | 7 |
| pCPP-31 | Original vector used for knockout construct preparation (5,620 bp) | 17 |
| pGEX-5X3 | Original vector used for knock-out construct preparation (4,974 bp) | Commercial source |
| pDG1662 | Integrational vector containing chloramphenicol acetyltransferase cassette (6,982 bp) | 7 |
| pYBXFK | pUC18 containing ybxF knockout construct (5,458 bp) | This study |
| pYBXFKs | pUC18 containing ybxF knockout construct − rpsL including the strA1 mutation (5,458 bp) | This study, reference 12 |
| pYMXCK | pGEX-5X3 containing the ymxC knockout construct (7,759 bp) | This study |
| pSG1154 | GFP-fusion vector containing the GFP cassette (7,600 bp) | 19 |
| pSG1154F11 | pSG1154 containing the ybxF gene (7,843 bp) | This study |
| pSG1154F11_K17A | pSG1154 containing the mutated ybxF gene (7,843 bp) | This study |
| pSG1154F11_K21A | pSG1154 containing the mutated ybxF gene (7,843 bp) | This study |
| pSG1154F11_K24A | pSG1154 containing the mutated ybxF gene (7,843 bp) | This study |
Media and transformation.
E. coli competent cells were prepared and transformations were carried out as described in reference 10. B. subtilis competent cells and transformation experiments were carried out as described in reference 1. Transformants were selected by plating onto LB agar plates supplemented with specific antibiotics (chloramphenicol, 5 μg/ml; streptomycin, 1 mg/ml; and spectinomycin, 100 μg/ml).
Media used for functional analysis of B. subtilis mutant cells were Spizizen minimal medium [composition per liter, 2 g (NH4)2SO4, 18.3 g K2HPO4·3H2O, 6 g K2HPO4, 1 g Na-citrate·2H2O, 0.2 g MgSO4·7H2O (plus tryptophan, final concentration of 50 μg/ml, for auxotrophic strains), various carbon sources; and Spizizen rich medium (composition per liter, 25 g tryptone, 5 g yeast extract, and minimal Spizizen medium).
Nucleic acid preparation and manipulation.
B. subtilis genomic DNAs were extracted and purified as described previously (21). Plasmid DNAs were prepared with the Plasmid Midi kit, and gel extractions were carried out with the gel extraction kit (both purchased from QIAGEN, Germany). Restriction mapping, agarose gel electrophoresis, and cloning of DNA fragments were performed by standard procedures (24). All constructs were verified by sequencing (Big Dye Terminator v3.1 cycle sequencing kit; Applied Biosystems).
Construction of ybxF knockout mutants.
The regions flanking ybxF (1,081 bp both upstream and downstream of ybxF) were PCR amplified using flanking primers K01E, K02, K16, and K04E (Table 2), and B. subtilis 168 genomic DNA. The cat reporter gene was amplified with PCR using primers K05 and K09 (Table 2), with pCPP31 plasmid DNA as a template. The primers contained, besides the target sequence, 20 nucleotides of a flanking sequence at their 5′ end for the subsequent annealing of the generated PCR fragments. The three PCR products were annealed and PCR amplified using terminal primers of the whole region (K01E and K04E). The product (2,772 bp) was cloned into pUC18, yielding pYBXFK and pYBXFKs (Table 1) Constructs were verified by sequencing. Plasmids pYBXFK and pYBXFKs were used to transform B. subtilis 168 and B. subtilis ALMT strains. ybxF-null mutants (168 ΔybxF cat, 168 ΔybxF cat str, ALMT ΔybxF cat, and ALMT ΔybxF cat str [see Table 1]) were selected on LB agar plates supplemented with chloramphenicol or with the combination of chloramphenicol and streptomycin. The colonies formed overnight and in similar numbers for all four genetic backgrounds as a control, excluding the possibility that the ΔybxF colonies grew because of an unknown suppressor. The authenticity of the integration event was confirmed by PCR and sequencing.
TABLE 2.
Primers used in this work
| Primer | Sequence 5′→3′a | Restriction enzyme |
|---|---|---|
| K01E | GGAATTCCCTCATGGTGTATGTAAACG | EcoRI |
| K02 | TTTGATATGCCTCCGAATTAACAGGTTTATTTCGAGCA | |
| K05 | CTCGAAATAAACCTGTTAATTCGGAGGCATATCAAATG | |
| K09 | GACGCAAAAACAAAAGTATATTATAAAAGCCAGTCATTAG | |
| K16 | GACTGGCTTTTATAATATACTTTTGTTTTTGCGTCAG | |
| K04E | GGAATTCCCTTCCTTATTAGGAAATTG | EcoRI |
| ybxFGFP for | GGGGTACCTCTTATGATAAAGTATCACAG | KpnI |
| ybxFGFP rev | GGGAATTCTAAAATAATGGCAACAGCGGC | EcoRI |
| Adaggio | CGGGATCCGCACAGATGATCCTGACG | BamHI |
| Moderato | CCCAAGCTTGAAACCATTCCATTCCAGAC | HindIII |
| Largo | CCGCTCGAGGAATAAGCTGATCAGCTTGC | XhoI |
| Cappricio | ATAAGAATGCGGCCGCCTTCATTATCAGGCTCTTCG | NotI |
| Spc for | GTCAGTAACTTCCACAGTAGTTC | |
| Spc rev | GGATATCGTGTTTCCACCATTTTTTC | EcoRV |
| lys17 for | ATTATTGGTACGGCGCAAACAGTG | |
| lys17 rev | CACTGTTTGCGCCGTACCAATAAT | |
| lys21 for | CAAACAGTGGCAGCTCTAAAGCG | |
| lys21 rev | CGCTTTAGAGCTGCCACTGTTTG | |
| lys24 for | GTGAAAGCTCTAGCGCGAGGTTCAG | |
| lys24 rev | CTGAACCTCGCGCTAGAGCTTTCAC |
Restriction enzyme portions of the sequences are underlined.
Functional analysis of the ybxF knockout mutants.
The growth (lag phase, growth rate, and maximum optical density at 600 nm [OD600]) of the mutants was tested in comparison with that of wild-type control strains Co1 to Co3 (Table 1) at 20°C, 37°C, and 45°C in rich and minimal Spizizen media (both on agar plates and in liquid media). Sporulation was tested as described by Schumann et al. (25). Sensitivity to inhibitors was tested on agar plates supplemented with various concentrations of ethanol (concentration range, 8 to 16%) or 1 M NaCl or sodium dodecyl sulfate (SDS; concentration range, 0.007 to 0.1%). Minimal inhibitory concentrations of streptomycin were also determined (1 mg for sensitive strains and 3 mg for resistant strains), with no difference between wild-type and mutant strains.
Construction of ybxF-gfp fusion strains.
The ybxF gene was prepared with PCR using primers ybxFGFP for and rev (Table 2) from B. subtilis 168 DNA and cloned into KpnI and EcoRI sites of pSG1154 (19) to fuse it to the 5′ end of gfp, resulting in pSG1154F11. Its identity was verified by sequencing. The construct was used to transform B. subtilis wild-type and ΔybxF strains. Transformants were selected on LB agar plates containing spectinomycin for the wild-type transformation (strain I [ybxF+ ybxF-gfp]) or a combination of chloramphenicol and spectinomycin for the ΔybxF transformation (strain II [ΔybxF ybxF-gfp]). The integration at the amyE locus was verified by a standard procedure (19) and by sequencing.
Strains III (ybxF+ gfp) and IV (ΔybxF gfp) were prepared by transforming B. subtilis 168 and B. subtilis 168 ΔybxF cat with plasmid pSG1154. Selection of transformants was performed as described above. The authenticity of the integration event was confirmed by PCR and sequencing.
Construction of ymxC knockout mutants.
The spc gene and the part preceding this gene (1,249 bp, including polylinker) were isolated from plasmid pDG1728 (PCR, primers Spc for and Spc rev [Table 2]) and cloned into pGEX-5X3 (BamHI plus EcoRV sites), yielding pGEX-5X3-SPC-1. The region preceding the gene ymxC (terminal part of the nusA gene and the ymxB gene; 717 bp) was prepared with PCR (primers Adaggio and Moderato [Table 2]) and cloned into pGEX-5X3-SPC-1 (BamHI plus HindIII sites), yielding pGEX-5X3-SPC-2. The region downstream from the ymxC gene (beginning of the infB gene; 684 bp) was prepared by PCR (primers Largo and Cappricio [Table 2]) and cloned into pGEX-5X3-SPC-2 (XhoI plus NotI sites), yielding pYMXCK (Table 1). Its identity was verified by sequencing. This plasmid was used to transform B. subtilis 168 and B. subtilis 168 ΔybxF cat, yielding B. subtilis 168 ΔymxC spc and B. subtilis 168 ΔybxF cat ΔymxC spc (Table 1). PCR and sequencing were used to verify the successful knockout of the ymxC gene.
Preparation of crude ribosomes.
One half of the volume of B. subtilis strains grown at 37°C in LB, supplemented with 1% xylose, was collected at an OD600 of ∼1.0 (log phase bacteria), and cells of the second half of the volume were left to grow overnight (stationary-phase bacteria, ∼10 h after the end of logarithmic phase) before collection. The cells were washed with buffer S (10 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 60 mM NH4Cl, 0.5 mM EDTA, 5 mM 2-mercaptoethanol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride) and rapidly frozen. All further operations were carried out at 4°C. The frozen cells (∼1 g) were suspended in buffer S and disrupted by sonication. After the removal of cell debris by centrifugation at 30,000 × g, the supernatant was centrifuged at 285,000 × g for 120 min. The sediment was dissolved in buffer S, aggregates and pigment (developed in stationary-phase bacteria) were removed by low-speed centrifugation, and 2-ml aliquots of the supernatant were layered onto 8 ml of buffer S containing a 30% (wt/vol) sucrose bed and centrifuged at 190,000 × g overnight. The sediment of ribosomes was dissolved in buffer S containing 1 M NH4Cl, aggregates were removed by low-speed centrifugation, and the supernatant was centrifuged at 285,000 × g for 3 h. Finally, the sediment of ribosomes was resuspended in buffer S, aggregates were removed by low-speed centrifugation at 20,000 × g for 20 min, and the supernatant containing ribosomes was divided into aliquots and stored at −70°C.
Ribosome dissociation into 30S and 50S subunits.
B. subtilis cells (log-phase bacteria), prepared as described in the above paragraph, were suspended in buffer S, disrupted by sonication, and centrifuged at 30,000 × g for 20 min to remove cell debris. The supernatant (0.2 ml) was subjected to 10 to 25% sucrose density gradient centrifugation in the presence of 0.3 mM Mg2+ using an SW41 Ti rotor in a Beckman Optima L-90K ultracentrifuge at 248,000 × g for 210 min. Sucrose gradients were divided into 15 fractions. After fractionation, absorbance at 260 nm of each fraction was used to determine the amounts of ribosomes.
Detection of YbxF-GFP fusion protein.
Crude ribosomes and ribosome fractions from sucrose gradient fractionations were precipitated with trichloroacetic acid (5% final concentration). The precipitates were dissolved in buffer A (1 ml 0.5 M Tris-HCl [pH 6.8], 0.8 ml glycerol, 1.6 ml 10% sodium dodecyl sulfate [SDS], 0.2 ml 0.05% bromphophenol blue, 1.2 ml 2-mercaptoethanol, 3.2 ml dH2O), separated by SDS-polyacrylamide gel electrophoresis on 12% gel (18), and analyzed by immunoblotting with anti-GFP antibody followed by horseradish peroxidase-conjugated goat anti-rabbit antibody (both Santa Cruz Biotechnology) and Western blotting chemiluminescent substrate detection system (Pierce).
Microscopy.
Fluorescence microscopy of 150-μl solutions of ribosomes of identical concentrations in Corning opaque 96-well plates (Sigma) was performed using a Leica Fluo III fluorescence stereomicroscope equipped with an Olympus C5050 digital camera and a GFP2 filter. Relative fluorescence was quantified with the ImageJ 1.34s program.
Fluorescence microscopy of B. subtilis cells was performed with an Olympus IX81 Cell-R system equipped with a Hamamatsu Orca-ER digital camera.
Modeling.
Sequences and coordinates of crystal structures of proteins used as templates for homology modeling were obtained from the Protein Data Bank (PDB code 1NMU chain D—yeast L30e; PDB code 1YSH chain C—wheat germ L30e; PDB code 1PXW chain A—Pyrococcus abyssii L7ae). Sequence alignments were created using the “align2d” module from the Modeler software package. Homology models were constructed using the “model” module from the Modeler software package.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out using mutagenic PCR as described in the QuikChange site-directed mutagenesis kit protocol from Stratagene. Plasmid pSG1154F11 was used as a template for PCRs. Primers carried nucleotide substitutions to introduce Ala at the desired positions (lys17 for and rev, lys21 for and rev, and lys24 for and rev [see Table 2]). After the amplification of the whole plasmid DNA, the template plasmid was digested by DpnI endonuclease (digesting methylated DNA only). The reaction mixture was used to transform E. coli followed by plasmid isolation and sequencing. The authenticity of the integration into B. subtilis chromosome at the amyE locus was verified by a standard procedure (19) and also by sequencing.
RESULTS AND DISCUSSION
Characterization of ybxF deletion strains.
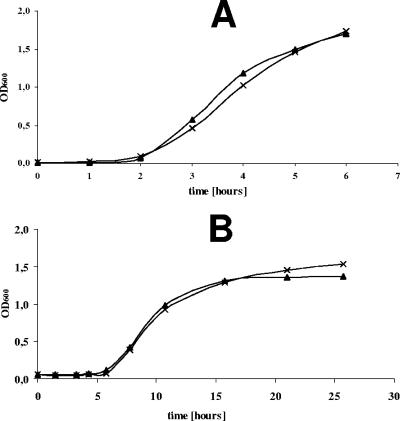
As a first approach to characterize YbxF's cellular functions, we deleted the ybxF gene from the chromosome of four different B. subtilis strains (for details, see Materials and Methods). The deletion of ybxF had no effect on viability and/or growth of the strains. This was proved by probing the ΔybxF strains in several functional assays; e.g., by determining the generation times and sporulation efficiencies at optimal, lowered, and elevated temperatures in rich and minimal media. Sensitivity to some inhibitors was also assayed. Figure 2 shows representative growth curves of wild-type and ΔybxF strains grown in rich and minimal media at 37°C. The ΔybxF strains did not differ from the wild-type parent strains in any respect.
FIG. 2.
Growth curves of B. subtilis ΔybxF (×) and wild-type (▴) strains (Table 1) in Spizizen rich (A) and Spizizen minimal (B) media with incubation at 37°C.
ymxC—a ybxF paralogue.
The ybxF gene has a paralogous counterpart in the B. subtilis genome, the ymxC gene (22% amino acid identity with YbxF). It is situated in the nusA-infB operon, and its function is unknown (26). Similarly as with ybxF, the deletion of ymxC had no effect on B. subtilis viability and/or growth. The double deletion strain (ΔybxF ΔymxC) was also viable and growing as well as the wild-type strain (data not shown).
Localization of YbxF.
To determine the localization and distribution of the YbxF protein in the B. subtilis cell, the ybxF gene was fused to the gfp gene, coding for GFP (for details, see Materials and Methods). The xylose-inducible fusion was integrated at the amyE site of the bacterial chromosome of both wild-type B. subtilis and ΔybxF B. subtilis strains, yielding strains I (ybxF+ ybxF-gfp) and II (ΔybxF ybxF-gfp). As controls, the same B. subtilis parent strains were transformed with constructs carrying gfp alone to monitor the distribution of free GFP in the cell (strains III [ybxF+ gfp] and IV [ΔybxF gfp]; see Table 1).
Cells producing both GFP alone and GFP fused to YbxF grew with generation times comparable to those of wild-type cells, and all emitted green fluorescence when illuminated by blue light. This indicated that the production of either GFP alone or YbxF-GFP fusion had no adverse effect on B. subtilis growth and viability. The ybxF-gfp fusion's integrity was verified by Western blotting (Fig. 3).
FIG. 3.
Western blot analysis of the B. subtilis sonicated cells of strain II (ΔybxF ybxF-gfp), IV (ΔybxF gfp), and 168 (ybxF+ parent strain [P]). M, marker.
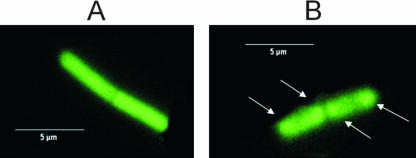
The fluorescent signals from cells growing in the log phase were stronger than those from the stationary-phase cells. No fluorescence was emitted from B. subtilis spores (data not shown). Fluorescence of the free GFP was distributed uniformly throughout the cell both in the logarithmic and stationary phases (Fig. 4A), in agreement with Mascarenhas (22). In contrast, the distribution of fluorescent signals from YbxF-GFP was not uniform in the exponentially growing strain II cells but localized predominantly toward the cell poles (Fig. 4B). Such bipolar distribution is characteristic for B. subtilis ribosomes due to their exclusion from the central area occupied by the nucleoid (13, 20, 22). The stationary-phase bacteria did not show this pattern.
FIG. 4.
GFP fluorescence of B. subtilis cells. Exponentially growing cells of strain IV (ΔybxF gfp) expressing GFP only (A) and exponentially growing cells of strain II (ΔybxF ybxF-gfp) expressing the YbxF-GFP fusion protein (B). Arrows indicate the GFP signal localized predominantly toward the cell poles.
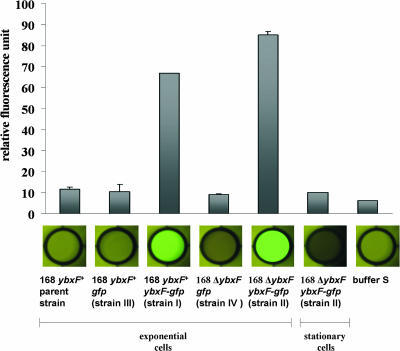
To examine whether YbxF is associated with ribosomes, ribosomes from cells of strains I to IV from log and stationary phases were purified and examined for green fluorescence. As shown in Fig. 5, only ribosomes isolated from strain I (ybxF+ ybxF-gfp) and strain II (ΔybxF ybxF-gfp) and harvested in log phase retained strong fluorescence after all washing and centrifugation procedures described in Materials and Methods, suggesting that YbxF is a ribosomal component. The fluorescent signal of ribosomes isolated from strain II (ΔybxF ybxF-gfp), lacking the wild-type ybxF gene, was stronger than that of ribosomes isolated from strain I (ybxF+ ybxF-gfp) with the original ybxF gene preserved (Fig. 5). The decreased fluorescence of strain I ribosomes (ybxF+ ybxF-gfp) was apparently the result of a competition between YbxF and YbxF-GFP for the same ribosomal binding site. The competitive effect of intact YbxF was rather low, in agreement with a high level of YbxF-GFP production.
FIG. 5.
GFP fluorescence of the B. subtilis ribosomes. Ribosomes were isolated from exponentially growing cells; GFP fluorescence of strain II (ΔybxF ybxF-gfp) ribosomes isolated from the stationary phase is also shown.
In a control experiment, ribosomes prepared according to the same protocol from logarithmically growing strains III (ybxF+ gfp) and IV (ΔybxF gfp), producing free GFP, were not fluorescent, excluding the possibility that YbxF binds to ribosomes via GFP and not by itself (Fig. 5).
The fluorescence of strain I (ybxF+ ybxF-gfp) and strain II (ΔybxF ybxF-gfp) ribosomes was no longer detectable when they were isolated from stationary-phase bacteria (Fig. 5), indicating that although the cells contain YbxF-GFP, this protein disappears from the ribosome during stationary phase. A similar growth phase-dependent association with the ribosome has also been reported recently for some other ribosomal proteins (23). Its physiological importance is poorly understood.
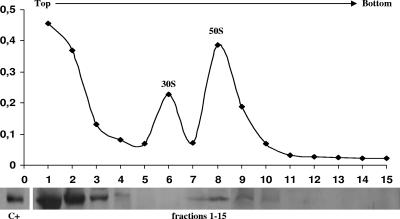
To identify to which ribosomal subunit YbxF binds, centrifugation of log-phase cell lysates of strain II (ΔybxF ybxF-gfp) was carried out in a sucrose density gradient in the presence of a low (0.3 mM) concentration of Mg2+ ions to dissociate ribosomes into 50S and 30S subunits. The majority of the cellular YbxF-GFP fusion protein was detected in the top supernatant fractions and a small amount in the 50S subunit fractions but not in the 30S subunit fractions (Fig. 6).
FIG. 6.
Ribosomal subunit profile and Western blot analysis of ribosome fractions of exponentially growing strain II (ΔybxF ybxF-gfp) using GFP antibody. C+, ribosomes of strain II (ΔybxF ybxF-gfp) isolated from exponentially growing cells.
3D structure of YbxF.
To address whether YbxF may be the bacterial counterpart of archeal/eukaryotic L7ae/L30e ribosomal proteins, in silico 3D modeling of B. subtilis YbxF was conducted using the available crystal structures of yeast L30e (4), wheat germ L30e (9), and Pyrococcus abyssii L7ae (5) as templates. The amino acid identities of yeast L30e, wheat germ L30e, and P. abyssii L7ae with B. subtilis YbxF are 19.5%, 23%, and 40%, respectively.
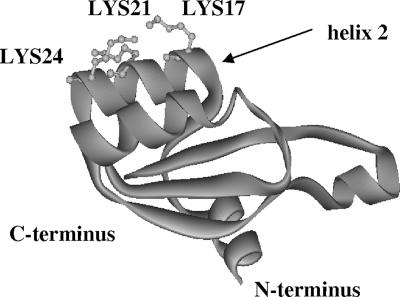
All three YbxF models, irrespective of the templates used, are highly similar in the overall structure and in some areas in particular. This is not surprising because the 3D structures of the three templates are already quite similar (not shown, PDB entries 1NMU, 1YSH, and 1PXW). All proteins have a hydrophobic core consisting of a four-stranded β-sheet surrounded by four α-helices. The second α-helix from the N-terminus deserves particular attention. It is composed of 12 amino acid residues with glycines at either end, giving the helix flexibility. The helix is the site of the highest amino acid identity in all four proteins. In wheat L30e and Haloarcula marismortui L7ae, the corresponding second α-helix, designated α-2, was identified as one of the most important interaction sites with the ribosome (3, 9, 15).
Helix 2 of YbxF has three basic amino acid residues (lysines 17, 21, and 24) facing the solvent (Fig. 7). Database searches have confirmed that the positively charged residues within helix 2 are highly conserved among YbxF (K17-K21-K24) and L30e (K-K-R) proteins identified to date (compare to the L7ae N34-K38-E41 motif). L30e and L7ae utilize the first basic amino acid residue of helix 2 to contact rRNA/mRNA. Residues corresponding to YbxF K21/K24 are directed toward the space occupied by proposed “unknown protein cluster II” (L30e PDB entry 1YSH) or ribosomal protein L15e (L7ae PDB entry 1S72).
FIG. 7.
3D homology model of YbxF obtained using as a template wheat germ L30e (PDB entry 1YSH, chain C). Basic amino acid residues of the second α-helix are indicated.
Site-directed mutagenesis.
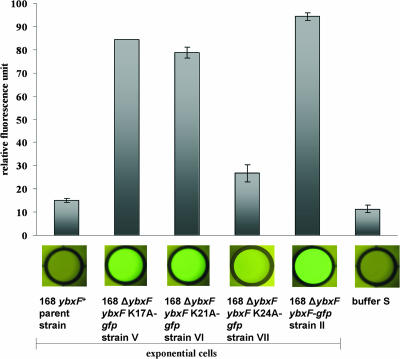
To test the importance of lysines 17, 21, and 24 for YbxF′s interaction with the ribosome, we created three ybxF-gfp constructs carrying single substitutions of the lysines for alanines: strains V (ΔybxF ybxF K17A-gfp), VI (ΔybxF ybxF K21A-gfp), and VII (ΔybxF ybxF K24A-gfp) (see Table 1).
We measured green fluorescence of ribosomes isolated from exponentially growing strains V to VII and strain II (a positive control). We observed strongly decreased fluorescence of ribosomes isolated from exponentially growing cells of strain VII (ΔybxF ybxF K24A-gfp) compared to the green fluorescence of ribosomes of strain II (ΔybxF ybxF-gfp) (Fig. 8). This finding suggests an important role of lysine 24 for the binding of YbxF to the ribosome. Green fluorescence of ribosomes isolated from strains V (ΔybxF ybxF K17A-gfp) and VI (ΔybxF ybxF K21A-gfp) was decreased only slightly relative to the positive control of strain II (ΔybxF ybxF-gfp), suggesting a less prominent role for the interaction of lysines 17 and 21. This result was confirmed by Western blot analysis of ribosomes prepared according to the same protocol, using the anti-GFP antibody (data not shown). In control experiments, expression levels of the three mutated YbxF-GFP fusion proteins (strains V, VI, and VII) were compared with expression levels of wild-type YbxF-GFP (strain II) to verify that the observed differences in binding of the proteins to the ribosome were not due to differences in their expression levels. Quantification of the YbxF-GFP levels in all four strains (analyzed by Western blotting of bacterial extracts using anti-GFP antibody) did not reveal any differences in YbxF-GFP expression levels (data not shown).
FIG. 8.
GFP fluorescence of B. subtilis ribosomes isolated from exponentially growing cells of the strains with mutated YbxF protein.
Concluding remarks.
Although L30e and L7ae are highly homologous in structure and the RNA kink-turn motif they recognize, they bind to different sites on the ribosome (3, 9). L30e was found to localize close to the ribosome subunit interface, contacting both ribosomal subunits (9). L7ae was reported to bind to the ribosomal 50S subunit (3). YbxF binds to the 70S ribosome, interacting with the 50S subunit. L30e proteins have a basic pI of ∼9. L7ae proteins are acidic (pI ∼4). YbxF′s pI is 9.51, and it does not bind to ribosomes from the stationary phase of growth. L30e is an essential protein (9), and YbxF is not. The function of L7ae has not been defined yet. This current state of knowledge does not allow us to decide whether YbxF is a bacterial counterpart of L30e or L7ae. We can hypothesize that L30e has become more important during evolution. Since the cellular roles of L30e and L7ae are still not understood, YbxF may also be a useful tool for comparative studies.
Acknowledgments
This work was supported by grant no. A5052206 from the Grant Agency of the Academy of Sciences of the Czech Republic and by project no. AVOZ 50520514 awarded by the Academy of Sciences of the Czech Republic.
We thank Leoš Valášek and Bela Szamecz for help with sucrose density gradient centrifugation, Ondřej Tolde and Ondřej Šebesta for help with microscopy, Hana Šanderová for discussion, and Zoltán Ferjentsik for technical assistance.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1960. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, E., T. Scholzen, W. Kromer, T. Hatakeyama, and M. Kimura. 1991. Primary structures of ribosomal proteins from the archaebacterium Halobacterium marismortui and the eubacterium Bacillus stearothermophilus. Biochimie 73:657-668. [DOI] [PubMed] [Google Scholar]
- 3.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 4.Chao, J. A., G. S. Prasad, S. A. White, C. D. Stout, and J. R. Williamson. 2003. Inherent protein structural flexibility at the RNA-binding interface of L30e. J. Mol. Biol. 326:999-1004. [DOI] [PubMed] [Google Scholar]
- 5.Charron, C., X. Manival, B. Charpentier, C. Branlant, and A. Aubry. 2004. Purification, crystallization and preliminary X-ray diffraction data of L7Ae sRNP core protein from Pyrococcus abyssii. Acta Crystallogr. D 60:122-124. [DOI] [PubMed] [Google Scholar]
- 6.Fučík, V., H. Grünerová, and S. Zadražil. 1982. Restriction and modification in Bacillus subtilis 168. Mol. Gen. Genet. 186:118-121. [DOI] [PubMed] [Google Scholar]
- 7.Guérout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 8.Gupta, R. S. 1998. What are archaebacteria: life′s third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Mol. Microbiol. 29:695-707. [DOI] [PubMed] [Google Scholar]
- 9.Halic, M., T. Becker, J. Frank, C. M. T. Spahn, and R. Beckmann. 2005. Localization and dynamic behavior of ribosomal protein L30e. Nat. Struct. Mol. Biol. 12:467-468. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Herwig, S., V. Kruft, and B. Wittmann-Liebold. 1992. Primary structures of ribosomal proteins L3 and L4 from Bacillus stearothermophilus. Eur. J. Biochem. 207:877-885. [DOI] [PubMed] [Google Scholar]
- 12.Hosoya, Y., S. Okamoto, H. Muramatsu, and K. Ochi. 1998. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 42:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt, A., J. P. Rawlins, H. B. Thomaides, and J. Errington. 2006. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 152:2895-2907. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, T., K. Takemoto, H. Mori, and T. Gojobori. 1999. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 16:332-346. [DOI] [PubMed] [Google Scholar]
- 15.Klein, D. J., P. B. Moore, and T. A. Steitz. 2004. The roles of ribosomal proteins in the structure, assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340:141-177. [DOI] [PubMed] [Google Scholar]
- 16.Koonin, E., and M. Y. Galperin. 1997. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr. Opin. Genet. Dev. 7:757-763. [DOI] [PubMed] [Google Scholar]
- 17.Krásný, L., T. Vacík, V. Fučík, and J. Jonák. 2000. Cloning and characterization of the str operon and elongation factor Tu expression in Bacillus stearothermophilus. J. Bacteriol. 182:6114-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101-109. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, P. J., S. D. Thaker, and J. Errington. 2000. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marmur, J. 1961. A procedure for isolation of deoxyribomucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 22.Mascarenhas, J., M. H. Weber, and P. L. Graumann. 2001. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2:685-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanamiya, H., G. Akanuma, Y. Natori, R. Murayama, S. Kosono, T. Kudo, K. Kobayashi, N. Ogasawara, S. M. Park, K. Ochi, and F. Kawamura. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52:273-283. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Schumann, W., S. D. Ehrlich, and N. Ogasawara. 2001. Functional analysis of bacterial genes. John Wiley and Sons. Ltd., New York, NY.
- 26.Shazand, K., J. Tucker, M. Grunberg-Manago, J. C. Rabinowitz, and T. Leighton. 1993. Similar organization of the nusA-infB operon in Bacillus subtilis and Escherichia coli. J. Bacteriol. 175:2880-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takami, H., Y. Takaki, K. Nakasone, C. Hirama, A. Inoue, and K. Horikoshi. 1999. Sequence analysis of a 32-kb region including the major ribosomal protein gene clusters from alkaliphilic Bacillus sp. strain C-125. Biosci. Biotechnol. Biochem. 63:452-455. [DOI] [PubMed] [Google Scholar]