Abstract
The branching order and coherence of the alphaproteobacterial orders have not been well established, and not all studies have agreed that mitochondria arose from within the Rickettsiales. A species tree for 72 alphaproteobacteria was produced from a concatenation of alignments for 104 well-behaved protein families. Coherence was upheld for four of the five orders with current standing that were represented here by more than one species. However, the family Hyphomonadaceae was split from the other Rhodobacterales, forming an expanded group with Caulobacterales that also included Parvularcula. The three earliest-branching alphaproteobacterial orders were the Rickettsiales, followed by the Rhodospirillales and then the Sphingomonadales. The principal uncertainty is whether the expanded Caulobacterales group is more closely associated with the Rhodobacterales or the Rhizobiales. The mitochondrial branch was placed within the Rickettsiales as a sister to the combined Anaplasmataceae and Rickettsiaceae, all subtended by the Pelagibacter branch. Pelagibacter genes will serve as useful additions to the bacterial outgroup in future evolutionary studies of mitochondrial genes, including those that have transferred to the eukaryotic nucleus.
The Alphaproteobacteria are a diverse class of organisms within the phylum Proteobacteria, with many important biological roles. They frequently adopt an intracellular lifestyle as plant mutualists or plant or animal pathogens (5). This has led to independent paths of genome reduction in several alphaproteobacterial lineages, but lineage-specific genome expansions are also apparent, with some genomes divided among multiple replicons that can include linear chromosomes (9). The Alphaproteobacteria include the most abundant of marine cellular organisms (20). A variety of metabolic strategies are found in the class, including photosynthesis, nitrogen fixation, ammonia oxidation, and methylotrophy. Stalked, stellate, and spiral morphologies are found. Developmental programs occur that switch between cell types, controlled by a web of regulatory systems (31).
Special interest attaches to the Alphaproteobacteria as the ancestral group for mitochondria. The Rickettsiales are most often cited as the alphaproteobacterial subgroup from which mitochondria arose, but there has been disagreement on this point (17, 19, 32). Proper placement of the mitochondrial branch on the alphaproteobacterial species tree benefits the study of eukaryotic nuclear genomes, which house many genes of mitochondrial origin.
Improvement in phylogenetic tree reconstruction can come from increasing the number of characters used, which whole-genome sequences have provided in abundance. However, even long character matrices can produce artifacts, for example, when taxon sampling is limited. Recently, the number of completely or nearly completely sequenced alphaproteobacterial genomes has become large, allowing for the assembly of a carefully selected character matrix that is both long and broad, in turn enabling robust phylogenetic inference. With such a matrix, we have generated a species tree for these bacteria, into which we have placed the mitochondrial branch. The reliability of the tree is indicated not only by its high Bayesian and bootstrap support values, which are generally very high for such large matrices, but by the agreement of maximum-likelihood (ML) and Bayesian methods, by convergence to this same tree for subsets of the full protein set, and by its good agreement with the highly supported bipartitions among the single-protein trees.
MATERIALS AND METHODS
Organisms.
We chose 72 alphaproteobacteria, 8 outgroup proteobacteria, and 8 mitochondria for study (see Table S1 in the supplemental material). This included all alphaproteobacterial strains with a completed genome sequence, or with an incomplete genome sequence in fewer than 100 contigs, that were available with annotation at NCBI on 1 May 2006. Maricaulis maris was included later due to special interest in the Hyphomonadaceae. Two strains, Agrobacterium tumefaciens C58 and Ehrlichia ruminantium Welgevonden, had each been sequenced in two independent projects; in these cases, we used data only from the project that had annotated the largest number of proteins (by the University of Washington and CIRAD, respectively). Because a recent genome-based study (13) including 63 proteobacteria showed that the sister group of the Alphaproteobacteria was the combined Betaproteobacteria and Gammaproteobacteria, seven outgroup species with completed genomes were chosen from these two classes, and an eighth outgroup species was selected from the Deltaproteobacteria, Geobacter sulfurreducens, which appeared to be the least derived of all Proteobacteria in that study. We included eight mitochondrial genomes that have been deemed primitive in various studies (summarized in reference 21) from two alveolates, four streptophytes, and two chlorophytes.
Key programs for phylogenetic analysis.
Unless otherwise stated, the following programs were employed. MUSCLE (15) was used with up to 100 iterations for sequence alignment. Gblocks (11) was used for masking gapped and other noisy portions of the alignments, with a minimum block length of 10 amino acids and gaps allowed in any alignment position for no more than half of the sequences. We performed ML phylogenetic analysis with PHYML v2.4.4 primed with the BIONJ tree (22), using the Whelan and Golding (WAG) amino acid substitution matrix, estimating all parameters, and using four substitution rate categories. We performed Bayesian phylogenetic analysis using MrBayes v3.1.2 (28) in model-jumping mode with a single chain and primed with the BIONJ tree, assessing burn-in (arrival at a likelihood plateau) as described previously (7).
rRNA gene sequences and trees.
The 16S and 23S rRNA gene sequences of the bacteria under study were identified using author annotations or homology searching and were trimmed or extended so that their endpoints corresponded to those of the E. coli RNAs. The initial trees showed that multiple versions of an rRNA gene within any genome were more closely related to each other than to those of any other genome (except for 23S rRNA gene sequences in Brucella melitensis 16 M and Brucella melitensis Abortus), so single representative 16S and 23S rRNA genes were taken from each genome. The rRNA gene sequences from all study genomes were aligned and guided by secondary structure, using ClustalW with a profile. Seed profiles came from structure-based prealignments: a 16S alignment for 59 ingroup and 8 outgroup species from the Ribosomal Database Project II (RDP) release 9.39 (14) and a 23S alignment for 20 species from the comparative RNA website (10). Masking by Gblocks (with a minimum block length of 5 nucleotides) left 1,337 and 2,449 positions for 16S and 23S sequences, respectively. The concatenation of these two masked rRNA alignments was also prepared.
Relationships among aligned sequences were examined by ML and Bayesian phylogeny inference analyses using the Hasegawa-Kishino-Yano substitution matrix. ML analysis employed 100 bootstrap resamplings. We employed Bayesian analysis with two MrBayes runs, each with four chains for 100,000 generations, taking the best (highest likelihood) and the consensus of trees after burn-in within 10,000 to 15,000 generations.
Bacterial protein families and trees.
Homology groups suitable for multiprotein phylogenetic analysis were identified using the GeneTrees database (29), which at the time covered 14 alphaproteobacteria and 254 other prokaryotes. Searching this large set of protein trees for subtrees spanning all 14 alphaproteobacteria and containing no more than four additional taxa yielded 216 seed groups. Hidden Markov models were built for each seed group (4) and used to search the full protein sets for all genomes under study. We filtered the resulting gene families, rejecting those missing an ortholog in more than four ingroup strains, those with more than four strains with multiple members, and those with the outgroup widely dispersed upon visual inspection of the initial trees. This resulted in a set of 115 homology groups that approximated the ideal of representing each strain once (see Table S2 in the supplemental material). TBLASTN helped to find 8 missing proteins that were previously unannotated and helped to reconstruct 22 missing or incomplete proteins that had a putative single-nucleotide frameshifting sequencing error.
Because the criteria for determining N termini can vary among genome projects, we used conservation as a criterion for their revision. All protein sequences were extended in the N-terminal direction until blocked by an in-frame stop codon. The N-extended sequences were aligned and masked. We used the N-terminal conserved sequence block to trim the extended sequences, identifying 225 genes in which the true N terminus may be further upstream than the annotated one; these trimmed N-extended sequences were used in further analysis.
The integrity of the 115 families with respect to possible paralogs was reexamined by first allowing each family to repopulate even with second-best BLAST hits. Each family member protein was used as a BLASTP query against the whole set of proteins for each strain. Any protein that was either a best hit for a query or a second-best hit with a bit score at least 80% of that of the best hit for that strain was added to the protein family. For the ingroup, this analysis left 80 families with no paralogs, 27 slightly expanded families (1 to 8 paralogs), and 8 greatly expanded families (41 or more paralogs). The sequences of the expanded families were aligned and masked, and the resulting neighbor-joining (BIONJ) trees were rooted with outgroup species. For the slightly expanded families, the cases of multiple-ortholog candidates for a strain were resolved by retaining the candidate with the shortest distance to the shared node if this distance was at least 20% shorter than any other; otherwise, no candidate was retained for that family. This simple technique always identified the candidate that best fit our emerging picture of the species tree and, for ingroup strains, resulted in 50 retentions of the original family member, five replacements of the original, and nine ambiguous cases in which neither candidate was retained. Among the greatly expanded families, the tree for one of them could be resolved into two subtrees corresponding to the σ32 and σ70 RNA polymerase subunit families and a third small group that was rejected. The trees for the other seven greatly expanded families could not be readily resolved, and these families were entirely rejected.
The tests of Novichkov et al. (25) were applied to identify protein families that might follow evolutionary models other than vertical inheritance. Distance scores were taken for each family alone and for the concatenation of all 109 remaining families, using TreePuzzle with the WAG amino acid substitution model and a setting of 1.0 for the shape parameter of the gamma distribution. As a tentative species tree, the neighbor-joining tree was prepared for the concatenated alignment by using PHYLIP with jumbling (18). For each family, distances were fit to each of three models: (i) no relationship between family member distances and species distances (noise), (ii) proportionality between family member distances and species distances (vertical inheritance), and (iii) a model for a single horizontal transfer evaluated for each branch of the tentative species tree. Five families were eliminated by these tests because they did not reach the critical value of the F distribution (95% confidence) for rejection of the noise model. The ability of this method to detect horizontal transfer within the Alphaproteobacteria was tested by switching a single protein within a family; not all such artificial transfers were detected.
The remaining 104 families closely approached 100% representation by the genomes, reaching 99.8% for the ingroup and 96.7% for the outgroup (see Table S2 in the supplemental material). No ingroup strain was missing more than 2 of the 104 families (see Table S1 in the supplemental material).
Ten amino acid substitution models were tested, and WAG was found to be ideal or nearly so for each family. For each masked alignment, two independent MrBayes runs proceeded in the model-jumping mode for 500,000 generations primed with the neighbor-joining tree for the same family. Most families (85) used the WAG matrix exclusively, and for four more families, WAG was preferred in more than 75% of the post-burn-in trees. For the remaining 15 families, which preferred a variety of other models, ML trees were generated with either the preferred substitution model or WAG. The log likelihood for the WAG-constrained ML tree was lower than for the preferred-model ML tree, on average only by 0.24% and by no more than 1.0%, justifying an expedient of the fixed use of the WAG substitution model in further ML analyses of these families.
Sets of families.
Several subsets of the 104 alignments were selected: the group of 16 protein families found in mitochondria (see below), 26 mutually exclusive groups of 4 randomly selected proteins, 5 mutually exclusive groups of 10 randomly selected proteins, 4 mutually exclusive groups of 26 randomly selected proteins, and the 5 groups based on similar alpha-versus-Pinvar values (see below). Alignments were concatenated for each subset, and BIONJ trees were prepared and used to prime Bayesian and ML analyses. A single MrBayes run proceeded for 200,000 generations, reaching burn-in at 10,000 to 80,000 generations.
To further explore the validity of concatenating protein alignments, parameters were evaluated for the plateau set of trees from a WAG-constrained MrBayes run for each family. In particular, the shape of the gamma distribution (alpha) had a 3.3-fold range (0.76 to 2.52) and the proportion of invariant sites (Pinvar) had a 15.6-fold range (0.016 to 0.25). Families were segregated into five partitions containing from 7 to 40 proteins that fell into approximately equal-sized areas in an alpha-versus-Pinvar plot.
Full concatenated alignment.
For the full concatenation of all 104 alignments, a neighbor-joining tree was determined using PHYLIP with jumbling. This was used to prime a MrBayes run of 100,000 generations for the partitioned alignment, unlinking the statefreq, alpha, and Pinvar parameters for the partitions; the topology reached by 8,180 generations was unchanged by the end of the run. A second MrBayes run for 100,000 generations with the unpartitioned alignment was primed with the ML tree for the concatenated rRNAs; the topology reached by 5,480 generations was identical to that of the previous run and was unchanged by the end of the run. Fifty bootstrap resamplings of the full concatenation were generated and subjected to ML analysis primed with the neighbor-joining tree. The extended majority-rule consensus tree matched the topology of the two MrBayes runs.
To examine concordance, the 5,320 highly supported bipartitions (found in ≥95% of post-burn-in trees) from each of the 106 single-gene (104 proteins and two rRNAs) MrBayes analyses were identified. All trees generated in the course of this work, in ML bootstrap and MrBayes runs for single genes and concatenations, comprising at least 2,200 different topologies, were evaluated by taking the bipartitions of the tree and counting the number of matches to the highly supported bipartitions from the single-gene trees.
Links between nodes on the species tree.
We used the EEEP program (6) to determine, when possible, the minimal edit paths between each MrBayes consensus protein tree (considering only nodes with ≥95% posterior probability) and the final species tree, applying the built-in ratchets when necessary. The same minimum number of edits may be shared by different edit paths such that two types of edits may occur for any tree comparison, obligate and nonobligate edits (6). The edits specify links between nodes on the species tree, and the links were scored by adding the counts of the corresponding obligate edits and the weighted nonobligate edits for all tree comparisons.
Mitochondrial protein families.
Sixteen of our 104 alphaproteobacterial protein families had homologs in at least one of the mitochondrial genomes, and each of the eight mitochondria were represented in at least 6 of these families. Mitochondrial homologs were added to each family, and sequences were aligned, masking ambiguous portions by using Gblocks in two steps. Two steps were applied because a single application of Gblocks either (i) trimmed too heavily, removing informative portions of the alphaproteobacterial alignment, or (ii) left unrelated portions of the more-divergent mitochondrial sequences (mainly at the termini) inappropriately aligned to the alphaproteobacterial sequence. The first Gblocks run was for ingroup Alphaproteobacteria and mitochondrial members of the alignment, and the two outermost endpoints of the resulting blocks were used for trimming only terminal portions of mitochondrial sequences. The second Gblocks run was for ingroup Alphaproteobacteria only, creating a mask for the ambiguous positions in all sequences in the alignment.
Based on the results of initial MrBayes runs for each family, the 16 families were segregated into five partitions containing one to five proteins that fell into approximately equal-sized areas in an alpha-versus-Pinvar plot. A BIONJ neighbor-joining tree was prepared and used to prime two parallel MrBayes runs of 200,000 generations, unlinking statefreq, alpha, and Pinvar parameters for the partitions, in the model-jumping mode, with burn-in by 80,000 generations. The BIONJ tree was also used to prime ML analysis for 100 bootstrap resamplings of the alignment.
RESULTS
Uncertainty among rRNA trees.
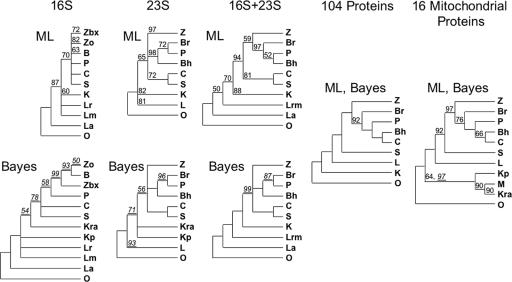
Alphaproteobacterial strains with complete or nearly complete genome sequences are now numerous; 72 of these and a large outgroup of eight diverse strains from the Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria were chosen for further analysis. Seven orders of Alphaproteobacteria were represented among these strains, with five represented by more than one strain. Sequences for both 16S and 23S rRNAs were collected and aligned based on secondary structure, and ambiguous regions of the alignment were masked. Trees were generated for the two masked alignments and for their concatenation, using both the ML algorithm implemented in PHYML and the Bayesian approach implemented in MrBayes. These trees were discordant, as illustrated by comparing the topologies of the basal branches that were well supported by either Bayesian posterior probability or ML bootstrap analysis (Fig. 1). The outgroup was consistently separated from the ingroup, and each of the five multiply represented alphaproteobacterial orders was retained intact in at least one of these trees. However, only one multiply represented order, Sphingomonadales, was retained intact in all the trees. Comparing the basal branching patterns of the pair of ML and Bayesian trees, we found one incompatibility for the 16S rRNA tree pair (regarding the integrity of the Rickettsiales), no incompatibilities for the 23S rRNA tree pair, and one incompatibility for the rRNA concatenation tree pair (regarding the affiliation of Parvularcula). Although the ML and Bayesian methods explore tree space in different ways, they do both evaluate likelihood by using the same substitution matrix and therefore could be expected to produce similar trees for a given alignment. When they instead produce trees whose highly supported nodes conflict, the common interpretation of support values as probabilities is called into question; these values may be artificially high. Comparing the trees among the three different rRNA alignments shows that none of their basal topologies are compatible. The 16S trees split the Acetobacteraceae from the other Rhodospirillales and the 23S trees group the Rhodospirillales but split the Hyphomonadaceae from the other Rhodobacterales, while the rRNA concatenation tree splits both the Rhodospirillales and Rhodobacterales. Discordance between 16S and 23S rRNA trees would be expected if the gene for one of the molecules had undergone lateral transfer, as has been invoked before for alphaproteobacteria (1, 30), but there are other explanations, such as imperfect sequence alignment and masking. With the incompatibilities between molecular phylogenies and between methods and the frequent failure to resolve basal nodes with high support, our rRNA analysis does not provide a robust species tree.
FIG. 1.
Basal branching patterns from various analyses. ML bootstrap and Bayesian (underlined) support values are shown when <100%, with nodes collapsed when support was <50%. Taxa: Z, Rhizobiales (b, Bradyrhizobiaceae; x, Xanthobacter; o, other Rhizobiales); B, Rhodobacterales (r, Rhodobacteraceae; h, Hyphomonadaceae); P, Parvularculales; C, Caulobacterales; S, Sphingomonadales; L, Rhodospirillales (r, Rhodospirillum; m, Magnetospirillum; a, Acetobacteraceae); K, Rickettsiales (r, Rickettsiaceae; a, Anaplasmataceae; p, Pelagibacter); M, mitochondria; O, outgroup Proteobacteria.
Collection of protein families.
We turned to collecting protein families that together might reveal a major vertical component of species phylogeny (27). Ideally, such families should represent each species once and only once, not mix members of paralogous families, and not show evidence of horizontal gene transfer. A search of GeneTrees (29), a large database of trees for prokaryotic protein families, identified 216 families in which the Alphaproteobacteria were found together in a subtree containing no more than four non-Alphaproteobacteria. This simple criterion was designed to exclude families affected by horizontal gene transfer from a nonalphaproteobacterial group into the Alphaproteobacteria; indeed, it excluded all eight protein families previously identified as singly represented in each alphaproteobacterial strain but with evidence of lateral gene transfer (25). The alignments for these seed groups were then used to build hidden Markov models with which to search for the homologs among all proteins of all the genomes under study. Expanded families that were missing an ortholog in more than four ingroup strains, had more than four strains with multiple members, or had the outgroup widely dispersed in initial trees were rejected, resulting in a set of 115 homology groups that approximated the ideal of representing each strain once and only once.
We used a novel approach to standardize the starting position for protein sequences by extending protein sequences N terminally according to genome sequence, realigning, and marking the N-terminal endpoint of conservation. As a second test of whether the remaining families might mix members of paralogous subgroups, the families were enlarged again by the addition of high-scoring second-best BLAST hits. Of eight greatly enlarged families, only one could be resolved into two orthologous subfamilies and the others were rejected. Among the families still under consideration, 27 had one or more paralogs. These were resolved, when possible, by a simple test based on tree branch lengths (55 of 64 cases); otherwise, no candidate was retained. The families were then subjected to tests for both noisy phylogenetic signal and possible horizontal gene transfer (25); five families were rejected due to noisy signal and none due to possible horizontal transfer.
The molecular functions of the 104 retained protein families were approximately equally distributed among ribosome structure, other protein synthesis roles, protein fate, RNA and DNA metabolism, and other metabolism; one family had no functional characterization (see Table S2 in the supplemental material).
Tree building.
For each protein family, sequences were aligned, ambiguous portions of the alignments were masked out, and trees were built using ML and Bayesian methods. As with the rRNA trees, tree topologies did not agree either between the two methods or between any two families and support for nodes was often weak. Of 7,968 nodes among the ML trees, 2,201 (28%) had <50% bootstrap support.
It was observed that the pair of tree topologies produced by the two methods had better agreement for the families with longer alignments (Fig. 2A). This suggested that part of the problem with single families is that they have insufficient information content and that trees from even longer alignments would be more reliable. Producing a longer alignment by the concatenation of alignments for multiple-protein families is justified if the families have evolved under similar parameters or if those parameters are allowed to vary independently in partitions of the family set. The WAG amino acid substitution matrix was found to be either the best available or near optimal in each of the single-protein analyses, so this matrix was used for analyzing concatenated alignments. Initial trees also showed that two parameters, the shape parameter of the gamma distribution and the proportion of invariant sites, varied somewhat among the protein families. The families were sorted into five subgroups according to their position in a plot of these two parameters, and the concatenation of all 104 alignments was partitioned accordingly. Bayesian analysis, primed with either a neighbor-joining tree for the concatenated alignment or an rRNA tree, quickly and stably settled upon a single tree topology. The consensus tree from ML analysis of bootstrap samples of the concatenated alignment had an identical topology. Thus, a single tree topology emerged from multiple analyses of the concatenated alignment, and the support values were very high: 100% for each node by Bayesian analysis and nearly so for ML bootstrap analysis. Additional measures described below provided further support for this topology, and we term the most likely Bayesian tree (Fig. 3) our “final tree.”
FIG. 2.
Topology convergence for trees from subsets of all single-molecule families. For each masked sequence alignment, the Robinson-Foulds symmetric difference was used to compare topologies between the Bayesian tree and either (A) the corresponding ML tree or (B) the final tree (Fig. 3). The data for the protein alignments were fit to the power law curves (A) y = 1,110x−0.70 (R2 = 0.61) and (B) y = 1,150x−0.55 (R2 = 0.74). The values (zero in both cases) for the final tree were not used in the curve fitting, but their positions are marked.
FIG. 3.
Tree for the Alphaproteobacteria. This tree was the most likely found by Bayesian analysis of the concatenation of masked alignments for 104 selected protein families (33,730 characters) and had topology identical to that of the ML bootstrap consensus tree. Note that unusual support values are displayed. Traditional support values were extremely high, with 100% posterior probability Bayesian and ML bootstrap support values for each node, except for the nodes marked by values in parentheses, which show bootstrap support when <100%. The main support values presented here instead show concordance with the 106 single-gene trees (104 proteins and two rRNAs), given as the percentage of single-gene trees with very high (≥95%) Bayesian support for the node. Node concordance is an extremely stringent criterion for support that should not be interpreted as the probability that the bipartition is true. The point at which the mitochondria branch in Fig. 5 is indicated. The dashed arrow is an edit (grouping the Caulobacterales/Parvularculales/Hyphomonadaceae with the Rhizobiales rather than with the Rhodobacterales) that would increase by 1 the total concordance, with highly supported nodes from single-gene trees. The taxon used to root the tree was Geobacter sulfurreducens from the Deltaproteobacteria.
Support for the tree.
Extremely high Bayesian and bootstrap support values such as those obtained here are common in studies with long concatenated protein alignments and have been regarded as misleading (26). Additional methods were applied to assess the reliability of the tree. Its overall concordance with the 106 single-molecule (104 proteins and two rRNAs) Bayesian trees was measured. For the 5,320 nodes in the single-molecule trees that had ≥95% support, 4,362 (82.0%) were in agreement with the final tree (7). Thus, the genes for these molecules show a strong trend toward the same pattern of vertical inheritance. Concordance was also used to assess each of the 77 nontrivial nodes in the final tree. Values for the percentage of single-molecule trees that showed high (≥95%) support for a node were low (53.6%) on average and ranged from 7 to 100% (Fig. 3). Node concordance should be considered an extremely stringent criterion for evaluating support within the tree and should not be interpreted as support values usually are, i.e., as the probability that a node is correct. Nonetheless, the values do appear to provide some measure of the reliability of the nodes, in that the nodes that have ML bootstrap values below 100% are among those with the lowest concordance values.
As a second approach to evaluating the reliability of the tree, we examined how well its topology was reproduced by subgroups of the full set of 104 proteins, in what might be considered whole-protein bootstrap analysis of the concatenated alignment (Fig. 2B). Random groups of 4, 10, or 26 of the proteins and other subgroups were taken, and their concatenated alignments were evaluated by ML and Bayesian methods. With longer alignments, the trees built by the two methods agreed better and also agreed better with the final tree. The curves fitting these two trends were extrapolated to the number of characters in the full concatenated alignment, showing that the full alignment was expected to produce a very reliable topology (Fig. 2). Comparison with the trends for the rRNA alignments indicates that the phylogenetic resolving power of nucleotide characters was six- to ninefold lower than for amino acids in our masked alignments.
Overall concordance with the single-molecule trees, a criterion quite different from the original one (likelihood), was used to reassess all the thousands of tree topologies encountered in all the MrBayes runs and ML studies for individual sequence alignments and concatenations. The final tree had the second-best concordance; another tree exceeded its concordance by 1 (agreeing with 4,363 of the highly supported nodes from the single-molecule trees). A single subtree prune-and-regraft operation (edit) to the final tree, grouping the Caulobacterales/Parvularculales/Hyphomonadaceae with the Rhizobiales rather than with the Rhodobacterales, was sufficient to produce the most concordant tree. No additional edits to the final tree that improved its concordance were identified among the next 10 most concordant topologies.
From tree to network.
When a single-protein tree disagrees with a species tree, a series of edits can often be proposed that bring the trees into agreement (6). An extreme interpretation of the minimal set of such edits is that they represent a minimum number of horizontal transfers in the history of the protein gene, although it should be noted that discordance between protein and species trees may have other causes, such as inappropriate alignment and masking. These edits create links between nodes of the species tree, converting the tree to a network, and the links can be ranked by the edit counts for all the protein trees under study. The systematic determination of such links, which have been termed “highways of gene sharing,” on a large prokaryotic species tree showed that most edits occurred within the major phylogenetic divisions but that a large number of edits occurred between divisions (7). We examined the minimal edit series that converted our final species tree into each of the single-protein trees of our study (except for 13 protein trees for which no series could be determined); the number of minimum edits averaged 7.5 per tree. The edits defined 556 links on the species tree, with 243 scoring at least 1.0. Figure 4 shows that the top links tend to cross the nodes with poorer concordance support. They illustrate some of the more likely phylogenetic avenues along which even the highly conserved genes in our collection may have been successfully transferred during the evolution of these genomes.
FIG. 4.
Links between nodes on species tree. The top 34 links, from an analysis of edit paths between single-protein family trees and the final tree, are mapped onto the final tree of Fig. 3. Noninteger scores for links were allowed because nonobligate edits were weighted and added to the count of obligate edits.
Placement of the mitochondrial branch.
Many studies have placed the ancestor of mitochondria within the Alphaproteobacteria, usually within the Rickettsiales, but one study suggested that it might have arisen in the Rhodospirillales (17, 19, 32). Most of the original gene complement of mitochondria has either been lost or migrated to the eukaryotic nuclear genome, such that the most primitive and gene-rich mitochondrial genome known, from the jakobid Reclinomonas americana, has only 67 protein-coding genes. Yet, 16 of these proteins are among the set of 104 in our alphaproteobacterial analysis, a number high enough that the concatenated alignment should have sufficient information content for reasonably accurate phylogenetic analysis. Seven additional mitochondrial genomes that have been considered primitive each had from 6 to 9 of these same proteins (and no others from the 104-protein set). These mitochondria do contain additional conserved protein families that could be useful if the goal were solely to produce a mitochondrial phylogeny. However, by restricting the analysis to the families that were already shown to be well behaved among Alphaproteobacteria alone, we sought to optimize the survey of alphaproteobacterial phylogeny for the point at which mitochondria diverged.
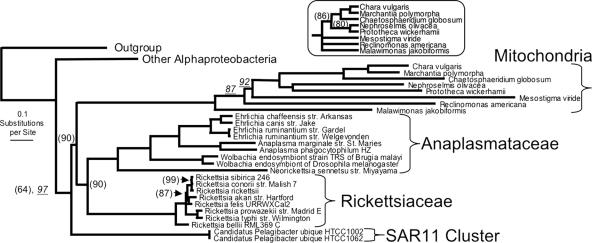
For these 16 protein families, the representative sequences from outgroup Proteobacteria, Alphaproteobacteria, and mitochondria were aligned, masked, and concatenated. Two trees were generated using MrBayes and ML bootstrap analyses; their topologies were compatible and, in the bacterial portion, matched that of the 104-protein tree. The mitochondria grouped together on a single branch that emerged from within the Rickettsiales, with the combined Anaplasmataceae/Rickettsiales as a sister group, and were subtended by the Pelagibacter branch (Fig. 5).
FIG. 5.
Mitochondrial branch. The portion for Rickettsiales is shown for the most likely tree found by Bayesian analysis of the concatenation of masked alignments for 16 selected protein families (4,830 characters), in which each node received 100% Bayesian support, except those indicated with underlined values. Identical topologies for bacterial strains arose as the ML bootstrap consensus, from which all nodes received 100% support, except those indicated in parentheses. The inset shows the topology for the mitochondrial branch from the ML bootstrap consensus, collapsing nodes with <50% support. The outgroup and alphaproteobacterial portions of the tree that are collapsed in this depiction had the same topology as those shown in Fig. 3.
DISCUSSION
The multiprotein tree presented here (Fig. 3) received perfect Bayesian support and nearly perfect bootstrap ML support. Such support values may be misleadingly high for long concatenated alignments, but this tree was also the second best among all the thousands of trees encountered in the course of this study, as judged by concordance with the highly supported nodes of the single-gene trees. The residual discordance between the species tree and the individual protein trees was used to explore non-tree-like aspects of the inheritance for even the “well-behaved” genes selected here (Fig. 4) that may have resulted from horizontal transfers (3).
This tree splits the Hyphomonadaceae from the Rhodobacterales and places them with the combined Caulobacterales and Parvularculales. The clustering of Hyphomonadaceae with Caulobacterales has been noted before. In one study, the Hyphomonadaceae were observed to cluster with the Caulobacterales in protein-based trees, yet with the Rhodobacterales in 16S rRNA trees (1). We obtained these same results; indeed, the unfavored Hyphomonadaceae/Rhodobacterales clustering was obtained for 16S rRNA trees whether we used (i) an alignment based on a profile from RDP and masked using Gblocks (Fig. 1), (ii) a masked alignment prepared directly by a server at RDP, or (iii) a de novo alignment by MUSCLE masked using Gblocks (data not shown). Horizontal transfer of the 16S rRNA gene has been invoked for other alphaproteobacteria and specifically for Hyphomonas (1, 30). However, the favored Hyphomonadaceae/Caulobacterales clustering did appear in a recent analysis of a manual alignment of 16S rRNA sequences (24), suggesting that horizontal transfer need not be invoked and that RDP-based and other 16S rRNA sequence alignments may have been misleading. Members of the Hyphomonadaceae and Caulobacterales share an unusual dimorphism, exhibiting both a nonmotile stalked reproductive cell type and a motile cell type, and comparison of Hyphomonas and Caulobacter genomes have revealed several close relationships. There have been calls to unite these groups within the order Caulobacterales (1, 2, 24). Our tree supports this unification and suggests that the order should additionally include the Parvularculaceae, with the abandonment of the order Parvularculales, which was designated mainly on the basis of 16S rRNA analysis (12). It can be noted that one characteristic linking the Hyphomonadaceae and Caulobacterales, a proliferation of TonB-dependent receptors (2), is shared by Parvularcula; 29 of its proteins are so annotated. However, this characteristic may not be highly distinctive; 18 of 34 non-Rickettsiales Alphaproteobacteria in a recent study had more than 10 TonB- dependent receptors, although only 6 had more than 25 (8). Comprehensive comparative genome analysis of the single Parvularcula genome will be an important next step in understanding its affiliation, especially since no candidate strain for a second genome project has been described that is both closely related and sufficiently distinct to provide a phylogenetic perspective. It has been further suggested that the remaining Rhodobacterales (exclusive of the Hyphomonadaceae) should be subsumed within the Caulobacterales (24); however, neither our data nor the data in that study strongly support this grouping.
Despite its success in grouping Hyphomonadaceae with Caulobacterales, the 16S tree used in the study of Lee et al. disagreed in several important ways with our multiprotein tree (24). It split the Rhodospirillales. It grouped Parvularcula with Sphingomonadales and Sphingomonadales with Rhizobiales. These discrepancies may be due to the usual problems with inference based on 16S rRNA gene sequences, possibly insufficient information content, or difficulties in proper alignment and masking. By concatenating masked alignments for multiple sequence families, the information content is increased and the effects of nonsystematic errors in alignment and masking are diluted.
The evolutionary branching order for six alphaproteobacterial orders inferred based on conservation patterns for indels in protein sequences (22) agrees with the branching order obtained for our tree, although that study did not resolve the branching order of the Rhodospirillales and Sphingomonadales as ours has. Our tree also confirms the deep split in the Rhizobiales noted frequently in previous studies (23, 24).
Much evidence supports the origin of mitochondria from within the Alphaproteobacteria, although various positions within the alphaproteobacterial phylogeny have been proposed. One study has pointed outside the Rickettsiales to Rhodospirillum (17), but most have placed the mitochondrial ancestor within or basal to the Rickettsiales, some specifically within the Rickettsiaceae (16), and others, like ours, as a sister to the combined Rickettsiaceae and Anaplasmataceae (19, 32). The availability of many recently sequenced genomes for our analysis adds new perspective to this placement, showing the free-living marine bacterium Pelagibacter closely subtending the mitochondria/Rickettsiaceae/Anaplasmataceae group. Thus, Pelagibacter will serve as a useful member of the outgroup in future phylogenetic analysis of mitochondrial genes.
Supplementary Material
Acknowledgments
This work was supported by the Virginia Bioinformatics Institute and by U.S. Department of Defense grant W911SR-04-0045 to B.W.S. and USDA CSREES grant 3602-22000-013-02 to A.W.D.
Footnotes
Published ahead of print on 4 May 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Badger, J. H., J. A. Eisen, and N. L. Ward. 2005. Genomic analysis of Hyphomonas neptunium contradicts 16S rRNA gene-based phylogenetic analysis: implications for the taxonomy of the orders ′Rhodobacterales’ and Caulobacterales. Int. J. Syst. Evol. Microbiol. 55:1021-1026. [DOI] [PubMed] [Google Scholar]
- 2.Badger, J. H., T. R. Hoover, Y. V. Brun, R. M. Weiner, M. T. Laub, G. Alexandre, J. Mrazek, Q. Ren, I. T. Paulsen, K. E. Nelson, H. M. Khouri, D. Radune, J. Sosa, R. J. Dodson, S. A. Sullivan, M. J. Rosovitz, R. Madupu, L. M. Brinkac, A. S. Durkin, S. C. Daugherty, S. P. Kothari, M. G. Giglio, L. Zhou, D. H. Haft, J. D. Selengut, T. M. Davidsen, Q. Yang, N. Zafar, and N. L. Ward. 2006. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 188:6841-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bapteste, E., E. Susko, J. Leigh, D. MacLeod, R. L. Charlebois, and W. F. Doolittle. 2005. Do orthologous gene phylogenies really support tree-thinking? BMC Evol. Biol. 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, R. D. Finn, and E. L. Sonnhammer. 1999. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batut, J., S. G. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2:933-945. [DOI] [PubMed] [Google Scholar]
- 6.Beiko, R. G., and N. Hamilton. 2006. Phylogenetic identification of lateral genetic transfer events. BMC Evol. Biol. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beiko, R. G., T. J. Harlow, and M. A. Ragan. 2005. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 102:14332-14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denance, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boussau, B., E. O. Karlberg, A. C. Frank, B. A. Legault, and S. G. Andersson. 2004. Computational inference of scenarios for α-proteobacterial genome evolution. Proc. Natl. Acad. Sci. USA 101:9722-9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 12.Cho, J. C., and S. J. Giovannoni. 2003. Parvularcula bermudensis gen. nov., sp. nov., a marine bacterium that forms a deep branch in the α-Proteobacteria. Int. J. Syst. Evol. Microbiol. 53:1031-1036. [DOI] [PubMed] [Google Scholar]
- 13.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283-1287. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The Ribosomal Database Project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emelyanov, V. V. 2001. Rickettsiaceae, rickettsia-like endosymbionts, and the origin of mitochondria. Biosci. Rep. 21:1-17. [DOI] [PubMed] [Google Scholar]
- 17.Esser, C., N. Ahmadinejad, C. Wiegand, C. Rotte, F. Sebastiani, G. Gelius-Dietrich, K. Henze, E. Kretschmann, E. Richly, D. Leister, D. Bryant, M. A. Steel, P. J. Lockhart, D. Penny, and W. Martin. 2004. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21:1643-1660. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 19.Fitzpatrick, D. A., C. J. Creevey, and J. O. McInerney. 2006. Genome phylogenies indicate a meaningful alpha-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol. Biol. Evol. 23:74-85. [DOI] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappe, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 21.Gray, M. W., B. F. Lang, and G. Burger. 2004. Mitochondria of protists. Annu. Rev. Genet. 38:477-524. [DOI] [PubMed] [Google Scholar]
- 22.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, R. S. 2005. Protein signatures distinctive of alpha proteobacteria and its subgroups and a model for alpha-proteobacterial evolution. Crit. Rev. Microbiol. 31:101-135. [DOI] [PubMed] [Google Scholar]
- 24.Lee, K. B., C. T. Liu, Y. Anzai, H. Kim, T. Aono, and H. Oyaizu. 2005. The hierarchical system of the ′Alphaproteobacteria': description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55:1907-1919. [DOI] [PubMed] [Google Scholar]
- 25.Novichkov, P. S., M. V. Omelchenko, M. S. Gelfand, A. A. Mironov, Y. I. Wolf, and E. V. Koonin. 2004. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J. Bacteriol. 186:6575-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips, M. J., F. Delsuc, and D. Penny. 2004. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 21:1455-1458. [DOI] [PubMed] [Google Scholar]
- 27.Rokas, A., B. L. Williams, N. King, and S. B. Carroll. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798-804. [DOI] [PubMed] [Google Scholar]
- 28.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 29.Tian, Y., and A. W. Dickerman. 2007. GeneTrees: a phylogenomics resource for prokaryotes. Nucleic Acids Res. 35:D328-D331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalainen, K. Lindstrom, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 185:2988-29898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viollier, P. H., and L. Shapiro. 2004. Spatial complexity of mechanisms controlling a bacterial cell cycle. Curr. Opin. Microbiol. 7:572-578. [DOI] [PubMed] [Google Scholar]
- 32.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C., Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.