Abstract
The gene for the extracytoplasmic function (ECF) sigma factor SigM was deleted from the chromosome of the gram-positive soil bacterium Corynebacterium glutamicum to elucidate the role of the SigM protein in the regulation of gene expression. Comparative DNA microarray hybridizations of the C. glutamicum wild type and sigM-deficient mutant C. glutamicum DN1 revealed 23 genes with enhanced expression in the sigM-proficient strain, encoding functions in the assembly of iron-sulfur clusters (suf operon), thioredoxin reductase (trxB), thioredoxins (trxC, trxB1), chaperones (groES, groEL, clpB), and proteins involved in the heat shock response (hspR, dnaJ, grpE). Deletion of the sigM gene rendered the C. glutamicum cells more sensitive to heat, cold, and the presence of the thiol oxidant diamide. Transcription of the sigM gene increased under different stress conditions, including heat shock, cold shock, and disulfide stress caused by diamide treatment, suggesting a regulatory role for SigM under thiol-oxidative stress conditions. Stress-responsive promoters were determined upstream of the suf operon and of the trxB, trxC, and trxB1 genes. The deduced SigM consensus promoter is characterized by the −35 hexamer gGGAAT and the −10 hexamer YGTTGR. Transcription of the sigM gene is apparently controlled by the ECF sigma factor SigH, since a sigH mutant was unable to enhance the expression of sigM and the SigM regulon under thiol-oxidative stress conditions. A typical SigH-responsive promoter was mapped upstream of the sigM gene. The ECF sigma factor SigM is apparently part of a regulatory cascade, and its transcription is controlled by SigH under conditions of thiol-oxidative stress.
Corynebacterium glutamicum is a gram-positive, nonsporulating soil bacterium that belongs to the order Actinomycetales, which also includes the genera Mycobacterium and Streptomyces. C. glutamicum is an industrially relevant producer of several l-amino acids (21, 40). Because of its biotechnological importance, the genetic and biochemical background of amino acid biosynthesis has been studied extensively. In recent years, these studies have been broadened by the completion of the genome sequence of the wild-type strain C. glutamicum ATCC 13032 (26, 30). The genome sequence allows the identification of genes by similarity to already known genes in other organisms. However, knowledge about gene expression and its regulation can only be derived from complementary studies. One attempt is based on the detection and classification of transcriptional regulators by bioinformatic methods (6), and the other is based on generating specific regulatory mutants and analyzing changes in gene expression by DNA microarray hybridization and real-time reverse transcription (RT)-PCR assays (25, 54, 36, 7). It is also essential to store this information on transcriptional regulators and their regulatory interactions in the form of specialized databases such as CoryneRegNet, a data warehouse for corynebacterial transcription factors and gene regulatory networks (4). Moreover, the sigma factors interacting with the RNA polymerase are of high relevance in understanding global changes in gene expression, since they regulate transcription at a higher hierarchical level and can influence the expression of specific target genes and of genes encoding transcription factors or even other sigma factors.
The bacterial RNA polymerase is composed of six subunits, comprising an α dimer; the β, β′, and ω subunits; and a σ factor (9). The sigma factor is responsible for promoter recognition by the RNA polymerase holoenzyme, and it thereby confers specificity on the process of transcription initiation by recognizing promoter elements of genes and operons (28). Under normal growth conditions, the essential sigma factor SigA is used. This sigma factor is apparently responsible for the transcription of housekeeping genes (18). Another category of sigma factors, termed alternative sigma factors, controls the transcription of specialized regulons that are active, for instance, during growth transitions, in the stationary phase, in response to environmental stress conditions, or during morphological development of the bacterial cell.
In 1994, a new alternative sigma factor subgroup designated the extracytoplasmic function (ECF) subfamily was described (42). The ECF sigma factors are small regulatory proteins that are strikingly divergent in amino acid sequence. They control the transcription of genes that are involved in cell envelope functions like protein transport and secretion processes and in response to extracytoplasmic stress conditions (20). ECF sigma factors can be regulated at the transcriptional, translational, and posttranslational levels (19). Transcriptional control can be complex and can be exerted by a hierarchical regulatory cascade of sigma factors. For instance, transcription of the ECF sigma factor SigJ is under the control of the stress-responsive sigma factor SigH in Streptomyces coelicolor (44). The most important regulation of ECF sigma factors, however, is reversible binding of the sigma factor to an anti-sigma factor, holding it in an inactive complex as long as the cognate environmental stimulus is absent. When changes in environmental conditions are sensed by the anti-sigma factor, the complex dissolves, rendering the sigma factor active, resulting in binding of the RNA polymerase core enzyme (24).
Because of the sequencing of more than 300 bacterial genomes in recent years, a large number of ECF sigma factor genes were detected, including 2 in Escherichia coli, 7 in Bacillus subtilis, 10 in Mycobacterium tuberculosis, about 50 in S. coelicolor (20), and 5 in C. glutamicum, named sigC, sigD, sigE, sigH, and sigM (30). Concerning the ECF sigma factors, only the sigH gene has been studied in more detail and was shown to play a prominent role in the corynebacterial response to heat stress (12) and oxidative stress (34). Furthermore, the nonessential sigma factor SigB of C. glutamicum has been characterized. It was shown that this sigma factor controls the expression of genes relevant for the transition phase of growth and the stationary phase (37). In this study, we analyzed the ECF sigma factor SigM of C. glutamicum in detail, including its gene regulatory network, the SigM-responsive promoters and the hierarchical regulation by the ECF sigma factor SigH.
MATERIALS AND METHODS
Media and growth conditions.
E. coli strains were routinely grown at 37°C in Luria-Bertani medium (56). C. glutamicum strains were cultivated at 30°C in minimal medium MM1 (MMYE without yeast extract; 32). Different stress conditions were applied to exponentially growing cultures (optical density at 600 nm of 7) of C. glutamicum RES167 (63). Where appropriate, diamide (N,N,N′,N′-tetramethylazodicarboxamide) was added to a final concentration of 2 mM. To determine the number of viable cells, dilutions of C. glutamicum cultures were plated on Luria-Bertani agar and incubated overnight at 30°C.
DNA techniques.
E. coli DH5αMCR (16) was used for standard cloning experiments. Vector DNA was prepared from E. coli cells by alkaline lysis with the QIAprep Spin Miniprep kit (QIAGEN, Hilden, Germany). DNA restriction fragments required for cloning were purified from agarose gels by means of the QIAEX II gel extraction kit (QIAGEN). All recombinant DNA techniques followed standard procedures (56). E. coli and C. glutamicum cells were transformed by electroporation (62, 63). Chromosomal DNA was extracted from C. glutamicum by an alkaline lysis technique with the QIAprep Spin Miniprep kit (QIAGEN) with a 2-h incubation of the cells at 37°C in resuspension buffer P1 containing 50 mg ml−1 lysozyme (185,000 U mg−1).
PCR techniques.
PCR experiments were carried out with the DNA Engine DYAD thermal cycler (MJ Research, Watertown, MA) and Pfu DNA polymerase. Initial denaturation was conducted at 94°C for 2 min, followed by 35 cycles of denaturation for 30 s, annealing for 30 s at a primer-dependent temperature, and extension at 72°C for 45 s, followed by a final extension step at 72°C for 3 min. PCR products were purified by with the QIAquick PCR purification kit (QIAGEN). Cloning of PCR products was performed with E. coli TOP10 cells by means of the Zero Blunt TOPO PCR cloning kit (Invitrogen, Karlsruhe, Germany).
Construction of sigM and sigH deletion mutants C. glutamicum DN1 and DN2.
Defined chromosomal deletions of the sigM gene and within the sigH coding region of C. glutamicum RES167 were constructed with the pK18mobsacB vector system, which helps to detect allelic exchange by homologous recombination (57). The respective plasmids (pDN1 and pDN2) were constructed by the GeneSOEing technique (23). Gene replacements in the chromosome of C. glutamicum RES167 were verified by PCR experiments.
RNA isolation from C. glutamicum cultures and DNA microarray hybridization.
Approximately 1 × 109 C. glutamicum cells were harvested from exponentially growing cultures by centrifugation at 11,000 × g for 15 s and subsequently transferred into liquid nitrogen. Purification of total RNA was carried out by means of the RNeasy Mini kit (QIAGEN) by following the manufacturer's instructions. The RNase-free DNase set (QIAGEN) was applied for on-column digestion of DNA. A second DNase I digestion of the RNA sample was performed with the DNase I kit (Sigma-Aldrich, Taufkirchen, Germany).
Global transcriptional profiling was performed with the C. glutamicum PCR product microarray (25). Synthesis and labeling of cDNA, as well as DNA microarray hybridization, signal detection, and data analysis, followed protocols described previously (25). Two biological replicates were hybridized by the application of a dye swap strategy in which the RNA samples of the two biological replicates were labeled with Cy3/Cy5 in one experiment and Cy5/Cy3 in the other. Since each DNA microarray contains four replicates per gene, a total of eight spots per gene were available to calculate changes in gene expression. Normalization and data evaluation were accomplished by the EMMA microarray data analysis software (11).
Real-time RT-PCR assays.
One-step real-time RT-PCR assays were performed with the LightCycler instrument (Roche Diagnostics, Mannheim Germany) and the QuantiTect SYBR green RT-PCR kit (QIAGEN). Analyses were carried out with 1 μg of total RNA as the template and the following cycler program: RT at 50°C for 20 min, initial activation at 95°C for 15 min, and three-step cycling with denaturation at 94°C for 15 s, annealing at 55°C for 40 s, and extension at 72°C for 60 s. Differences in gene expression were determined by comparing the crossing points of two samples measured in eight replicates. The crossing point is defined as the PCR cycle at which the maximum increase in fluorescence within the log-linear phase of the amplification curve occurs. It was calculated by determining the second derivative maximum of the amplification curve by use of the LightCycler software, version 3 (Roche Diagnostics).
Mapping of transcriptional start sites by rapid amplification of cDNA ends (RACE)-PCR assays.
Total RNA samples of C. glutamicum RES167 grown in MM1 medium and in MM1 medium containing 2 mM diamide were used to determine transcriptional start sites by means of the 5′/3′ RACE kit second generation (Roche Diagnostics). RACE-PCRs were carried out as recommended by the supplier, with 1 μg of total RNA. Resulting PCR products were ligated into the vector pCR2.1 by using the TOPO TA cloning system and chemically competent E. coli TOP10 cells (Invitrogen). Sequencing of cloned RACE products was carried out by IIT Biotech (Bielefeld, Germany).
RESULTS
The sigM gene is located adjacent to trx genes in the genomes of almost all actinobacteria.
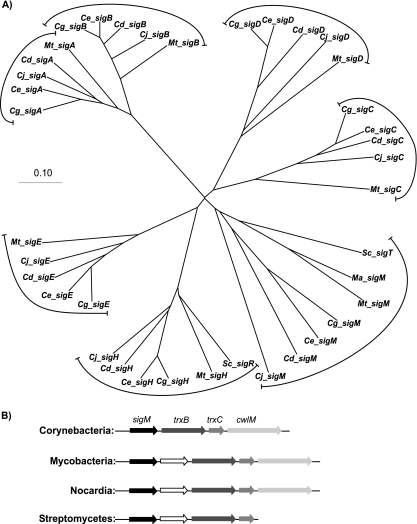
The genome of C. glutamicum ATCC 13032 encodes seven sigma factors, including the essential sigma factor SigA, the nonessential sigma factor SigB, and five alternative sigma factors (SigC, SigD, SigE, SigH, and SigM) that belong to the ECF subgroup (49, 30). The sigma factors of C. glutamicum were named according to their closest orthologs in other actinobacteria, in particular, the designations used in the close taxonomic relative M. tuberculosis. An unrooted phylogenetic tree of actinobacterial sigma factors was constructed by using different bioinformatic tools. First, a multiple alignment of the amino acid sequences of selected sigma factors was performed with the DIALIGN2 software (45). The unrooted phylogenetic tree was then obtained by using the neighbor-joining algorithm (55) integrated in the CLUSTAL_X software package (64) (Fig. 1A). From the phylogenetic tree, it became apparent that all of the sigma factors of C. glutamicum have close orthologs in M. tuberculosis and other corynebacteria, with the exception of SigM. The different SigM proteins showed only weak amino acid sequence similarities to each other. This is reflected by the deep branching of the different SigM proteins within the phylogenetic tree. Despite this amino acid sequence divergence, genes homologous to sigM are located together with genes encoding thioredoxin (trxC) and thioredoxin reductase (trxB) in a conserved cluster in almost all sequenced actinobacteria, including mycobacteria and streptomycetes (Fig. 1B). In order to learn more about the SigM subfamily, the regulatory role in gene expression of the sigma factor SigM was investigated in C. glutamicum.
FIG. 1.
Comparative analysis of sigma factor proteins and sigM gene regions of actinobacterial species. (A) Phylogenetic tree of actinobacterial sigma factors including the seven sigma factors of C. glutamicum ATCC 13032. The unrooted phylogenetic tree for different actinobacterial sigma factors is shown. Abbreviations: Cg, C. glutamicum; Ce, C. efficiens; Cd, C. diphtheriae; Cj; C. jeikeium; Ma, M. avium; Mt, M. tuberculosis; Sc, S. coelicolor. Scale bar, 0.1% amino acid substitution. (B) Comparison of the sigM gene regions of different actinobacteria. For corynebacteria, mycobacteria, nocardia, and streptomycetes, the organization of the sigM gene region is shown. The locations of the genes sigM, trxB, trxC, and cwlM are indicated. In mycobacteria, streptomycetes, and nocardia, a further gene of unknown function is located downstream of sigM.
A sigM-deficient C. glutamicum mutant showed significant differences in global gene expression in comparison to its sigM-proficient parental strain.
To elucidate the regulatory role of SigM, a defined sigM deletion mutant was constructed by gene replacement in C. glutamicum RES167 and designated C. glutamicum DN1. The chromosomal deletion comprised 618 bp and removed the complete sigM coding region (data not shown). To monitor changes in the global gene expression of the sigM mutant, DNA microarray hybridizations were performed with a genome-wide C. glutamicum microarray (25). C. glutamicum RES167 and DN1 cells were grown in minimal medium MM1 at 30°C and harvested during the exponential growth phase to purify total RNA. The RNA samples were prepared from two independently grown cultures of each strain, and the preparations were used for DNA microarray hybridization by label swapping. Differential gene expression was therefore determined by two DNA microarray hybridizations. Since each DNA microarray contains four replicates per gene, a total of eight replicates per gene were available to calculate changes in gene expression. Labeling of samples, DNA microarray hybridization, and data evaluation were carried out as described previously (25). Normalization by the LOWESS function and t test statistics were accomplished by the EMMA microarray data analysis software (11). In all experiments, an m value cutoff of ± 1.0 was applied, corresponding to relative expression changes of ≥2 or ≤0.5. The expression changes were regarded as significant if the P values were smaller than 0.05 (25).
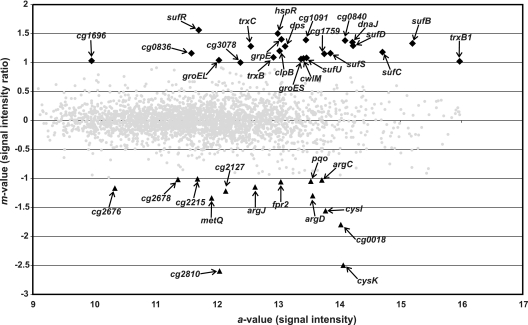
The transcriptome of C. glutamicum RES167 was compared with that of C. glutamicum DN1 to reveal genes that are transcribed with the help of SigM, according to their enhanced expression in the RES167 strain. The results obtained by DNA microarray hybridization are shown in a ratio-versus-intensity plot (Fig. 2). Genes with enhanced expression were of great interest because sigma factors, in principle, function as transcriptional activators of gene expression. The DNA microarray experiment revealed 23 genes with enhanced expression in C. glutamicum RES167, compared with the sigM-deficient mutant C. glutamicum DN1. These genes were therefore considered to represent candidate genes under the transcriptional control of SigM. The differentially expressed genes were classified into the categories (i) disulfide stress-related genes, (ii) heat stress-related genes, and (iii) genes with other functions.
FIG. 2.
Ratio-versus-intensity plot of the DNA microarray hybridization comparing the gene expression of C. glutamicum parental strain RES167 with that of sigM mutant C. glutamicum DN1. Genes were regarded as differentially expressed with m values of ≥1 or ≤−1. The m value is the log2 normalized expression ratio. Genes with enhanced expression in C. glutamicum RES167 are marked by black diamonds; genes with decreased expression are shown as black triangles. Differentially expressed genes of C. glutamicum RES167 are labeled by names or identifiers; genes not differentially expressed are shown as gray dots.
Of interest are disulfide stress-related genes (Table 1, disulfide stress-related genes), e.g., the genes sufR, sufB, sufD, sufC, sufS, sufU, and cg1759. The gene products of the suf cluster are responsible for the assembly, formation, and repair of iron-sulfur clusters in proteins (29, 38, 48, 68). As known from other organisms, like cyanobacteria (68), the sufR gene of C. glutamicum encodes the repressor of the suf gene cluster, as deduced from the analysis of a sufR deletion mutant (data not shown). Another regulated gene cluster is located directly adjacent to the sigM gene (Fig. 1B). It contains genes that code for a thioredoxin reductase (trxB), a thioredoxin (trxC), and a putative cell wall hydrolase (cwlM) (10). A further thioredoxin gene, trxB1, was also found to be differentially expressed. All of the gene products apparently provide protective functions and are important for the bacterial cell to cope with thiol-oxidizing conditions or misfolded proteins. In particular, the trx genes encode repair functions for oxidized thiol groups in proteins (22) and both thioredoxin and thioredoxin reductase are known to be induced by disulfide stress in different actinobacteria (50, 65). Altogether, 14 genes showed enhanced expression in the mutant strain. These genes encode proteins involved in arginine (argC, argJ) and cysteine (fpr2, cysI, cysK) biosynthesis, a methionine transporter (metQ), a pyruvate:quinone oxidoreductase (pqo), and several proteins of hitherto-unknown function.
TABLE 1.
Genes with enhanced expression in parental strain C. glutamicum RES167 compared with sigM mutant C. glutamicum DN1
| Coding sequence | Gene | Predicted function | m valuea | a valueb |
|---|---|---|---|---|
| Disulfide stress-related genes | ||||
| cg1765 | sufR | Transcriptional regulator of suf operon | 1.56 | 11.71 |
| cg1764 | sufB | Fe-S cluster assembly protein | 1.33 | 15.20 |
| cg1763 | sufD | Fe-S cluster assembly membrane protein | 1.18 | 14.71 |
| cg1762 | sufC | Fe-S cluster assembly ATPase | 1.29 | 14.23 |
| cg1761 | sufS | Fe-S cluster assembly protein | 1.16 | 13.86 |
| cg1760 | sufU | Cysteine desulfhydrase | 1.08 | 13.47 |
| cg1759 | Fe-S cluster assembly protein | 1.15 | 13.76 | |
| cg3299 | trxB1 | Thioredoxin | 1.02 | 15.97 |
| cg3422 | trxB | Thioredoxin reductase | 1.09 | 12.93 |
| cg3423 | trxC | Thioredoxin | 1.28 | 12.56 |
| cg3424 | cwlM | Cell wall hydrolase (autolysin) | 1.07 | 13.42 |
| Heat stress-related genes | ||||
| cg0690 | groES | Chaperone | 1.06 | 13.38 |
| cg0691 | groEL | Chaperone | 1.04 | 12.04 |
| cg3079 | clpB | Chaperone | 1.20 | 13.03 |
| cg3078 | Hypothetical protein | 1.00 | 12.39 | |
| cg3099 | grpE | Chaperone | 1.40 | 13.06 |
| cg3098 | dnaJ | Chaperone | 1.35 | 14.22 |
| cg3097 | hspR | Transcriptional regulator | 1.50 | 13.00 |
| Genes with other functions | ||||
| cg0840 | Hypothetical protein | 1.38 | 14.10 | |
| cg0836 | Hypothetical protein | 1.16 | 11.59 | |
| cg1091 | Hypothetical protein | 1.39 | 13.46 | |
| cg1696 | Putative permease | 1.03 | 9.96 | |
| cg3327 | dps | Starvation-induced DNA-protecting protein | 1.28 | 13.12 |
m value, log2 signal intensity ratio.
a value, signal intensity.
Genes that take part in the heat shock response of C. glutamicum were also detected as differentially expressed (Table 1, heat stress-related genes). These genes are groES, groEL, dnaJ, grpE, and clpB, which encode chaperones and hspR, which encodes a transcriptional regulator of the heat shock response system (3, 12, 67). Moreover, four genes that encode hypothetical proteins, as well as a putative starvation-induced, DNA-protecting protein, were found to be differentially expressed (Table 1, genes with other functions).
The sigM-deficient mutant C. glutamicum DN1 was impaired in survival after oxidative and temperature stress.
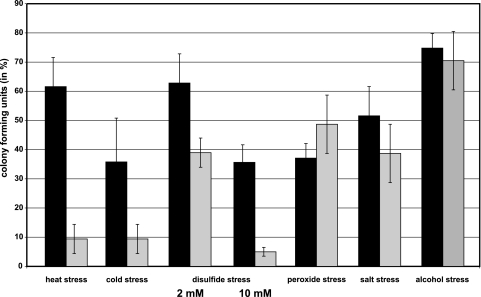
The sigM-deficient strain C. glutamicum DN1 and the sigM-proficient parental strain C. glutamicum RES167 were grown in minimal medium MM1. Different stress conditions were applied to exponentially growing cultures for 15 min. These stress conditions were heat shock at 50°C, cold shock at 10°C, disulfide stress caused by the addition of diamide to a concentration of 2 or 10 mM, oxidative stress caused by the addition of hydrogen peroxide to a concentration of 1%, salt stress caused by the addition of NaCl to a concentration of 1 M, and alcohol stress caused by the addition of ethanol to a concentration of 10%. The cells were washed after stress application, and serial dilutions were plated on Luria-Bertani agar (56). After incubation at 30°C for 24 h, the numbers of CFU were determined for both strains. It turned out that the sigM mutant showed decreased cell viability after heat, cold, and disulfide stress (Fig. 3), suggesting that SigM is involved in the respective stress responses of C. glutamicum. It was also apparent that the higher diamide concentration (10 mM) had a more severe effect on survival of the sigM mutant DN1 than the lower concentration (2 mM), obviously by causing a stronger disulfide stress. Disulfide stress is generally regarded as a subcategory of oxidative stress and is induced by thiol oxidation (39). Moreover, temperature stress and oxidative stress are related, since the induction of both types of stress response may be due to the incorrect folding of proteins which occurs under heat, cold, and oxidative stress conditions (67).
FIG. 3.
CFU of C. glutamicum parental strain RES167 and sigM mutant C. glutamicum DN1 after the application of different stress conditions. Cultures of C. glutamicum RES167 and DN1 were grown in minimal medium (MM1), and cells were harvested during exponential growth at an optical density of 7. The following stress conditions were applied for 15 min: heat shock at 50°C, cold shock at 10°C, disulfide stress caused by 2 or 10 mM diamide, oxidative stress caused by 1% hydrogen peroxide, salt stress caused by 1 M NaCl, and alcohol stress caused by 10% ethanol. Following stress treatment, the colony-forming ability of the cultures was determined with three biological and two technical replicates. CFU percentages are represented by black (RES167) and gray (DN1) columns.
The C. glutamicum sigM gene showed enhanced expression in response to stress conditions.
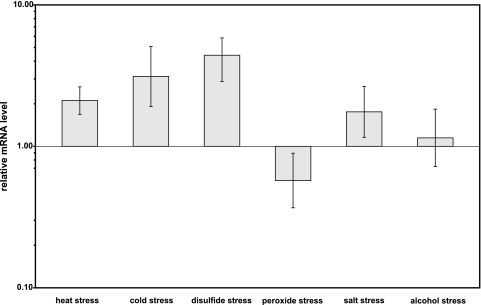
To explore the role of sigM in more detail, gene expression analyses under different stress conditions were performed by real-time RT-PCR. For this purpose, the C. glutamicum RES167 strain was grown in minimal medium MM1 and the already mentioned stress conditions were applied to exponentially growing cultures for 15 min. After stress application, the cells were washed with MM1 medium and total RNA was extracted. The relative amounts of sigM transcripts were subsequently determined by real-time RT-PCR and calculated in relation to that of an untreated control culture (Fig. 4). Transcription of the sigM gene increased significantly when heat, cold, or disulfide stress was applied. In contrast, transcription of the sigM gene was not changed significantly following peroxide, salt, and alcohol stress. These results support the view that the sigma factor SigM is involved in the transcriptional activation of genes engaged in the cellular response to heat, cold, and disulfide stress.
FIG. 4.
Relative mRNA levels of the sigM gene of a C. glutamicum culture following the application of different stress conditions. The stress conditions were heat shock at 50°C, cold shock at 10°C, disulfide stress caused by 2 mM diamide, oxidative stress caused by 1% hydrogen peroxide, salt stress caused by 1 M NaCl, and alcohol stress caused by 10% ethanol. After stress application, the cells were washed with MM1 medium and total RNA was extracted. The relative amounts of sigM transcripts were subsequently determined by real-time RT-PCR and calculated in relation to that of an untreated control culture. The measurements were carried out with three biological and two technical replicates.
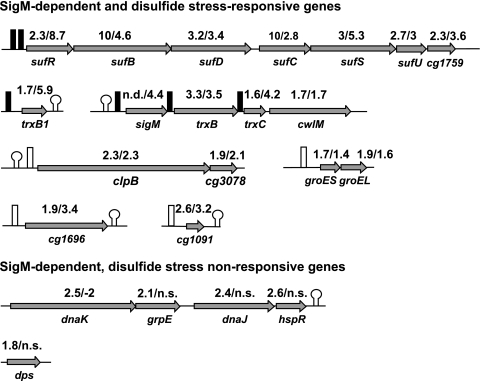
Most of the SigM-dependent genes are involved in the disulfide stress response of C. glutamicum.
Since disulfide stress caused the greatest change in the expression of the sigM gene, the transcription of SigM-dependent genes, as listed in Table 1, was analyzed as follows. First, the results of the DNA microarray hybridizations, as presented in Fig. 2, were checked by an RT-PCR experiment. For this reason, the mRNA levels of the respective genes (Table 1) of sigM-proficient strain RES167 and sigM-deficient mutant DN1 were determined and compared by performing RT-PCR assays. The results obtained are incorporated into Fig. 5, presenting most of the SigM-dependent genes of Table 1 in their genomic environment. The results confirmed the findings deduced from the DNA microarray hybridizations shown in Table 1. The main experiment, however, dealt with the question of whether disulfide stress induces the same group of SigM-dependent genes as listed in Table 1. This question was explored by analyzing the gene expression of two C. glutamicum RES167 cultures. One culture was treated for 15 min with diamide, whereas the other one was kept untreated and served as a control. The mRNA levels of the genes listed in Table 1 were then compared by RT-PCR experiments. The relative gene expression is incorporated into Fig. 5. Comparing the relative transcript levels from both experiments, it is obvious that nearly all of the genes identified as transcribed SigM dependently also have increased mRNA levels under disulfide stress. It is well established that dnaK, dnaJ, grpE, hspR, groEL, groES, and clpB are involved in the bacterial heat shock response (3, 46, 67) and that the Dps protein protects the DNA from oxidative damage (8). This suggests that the C. glutamicum sigma factor SigM plays a major role in the response to disulfide stress.
FIG. 5.
Classification of SigM-dependent genes differing in their responses to disulfide stress. The genetic organization of the respective gene regions is shown. Stem-loops denote rho-independent transcriptional terminators (13). Black boxes indicate experimentally mapped promoters, and white boxes indicate promoters that where found by sequence similarity. RT-PCR ratios were deduced from comparisons of gene expression in C. glutamicum RES167 and sigM mutant C. glutamicum DN1 (first number above the genes), as well as from comparison of C. glutamicum RES167 treated with 2 mM diamide and a C. glutamicum RES167 control (second number above the genes). n.s., no significant change in gene expression.
The transcription of the dnaK operon, consisting of the genes dnaK, grpE, dnaJ, and hspR, is not induced by disulfide stress (Fig. 5). This means that there is a second class of SigM-dependent genes which do not respond to disulfide stress. Also, the C. glutamicum dps gene belongs to this second class.
Determination of SigM-dependent promoters.
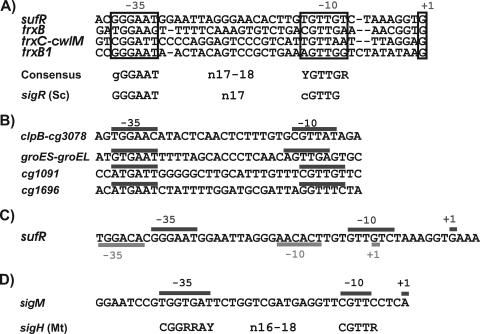
RACE-PCR experiments were conducted to identify SigM-dependent and disulfide stress-responsive promoters of differentially expressed genes detected in C. glutamicum RES167. The assays were performed with primers addressing the upstream regions of sufR, trxB, trxC, and trxB1 (Fig. 6A), with total RNA obtained from a diamide-containing C. glutamicum RES167 culture. These assays allowed the mapping of transcriptional start sites in the upstream regions of sufR, trxB, trxC, and trxB1 (Fig. 6A). By RT-PCR assays, it was shown that the genes of the suf cluster form an operon, that the sigM and trxB transcripts are monocistronic, and that trxC and cwlM form a bicistronic operon (data not shown). A multiple DNA sequence alignment performed with the CLUSTAL_X tool (64) showed that the −35 and −10 regions of the deduced promoters have similar sequences, enabling the prediction of the promoter consensus sequences GGGAAT for the −35 hexamer and YGTTGR for the −10 hexamer (Fig. 6A). The two hexamers are separated by a spacer sequence of 17 or 18 bp. All of the mapped transcriptional start sites are characterized by a guanine residue located 8 to 10 nucleotides downstream of the −10 box. These promoters were not detected by RACE-PCR experiments in the sigM mutant C. glutamicum DN1 (data not shown).
FIG. 6.
Promoter sequences of disulfide stress-responsive genes. (A) Alignment of SigM-dependent promoter regions of differentially expressed genes of C. glutamicum RES167. The transcriptional start sites (+1) were determined by RACE-PCR, with total RNA from a C. glutamicum RES167 culture treated with diamide. sigR (Sc), consensus sequence of SigR-dependent promoters from S. coelicolor (50). (B) Promoter sequences of other C. glutamicum genes potentially transcribed with the help of SigM. The promoter regions (−10 and −35) were detected by similarities to the consensus sequence shown in panel A. (C) Nucleotide sequence of the suf operon promoter region of C. glutamicum. The transcriptional start sites (+1) and deduced promoter elements (−10 and −35) of the housekeeping promoter (underlined) and the stress-responsive SigM promoter (overlined) were determined by RACE-PCR. (D) Mapped promoter of the sigM gene of C. glutamicum. sigH (Mt), consensus sequence of SigH-dependent promoters from M. tuberculosis (33). Abbreviations: R, A or G; Y, C or T.
By searching with the deduced consensus sequence in the upstream regions of differentially expressed genes, four similar putative promoters were detected in front of groES-groEL, clpB-cg3078, cg1091, and cg1696 (Fig. 6B). These data supported the assumption that the deduced consensus sequence describes the SigM-dependent disulfide stress-responsive promoter of C. glutamicum. The SigM consensus promoter of C. glutamicum revealed similarities to ECF sigma factor-dependent promoters of other actinobacteria, especially to that of the ECF-type sigma factor SigR from S. coelicolor (Fig. 6A). SigR is known to be responsible for the transcriptional response to disulfide stress in this organism (50, 51).
The transcriptional start site of the sufR gene was furthermore mapped with total RNA from C. glutamicum RES167 cells harvested during exponential growth in minimal medium MM1 without added diamide. The deduced promoter of the suf operon had a sequence different from that of the promoter that was mapped by using RNA from a diamide-treated culture (Fig. 6C). Closer inspection of the promoter elements revealed that the second promoter of the suf operon closely resembles housekeeping promoters of C. glutamicum (52). Housekeeping promoters of C. glutamicum are defined by the consensus sequences TTGACA for the −35 hexamer and TATAAT for the −10 region (52). The two hexamers are generally separated by 17 bp. The housekeeping promoter in front of the suf operon matched this consensus sequence well, since five nucleotides matched the −35 hexamer and three matched the −10 hexamer. This result confirms the view that promoter mapping with RNA samples from diamide-treated cultures revealed a distinct set of promoters involved in the stress response of C. glutamicum.
The sigma factor SigM of C. glutamicum is part of a hierarchical regulatory network and is under control of the ECF sigma factor SigH.
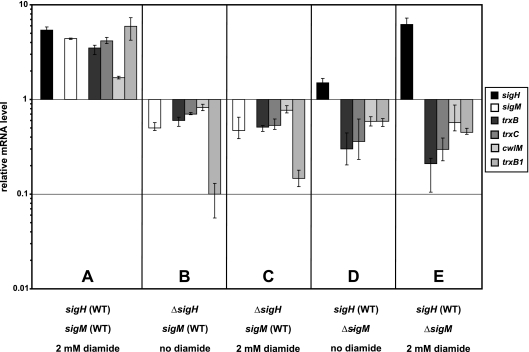
The next question to be addressed was how sigM expression is elicited during the disulfide stress response of the corynebacterial cell. In M. tuberculosis, the ECF sigma factor SigH plays a major role in the cellular stress response (43). To analyze the effect of the C. glutamicum sigma factor SigH on the expression of the sigM gene, a sigH deletion mutant of C. glutamicum RES167 was constructed by gene replacement, removing an internal fragment of 456 bp from the sigH coding region, which is 620 bp in size (data not shown). This mutant was designated C. glutamicum DN2. Subsequently, RT-PCR assays were performed to test the hypothesis that sigM is transcribed with the help of SigH. Relative expression levels detected in C. glutamicum DN1 and C. glutamicum DN2 were compared with those measured in C. glutamicum RES167 (Fig. 7).
FIG. 7.
Relative mRNA levels measured by RT-PCR of differentially expressed genes upon disulfide stress caused by diamide (2 mM) in C. glutamicum RES167 (WT [wild type]), the sigH-deficient mutant C. glutamicum DN2, and the sigM-deficient mutant C. glutamicum DN1. The mRNA level of an untreated C. glutamicum RES167 culture was used as a reference and set to 1. A, diamide-treated C. glutamicum parental strain RES167; B, untreated sigH-deficient mutant DN2; C, diamide-treated sigH-deficient mutant DN2; D, untreated sigM-deficient mutant DN1; E, diamide-treated sigM-deficient mutant DN1.
Transcription of sigH, sigM, trxB, trxC, cwlM, and trxB1 increased after diamide treatment of a C. glutamicum RES167 culture (Fig. 7A). On the other hand, transcription of sigM, trxB, trxC cwlM, and trxB1 was significantly lower in the sigH mutant C. glutamicum DN2 (Fig. 7B), as was the case when a C. glutamicum DN2 culture was treated with diamide (Fig. 7C). Likewise, transcription of trxB, trxC, cwlM, and trxB1 decreased in the sigM mutant C. glutamicum DN1 but that of the sigH gene did not (Fig. 7D). Gene expression was also lower in the sigM mutant treated with diamide, with the exception of the sigH gene, which showed an increased mRNA level (Fig. 7E). These results suggested that the sigM gene is under the control of and most probably transcribed with the help of the ECF sigma factor SigH and that SigM then initiates the transcription of its regulon. In order to find further evidence for a regulatory cascade involving SigH and SigM, the promoter of the sigM gene was determined by RACE-PCR. The transcriptional start site was mapped at an adenine residue (Fig. 6D), and the deduced −35 and −10 regions were specified by the sequences TGGTGAT and CGTTC, respectively. These −35 and −10 regions are similar to those of the SigH consensus promoter of M. tuberculosis, which consists of promoter elements with a CGGRRAY sequence in the −35 region and a CGTTR sequence in the −10 region (33).
DISCUSSION
The sigma factor SigM is involved in the oxidative stress response of C. glutamicum.
The viability of C. glutamicum RES167 and the sigM-deficient strain C. glutamicum DN1 after the application of different stresses was measured by determining the numbers of CFU. These experiments revealed that the deletion of the sigM gene caused a reduction of the viable cell numbers upon the application of heat, cold, and disulfide stress. Moreover, real-time RT-PCR experiments showed that the transcription of sigM increased significantly after the application of heat, cold, and disulfide stress. Both assays suggested that SigM is involved in the oxidative stress response of C. glutamicum, especially in the disulfide stress response. Disulfide stress is a subcategory of oxidative stress and is defined as the accumulation of nonnative disulfide bonds in the cytoplasm (2). In this study, disulfide stress was induced by diamide generally reacting with free thiols. The products of this chemical reaction are a disulfide bond and a hydrazine derivative. Heat, cold, and oxidative stresses are mechanistically related because they cause incorrect protein folding (2, 67). However, sigM expression did not increase after application of oxidative stress by hydrogen peroxide (H2O2). It is likely that the response to hydrogen peroxide stress is controlled not directly by an ECF sigma factor but by a DNA-binding transcriptional regulator. In E. coli and S. coelicolor, the transcriptional activator OxyR controls the cellular response to hydrogen peroxide stress (17, 60). An OxyR homologue is also encoded in the C. glutamicum genome (cg2109), but up to now the physiological function of this putative H2O2-sensing transcriptional regulator has not been determined.
The SigM-dependent regulon of C. glutamicum comprises genes that encode protein maintenance and repair functions.
To elucidate the SigM-dependent regulon of C. glutamicum, comparative DNA microarray hybridizations of sigM-proficient and sigM-deficient strains were performed. According to our definition, the SigM regulon comprises genes that appear with enhanced expression in C. glutamicum RES167 compared with that in the C. glutamicum DN1 mutant. Accordingly, 23 genes were identified as candidates for SigM-dependent transcription. The screening was validated by the more sensitive real-time RT-PCR assays comparing the sigM-deficient strain with the sigM-proficient strain and the untreated sigM-proficient strain with a diamide-treated sigM-proficient strain and also by the determination of SigM-responsive promoters. Components of the SigM-dependent disulfide stress-responsive regulon in C. glutamicum are, among others, the genes of the suf operon that encode seven proteins, i.e., a transcriptional regulator of the ArsR family, named SufR (6), that functions as a repressor of the suf operon (data not shown) and six proteins, SufBDCUS and Cg1759, which are generally required for the assembly and formation of iron-sulfur (Fe-S) clusters and their insertion into various proteins (15). Fe-S clusters are cofactors of different enzymes providing them with the ability to transfer electrons but are otherwise very sensitive to oxidative stress conditions (27). The gene products encoded by the suf operon apparently function in the assembly of Fe-S clusters under oxidative stress conditions (47, 61). When Fe-S clusters are damaged by disulfide stress, the expression of the biosynthesis genes increases, thereby enabling the bacterial cell to repair damaged or to assemble new Fe-S clusters.
The genes located directly adjacent to sigM are also regulated by the sigma factor SigM in response to disulfide stress. These genes are trxB, which encodes a thioredoxin reductase, and trxC, which encodes a thioredoxin. Another thioredoxin encoded by trxB1 that is located elsewhere in the genome of C. glutamicum is also part of the SigM regulon. It is known from B. subtilis (39), S. coelicolor (50), and Staphylococcus aureus (66) that thioredoxins and thioredoxin reductase take part in the disulfide stress response. Thioredoxins reduce disulfides in proteins and are therefore oxidized themselves. The oxidized form of thioredoxin is then reduced by thioredoxin reductase. The gene located adjacent to sigM, trxB, and trxC is cwlM, which encodes a potential cell wall autolysin. Up to now, little was known about cell wall autolysins, although the M. tuberculosis CwlM protein is able to lyse whole mycobacterial cells (10).
The heat stress-related genes groES, groEL, and clpB, which encode chaperones, are also part of the SigM regulon. These genes are known to take part in the heat shock response of C. glutamicum (3). The integration into the SigM regulon of heat stress-related genes is apparent when considering that the formation of disulfide bonds of cysteine side chains leads to misfolded proteins, as is the case under heat shock conditions (39, 14). The response of C. glutamicum to disulfide stress is probably addressing different regulatory networks of the cell, because redox-sensing cysteines play a key role in different signaling pathways, sensing oxidative stress and heat stress. A SigM promoter sequence was also found upstream of the genes cg1091 and cg1696, providing evidence that they are part of the SigM regulon of C. glutamicum.
Altogether, 14 genes revealed enhanced transcription in the mutant strain C. glutamicum DN1. This set of genes includes, among others, genes involved in cysteine and arginine biosynthesis. Since the mutant is defective in iron-sulfur cluster biosynthesis, more cysteine might be needed to compensate for defective Fe-S clusters. The sulfur for Fe-S clusters is generally mobilized from l-cysteine (69), and cysteine is the most common protein ligand, in accordance with the strong affinity of iron for thiolate residues. Although arginine residues in proteins are possible ligands for Fe-S clusters (5), the connection between oxidative stress and arginine synthesis is not clear.
Genes belonging to the SigM regulon are transcribed from a SigM-dependent promoter.
A new type of C. glutamicum promoter was recognized upstream of several disulfide-responsive genes, and a consensus sequence for the −35 (gGGAAT) and −10 (YGTTGR) hexamers was deduced from the mapped promoter regions. The consensus sequence of the SigM promoter is very similar to that of SigR from S. coelicolor, specified by the −35 region GGAAT and the −10 region GTT (51). The C. glutamicum SigM protein revealed 27% amino acid sequence similarity to SigR and 33% similar amino acid sequence similarity to SigT from S. coelicolor. Both sigma factors SigR and SigT are involved in the disulfide stress response (20). It is unknown which genes are regulated by SigT upon disulfide stress, whereas the SigR regulon of S. coelicolor comprises, among others, genes that encode thioredoxin, thioredoxin reductase, a glutaredoxin-like protein, and some ribosome-associated products, suggesting that the translational machinery of S. coelicolor is also modified in response to disulfide stress (51).
An additional housekeeping promoter apparently recognized by SigA was detected upstream of the suf operon of C. glutamicum. This indicates that the enzymatic functions encoded by the suf operon are necessary under standard growth conditions, under which the operon may be transcribed by the help of SigA. The growing requirement for repair and maintenance of iron-sulfur clusters under environmental stress conditions is apparently satisfied by enhanced expression of the suf operon, with the ECF sigma factor SigM. The use of different sigma subunits by RNA polymerase seems to be a general principle by which to ensure efficient expression of stress-related genes, since ECF sigma factor promoters and housekeeping sigma factor promoters appear together also upstream of other stress-responsive genes, for instance, upstream of the dnaK operon of C. glutamicum (3).
Enhanced transcription of the sigM gene is dependent on the ECF sigma factor SigH.
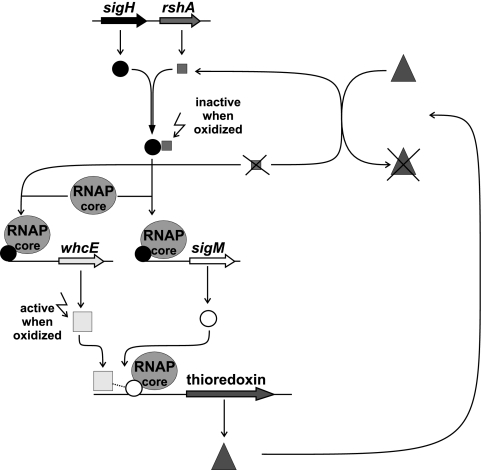
When a C. glutamicum culture was treated with diamide, transcription of the sigH gene was enhanced in both the sigM-proficient and the sigM-deficient strains, whereas enhanced expression of sigM was not detected in a sigH mutant. Although a self-activating function of SigM could not be ruled out completely by the experiments performed, the detection of a SigH-type promoter upstream of sigM supported the view that SigM is part of a cascade and under the direct transcriptional control of SigH. In M. tuberculosis, SigH plays a prominent role in survival following heat shock and oxidative stress conditions (14), since the expression of other sigma factor genes, like sigB and sigE, is controlled by SigH in this organism (43). Many ECF sigma factors are regulated moreover at the protein level by reversible binding to an anti-sigma factor (24). In particular, the activity of oxidative-stress-responsive sigma factors can be controlled effectively in this way. The anti-sigma factor possesses cysteine residues in its protein sequence that are susceptible to oxidation. In the case of oxidative or disulfide stress, the cysteinyl residues form disulfide bridges, leading to a conformational change in the protein and rendering the anti-sigma factor unable to bind the sigma factor. Release of the sigma factor and its binding to the RNA polymerase core enzyme then cause activation of the transcription of its gene regulatory network. The SigR protein of S. coelicolor contains no cysteines that would enable the protein to respond to thiol oxidation and reduction. The activity of the SigR protein is therefore regulated by interaction with the anti-sigma factor RsrA (31). The activity of the M. tuberculosis sigma factor SigH is also regulated by its anti-sigma factor, named RshA (58). In this organism, the sigH gene forms an operon together with the gene rshA, which encodes the anti-sigma factor (58). Likewise, the C. glutamicum SigH protein contains no cysteine residues and the sigH gene is located together with the rshA gene, which encodes a putative anti-sigma factor. The ensemble of SigH and RshA probably senses disulfide stress by oxidation of cysteine residues in the RshA protein, releasing SigH to activate its regulon, including the sigM gene. Then, SigM is expressed and able to activate its own regulon. Among others, the SigM regulon encodes repair functions like the thioredoxin system encoded by trxB1 and trxBC, leading to the reduction of the oxidized cysteinyl groups in RshA, and consequently, SigH can be bound again by the anti-sigma factor protein. Such a reactivation has been described for the S. coelicolor sigma factor SigR (51) and represents a regulatory mechanism by which to switch off the cellular response to environmental stress conditions.
The regulatory network structure of the C. glutamicum disulfide stress response is different from that of M. tuberculosis and S. coelicolor.
Although the regulatory role in gene expression of the C. glutamicum SigH protein may be similar to those of SigH and SigR from M. tuberculosis and S. coelicolor, respectively, the regulatory network structure of the disulfide stress response is different in C. glutamicum. In particular, the thioredoxin genes are transcribed with the help of SigH in M. tuberculosis (43), and trx gene expression is controlled directly by SigR in S. coelicolor (50). In contrast to the observations described for M. tuberculosis and S. coelicolor, C. glutamicum regulates the disulfide stress response and trx gene expression through a hierarchical network comprising the sigma factors SigH and SigM (Fig. 8). The data presented in this study unraveled a new physiological role for SigM, since the SigM protein of M. tuberculosis was shown to regulate genes required for the synthesis of surface or excreted molecules (53, 1). Moreover, expression of the sigM gene in M. tuberculosis was found to be induced only after heat shock and in the stationary phase and not under cold shock and disulfide stress conditions (53, 1). Furthermore, the promoter elements recognized by the SigM proteins differ in the deduced −10 region when considering the consensus sequence GGGAACC-N17-GTCCGA of the M. tuberculosis SigM protein (53).
FIG. 8.
The regulatory network of the C. glutamicum disulfide stress response. The sigma factor SigH is bound to its anti-sigma factor RshA until the anti-sigma factor senses an oxidative stress by being oxidized and releases the sigma factor. SigH binds to the RNA polymerase and initiates the transcription of the whcE and sigM genes. SigM binds to the RNA polymerase, interacts with WhcE, and initiates the transcription of the SigM-dependent disulfide stress-responsive genes, including thioredoxin-encoding genes. Thioredoxin is able to reduce oxidized groups in RshA, and therefore RshA is again functional and binds SigH.
Regulation of trx gene expression in C. glutamicum is even more complex, since the WhcE protein, a DNA-binding regulator of the WhiB family, is involved in the transcriptional control of trx genes and the whcE gene is expressed only in the presence of the sigma factor SigH (34). The WhcE protein apparently functions as an activator of trx gene expression under heat and disulfide stress conditions (35). The WhiB3 protein of M. tuberculosis was demonstrated in a yeast two-hybrid screen to interact directly with the 4.2 region of the principal sigma factor (59) which is involved in recognizing the −35 promoter region (41). It is possible that WhcE interacts with the SigM subunit of the C. glutamicum RNA polymerase to trigger the transcription of the SigM regulon under disulfide stress conditions (Fig. 8). This conclusion provides an attractive hypothesis for further examination of the disulfide stress response of C. glutamicum.
Acknowledgments
This study was supported by Degussa GmbH (Düsseldorf, Germany).
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Agarwal, N., S. C. Woolwine, S. Tyagi, and W. R. Bishai. 2007. Characterization of the Mycobacterium tuberculosis sigma factor SigM by assessment of virulence and identification of SigM-dependent genes. Infect. Immun. 75:452-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslund, F., and J. Beckwith. 1999. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell 96:751-753. [DOI] [PubMed] [Google Scholar]
- 3.Barreiro, C., E. Gonzalez-Lavado, M. Pátek, and J. F. Martín. 2004. Transcriptional analysis of the groES-groEL1, groEL2, and dnaK genes in Corynebacterium glutamicum: characterization of heat shock-induced promoters. J. Bacteriol. 186:4813-4817. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Baumbach, J., K. Brinkrolf, L. F. Czaja, S. Rahmann, and A. Tauch. 2006. CoryneRegNet: an ontology based data warehouse of corynebacterial transcription factors and regulatory networks. BMC Genomics 7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkovitch, F., Y. Nicolet, J. T. Wan, J. T. Jarrett, and C. L. Drennan. 2004. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303:76-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune, I., K. Brinkrolf, J. Kalinowski, A. Pühler, and A. Tauch. 2005. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brune, I., H. Werner, A. T. Hüser, J. Kalinowski, A. Pühler, and A. Tauch. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceci, P., A. Ilari, E. Falvo, L. Giangiacomo, and E. Chiancone. 2005. Reassessment of protein stability, DNA binding, and protection of Mycobacterium smegmatis Dps. J. Biol. Chem. 280:34776-34785. [DOI] [PubMed] [Google Scholar]
- 9.Darst, S. A. 2001. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 11:155-162. [DOI] [PubMed] [Google Scholar]
- 10.Deng, L. L., D. E. Humphries, R. D. Arbeit, L. E. Carlton, S. C. Smole, and J. D. Carroll. 2005. Identification of a novel peptidoglycan hydrolase CwlM in Mycobacterium tuberculosis. Biochim. Biophys. Acta 1747:57-66. [DOI] [PubMed] [Google Scholar]
- 11.Dondrup, M., A. Goesmann, D. Bartels, J. Kalinowski, L. Krause, B. Linke, O. Rupp, A. Sczyrba, A. Pühler, and F. Meyer. 2003. EMMA: a platform for consistent storage and efficient analysis of microarray data. J. Biotechnol. 106:135-146. [DOI] [PubMed] [Google Scholar]
- 12.Engels, S., J.-E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor σH. Mol. Microbiol. 52:285-302. [DOI] [PubMed] [Google Scholar]
- 13.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27-33. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes, N. D., Q.-L. Wu, D. Kong, X. Puyang, S. Garg, and R. N. Husson. 1999. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazzon, J., J. R. Fick, and D. R. Dean. 2002. Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem. Soc. Trans. 30:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn, J.-S., S.-Y. Oh, and J.-H. Roe. 2002. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184:5214-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmann, J. D., and M. J. Chamberlin. 1988. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 57:839-872. [DOI] [PubMed] [Google Scholar]
- 19.Helmann, J. D. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2:135-141. [DOI] [PubMed] [Google Scholar]
- 20.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 21.Hermann, T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104:155-172. [DOI] [PubMed] [Google Scholar]
- 22.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 25.Hüser, A. T., A. Becker, I. Brune, M. Dondrup, J. Kalinowski, J. Plassmeier, A. Pühler, I. Wiegräbe, and A. Tauch. 2003. Development of a Corynebacterium glutamicum DNA microarray and validation by genome-wide expression profiling during growth with propionate as carbon source. J. Biotechnol. 106:269-286. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99-109. [DOI] [PubMed] [Google Scholar]
- 27.Imlay, J. A. 2006. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59:1073-1082. [DOI] [PubMed] [Google Scholar]
- 28.Jishage, M., A. Iwata, S. Ueda, and A. Ishihama. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 30.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmann, L. Gaigalat, A. Goesmann, M. Hartmann, K. Hutmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegräbe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 31.Kang, J.-G., M. S. B. Paget, Y.-J. Seok, M.-Y. Hahn, J.-B. Bae, J.-S. Hahn, C. Kleanthous, M. J. Buttner, and J.-H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katsumata, R., A. Ozaki, T. Oka, and A. Furuya. 1984. Protoplast transformation of glutamate-producing bacteria with plasmid DNA. J. Bacteriol. 159:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative σ factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, T.-H., H.-J. Kim, J.-S. Park, J. Kim, P. Kim, and H.-S. Lee. 2005. Functional analysis of sigH expression in Corynebacterium glutamicum. Biochem. Biophys. Res. Commun. 331:1542-1547. [DOI] [PubMed] [Google Scholar]
- 35.Kim, T.-H., J.-S. Park, H.-J. Kim, J. Kim, P. Kim, and H.-S. Lee. 2005. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337:757-764. [DOI] [PubMed] [Google Scholar]
- 36.Koch, D. J., C. Rückert, A. Albersmeier, A. T. Hüser, A. Tauch, A. Pühler, and J. Kalinowski. 2005. The transcriptional regulator SsuR activates expression of the Corynebacterium glutamicum sulphonate utilization genes in the absence of sulphate. Mol. Microbiol. 58:480-494. [DOI] [PubMed] [Google Scholar]
- 37.Larisch, C., D. Nakunst, A. T. Hüser, A. Tauch, and J. Kalinowski. 2007. The alternative sigma factor SigB of Corynebacterium glutamicum modulates global gene expression during transition from exponential growth to stationary phase. BMC Genomics 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J.-H., W.-S. Yeo, and J.-H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51:1745-1755. [DOI] [PubMed] [Google Scholar]
- 39.Leichert, L. I. O., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leuchtenberger, W., K. Huthmacher, and K. Drauz. 2005. Biotechnological production of amino acids and derivatives: current status and prospects. Appl. Microbiol. Biotechnol. 69:1-8. [DOI] [PubMed] [Google Scholar]
- 41.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 44.Mazurakova, V., B. Sevcikova, B. Rezuchova, and J. Kormanec. 2006. Cascade of sigma factors in streptomycetes: identification of a new extracytoplasmic function sigma factor σJ that is under the control of the stress-response sigma factor σH in Streptomyces coelicolor A32. Arch. Microbiol. 186:435-446. [DOI] [PubMed] [Google Scholar]
- 45.Morgenstern, B. 1999. DIALIGN2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15:211-218. [DOI] [PubMed] [Google Scholar]
- 46.Muffler, A., S. Bettermann, M. Haushalter, A. Hörlein, U. Neveling, M. Schramm, and O. Sorgenfrei. 2002. Genome-wide transcription profiling of Corynebacterium glutamicum after heat shock during growth on acetate and glucose. J. Biotechnol. 98:255-268. [DOI] [PubMed] [Google Scholar]
- 47.Nachin, L., M. E. Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39:960-972. [DOI] [PubMed] [Google Scholar]
- 48.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oguiza, J. A., A. T. Marcos, M. Malumbres, and J. F. Martín. 1996. Multiple sigma factor genes in Brevibacterium lactofermentum: characterization of sigA and sigB. J. Bacteriol. 178:550-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paget, M. S. B., J.-G. Kang, J.-H. Roe, and M. J. Buttner. 1998. σR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 19:5776-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paget, M. S. B., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A32: identification of the σR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 52.Pátek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104:311-323. [DOI] [PubMed] [Google Scholar]
- 53.Raman, S., X. Puyang, T.-Y. Cheng, D. C. Young, D. B. Moody, and R. N. Husson. 2006. Mycobacterium tuberculosis SigM positively regulates Esx secreted protein and nonribosomal peptide synthetase genes and down regulates virulence-associated surface lipid synthesis. J. Bacteriol. 188:8460-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rey, D. A., S. S. Nentwich, D. J. Koch, C. Rückert, A. Pühler, A. Tauch, and J. Kalinowski. 2005. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism in Corynebacterium glutamicum ATCC 13032. Mol. Microbiol. 56:871-887. [DOI] [PubMed] [Google Scholar]
- 55.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, second ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 58.Song, T., S. L. Dove, K. H. Lee, and R. N. Husson. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50:949-959. [DOI] [PubMed] [Google Scholar]
- 59.Steyn, A. J. C., D. M. Collins, M. K. Hondalus, W. R. Jacobs, Jr., R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 62.Tauch, A., O. Kirchner, L. Wehmeier, J. Kalinowski, and A. Pühler. 1994. Corynebacterium glutamicum DNA is subjected to methylation-restriction in Escherichia coli. FEMS Microbiol. Lett. 123:343-347. [DOI] [PubMed] [Google Scholar]
- 63.Tauch, A., O. Kirchner, B. Löffler, S. Götker, A. Pühler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 64.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ung, K. S. E., and Y. Av-Gay. 2006. Mycothiol-dependent mycobacterial response to oxidative stress. FEBS Lett. 580:2712-2716. [DOI] [PubMed] [Google Scholar]
- 66.Uziel, O., I. Borovok, R. Schreiber, G. Cohen, and Y. Aharonowitz. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ventura, M., C. Canchaya, Z. Zhang, V. Bernini, G. F. Fitzgerald, and D. van Sinderen. 2006. How high G+C gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30:734-759. [DOI] [PubMed] [Google Scholar]
- 68.Wang, T., G. Shen, R. Balasubramanian, L. McIntosh, D. A. Bryant, and J. H. Golbeck. 2004. The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J. Bacteriol. 186:956-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]