Abstract
Upstream of the gene for flavin adenine dinucleotide (FAD)-dependent d-sorbitol dehydrogenase (SLDH), sldSLC, a putative transcriptional regulator was found in Gluconobacter frateurii THD32 (NBRC 101656). In this study, the whole sboR gene and the adjacent gene, sboA, were cloned and analyzed. sboR mutation did not affect FAD-SLDH activity in the membrane fractions. The SboA enzyme expressed and purified from an Escherichia coli transformant showed NADPH-dependent l-sorbose reductase (NADPH-SR) activity, and the enzyme was different from the NADPH-SR previously reported for Gluconobacter suboxydans IFO 3291 in molecular size and amino acid sequence. A mutant defective in sboA showed significantly reduced growth on l-sorbose, indicating that the SboA enzyme is required for efficient growth on l-sorbose. The sboR mutant grew on l-sorbose even better than the wild-type strain did, and higher NADPH-SR activity was detected in cytoplasm fractions. Reverse transcription-PCR experiments indicated that sboRA comprises an operon. These data suggest that sboR is involved in the repression of sboA, but not in the induction of sldSLC, on d-sorbitol and that another activator is required for the induction of these genes by d-sorbitol or l-sorbose.
Gluconobacter strains are known to have the ability to oxidize various kinds of sugar-related compounds and alcohols to accumulate the corresponding oxidized products (3); however, in most cases, oxidation is not completed with water and carbon dioxide, since the strains possess an incomplete citrate cycle lacking the succinate dehydrogenase gene in the genome (17). Therefore, the growth efficiencies of these microorganisms are quite low compared to those of other aerobic bacteria. Their high oxidation ability and low biomass yield make them suitable for industrial applications for bioconversion to obtain a variety of valuable products, such as l-sorbose, keto-d-gluconates, l-erythrulose, and dihydroxyacetone (3). Such oxidation reactions are called oxidative fermentation, which is mostly performed using membrane-bound dehydrogenases with pyrroloquinoline quinine (PQQ) or flavin adenine dinucleotide (FAD) as a prosthetic group. These membrane-bound dehydrogenases localize on the outer surface of the cytoplasmic membrane and link to the respiratory chain of the organism to generate bioenergy during oxidation (14). On the other hand, there are also various kinds of NAD(P)-dependent dehydrogenases in the cytoplasm of Gluconobacter strains, many of which show similar reactions under different conditions to those with the enzymes found in the cytoplasmic membrane (1, 2); for example, NADP-dependent d-glucose dehydrogenase, producing d-gluconolactone from d-glucose, is found in the cytoplasm, whereas d-glucose dehydrogenase in the cytoplasmic membrane is found as a PQQ-dependent enzyme. These NAD(P)-dependent dehydrogenases are proposed to work as reductases in vivo to assimilate the oxidized products produced by membrane-bound dehydrogenases (14).
From d-sorbitol, Gluconobacter strains effectively produce l-sorbose, an important intermediate in industrial vitamin C production, with strong activities of two membrane-bound d-sorbitol dehydrogenases, FAD-SLDH and PQQ-SLDH (25). Both are categorized as EC 1.1.99.21. The latter enzyme is also known as a versatile polyol dehydrogenase, reacting with various sugar alcohols and d-gluconate but strictly obeying the so-called Bertrand-Hudson rule (15), and we propose to call this enzyme PQQ-glycerol dehydrogenase (PQQ-GLDH) because it is its historical name and because glycerol is the most abundant substrate in nature. On the other hand, two cytosolic NAD(P)-dependent enzymes capable of oxidizing d-sorbitol have been reported for several Gluconobacter strains; they are d-sorbitol:NAD oxidoreductase (EC 1.1.1.14), producing d-fructose (1), and d-sorbitol:NADP oxidoreductase (EC 1.1.1.289), producing l-sorbose. The latter enzyme is also known as an NADPH-dependent l-sorbose reductase (NADPH-SR), and two different NADPH-SRs were purified and characterized, including one with two identical subunits of 30 kDa from Gluconobacter melanogenus IFO 3294 (2) and another with a subunit of 60 kDa from G. melanogenus N44-1 (22). In addition, a gene encoding an NADPH-SR with a 60-kDa subunit from Gluconobacter oxydans G624 was cloned and characterized, having one subunit with a molecular mass of 53,634 Da (19). A recent study revealed that a similar NADPH-SR with a calculated molecular mass of 53,541 Da plays the main role in l-sorbose assimilation in Gluconobacter suboxydans IFO 3291 (21).
It has been reported that l-sorbose is incorporated into cells of Agrobacterium tumefaciens, Neurospora crassa, Aspergillus nidulans, and Candida albicans without phosphorylation (8). In contrast, in Escherichia coli, Lactobacillus casei, and Klebsiella pneumoniae, l-sorbose is phosphorylated to l-sorbose-1-phosphate at the cell surface during transport into the cell and reduced to d-sorbitol-6-phosphate by l-sorbose-1-phosphate reductase (26, 27, 30). In either case, an NAD(P)H-dependent reductase catalyzing the reduction of l-sorbose or l-sorbose-1-phosphate is essential for l-sorbose utilization in these microorganisms (8). The metabolic pathways of d-sorbitol, l-sorbose, and their metabolites in Gluconobacter strains have been studied previously (9, 20, 22), and these C sources were shown to be utilized through a nonphosphorylating pathway; however, no transcriptional analysis has yet been performed.
In a previous study, the structural genes for FAD-SLDH (sldSLC) were cloned from Gluconobacter frateurii THD32, a thermotolerant strain isolated in Thailand (25). Part of a putative transcriptional regulator transcribed divergently was found upstream of sldSLC and was thought to be involved in the transcriptional activation of sldSLC. In this study, the complete gene for this transcriptional regulator, sboR, and the flanking downstream region were obtained. Another gene, sboA, downstream of sboR, carries an open reading frame (ORF) showing similarity to those for many proteins belonging to the short-chain dehydrogenase/reductase family, and it is revealed here that SboA is an NADPH-SR with a 29-kDa subunit. To confirm the physiological roles of sboR and sboA, mutants with gene disruption in each gene were examined for cell growth and enzyme activities, and characterization of the sboA gene product was also performed.
MATERIALS AND METHODS
Chemicals.
NAD, NADP, NADH, NADPH, and yeast extract were a kind gift from the Oriental Yeast Co., Tokyo, Japan. The other chemicals used were of guaranteed grade from commercial sources. Plasmid pSA19 (24) was kindly provided by Ajinomoto Co. Inc.
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Reference or source |
|---|---|---|
| Strains | ||
| G. frateurii strains | ||
| THD32 | Wild-type strain; NBRC 101656 | 25 |
| sboR::Tcr | THD32 defective in sboR by insertion of Tcr cassette | This study |
| sboA::Tcr | THD32 defective in sboA by insertion of Tcr cassette | This study |
| sboR::Tcr(pSAsboR) | sboR disruptant harboring the plasmid pSAsboR | This study |
| sboA::Tcr(pSABsboA) | sboA disruptant harboring the plasmid pSABsboA | This study |
| sboA::Tcr(pSAEsboA) | sboA disruptant harboring the plasmid pSAEsboA | This study |
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 recA1 supE44 thi-1 φ80dlacZΔM15 | Toyobo |
| BL21(DE3) | F−dcm ompT hsdSB(rB− mB−) gal | Novagen |
| Plasmids | ||
| pGEM-T Easy vector | E. coli cloning vector; Ampr | Promega |
| pUC119 | E. coli cloning vector; Ampr | 29 |
| pKRP12 | Tetracycline cassette; Tcr | 10 |
| pET-28a(+) | E. coli T7 system expression vector; Kmr | Novagen |
| pSA19 | Gluconobacter cloning vector; Ampr | 24 |
| pUC46 | Carries 3.5-kb DNA fragment at SphI site of pUC119; Ampr | This study |
| pSAsboR | Carries 0.8 kb of sboA at SmaI and SalI sites of pSA19; Ampr | This study |
| pUCSDRTc | Insertion of Tcr cassette at HincII site of pUCSDR; Tcr Ampr | This study |
| pUCsboRTc | Insertion of Tcr cassette at HincII site of pUCsboR; Tcr Ampr | This study |
| pETsboA | pET-28a(+) with the complete coding sequence of sboA | This study |
| pSABsboA | pSA19 with the complete coding sequence of sboA downstream of the lac promoter | This study |
| pSAEsboA | pSA19 with the complete coding sequence of sboA in the opposite direction from that of the lac promoter | This study |
Culture media and growth conditions.
Gluconobacter strains were maintained on potato-CaCO3 agar slants, which were prepared by adding 2% agar and 0.5% CaCO3 to a potato medium consisting of 5 g of d-glucose, 10 g of yeast extract, 10 g of polypeptone, 20 g of glycerol, and 100 ml of potato extract, filled to 1 liter with tap water. Preculture was performed in 5 ml of potato medium with shaking for 24 h at 30°C, and precultures were transferred to 100 ml of the appropriate medium in a 500-ml Erlenmeyer flask and then cultured for another 24 h. Bacterial growth was monitored with a Klett-Summerson photoelectric colorimeter with a red filter. Escherichia coli for plasmid construction and DNA sequencing was cultured in Luria-Bertani (LB) medium containing 50 μg/ml ampicillin, 50 μg/ml kanamycin, or 25 μg/ml tetracycline when required.
Preparation of membrane and soluble fractions.
Cells were harvested by centrifugation at 9,000 rpm for 10 min and washed once with distilled water. The washed cells were resuspended in 10 mM phosphate buffer (KPB; pH 6.0) at a concentration of 3 ml/g wet cells and passed twice through a French pressure cell (American Instruments Co., Silver Spring, MD) at 16,000 lb/in2. After centrifugation at 6,000 rpm for 10 min to remove the intact cells, the supernatants were ultracentrifuged at 40,000 rpm for 60 min. The resultant precipitate was resuspended in McIlvaine buffer (pH 5.0) and used as a membrane fraction, while the supernatant was used as the soluble fraction.
Qualitative and quantitative analyses of ketohexose.
For quantitative measurement of the total amount of ketohexose, resorcinol was used as described previously (11).
Enzyme assay.
Dehydrogenase activities in the membrane fraction with d-sorbitol, glycerol, d-mannitol, and d-arabitol were measured by the ability to reduce potassium ferricyanide, as described previously (5). One unit of enzyme activity was defined as the amount of enzyme which catalyzed 1 μmol of substrate oxidation per min under the above conditions, which was equivalent to an absorbance of 4.0 at 660 nm. The enzyme activity of NAD(P)-dependent dehydrogenase was measured following the increase of NAD(P)H at 340 nm in a reaction mixture (1 ml) containing 100 μmol of substrate, 0.1 μmol of NAD(P), and the appropriate amount of enzymes in 50 mM KPB (pH 6.0). For the reduction reaction, enzyme activity was measured according to decreasing NAD(P)H at 340 nm in a reaction mixture (1 ml) containing 100 μmol of substrate, 0.1 μmol of NAD(P)H, and the appropriate amount of enzymes in 100 mM Tris-HCl (pH 9.0). One unit of enzyme activity was defined as the amount of enzyme catalyzing 1.0 μmol of substrate.
Determination of protein concentration.
The protein concentration was measured by a modification of the Lowry method (6). Bovine serum albumin was used as the standard protein.
Measurement of molecular mass.
The molecular mass of the native enzyme was determined using high-performance liquid chromatography with gel filtration column chromatography (Superdex S-200; Amersham Biosciences). Glutamate dehydrogenase (290 kDa), lactate dehydrogenase (142 kDa), enolase (67 kDa), myokinase (32 kDa), and cytochrome c (12.4 kDa) were used as standard proteins.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 12.5% (wt/vol) acrylamide slab gels by the method described by Laemmli (12). The following calibration proteins with the indicated molecular masses were used as references for the measurement of molecular mass: phosphorylase b (94 kDa), bovine serum albumin (68 kDa), ovalbumin (43 kDa), carbonic anhydrase (31 kDa), and lysozyme (14.4 kDa). Proteins were stained with 0.1% Coomassie brilliant blue R-250.
Determination of N-terminal amino acid sequences.
After SDS-PAGE, the proteins in the gel were transferred electrophoretically to a polyvinylidene difluoride membrane at 100 mA for 4 h. The proteins were stained with Coomassie brilliant blue R-250, destained with 50% methanol, and dried, and then the stained bands were cut off. The N-terminal amino acid sequence was analyzed with a PPSQ-21 protein sequencer (Shimadzu).
DNA manipulations.
Restriction enzyme digestion, DNA ligation, and other DNA modifications were performed according to the vendor's recommendations. The preparation of plasmid DNA from E. coli strains and other general molecular biology techniques were performed as described previously (18). Genomic DNAs of Gluconobacter strains were isolated from cells grown to the mid-exponential phase in d-sorbitol medium by modification of the method of Marmur (13). PCR was performed using a Ready-To-Go PCR bead kit (Amersham Biosciences). DNA fragments obtained by PCR were isolated by agarose gel electrophoresis, purified with a QIAquick gel extraction kit (QIAGEN), and then cloned into the pGEM-T Easy vector (Promega). G. frateurii THD32 was transformed by the electroporation method (25).
RNA isolation.
Total RNA was isolated from the G. frateurii THD32 wild-type strain grown on d-sorbitol medium for 18 h (corresponding to the late exponential phase). RNAs were prepared quickly according to the hot phenol method (4). After phenol-chloroform treatment and ethanol precipitation, the resultant RNAs were resuspended in 100 mM sodium acetate (pH 5.5)-50 mM MgSO4 and treated with 10 units of RNase-free DNase at room temperature for 1 h. RNAs were then recovered after phenol-chloroform and ethanol precipitation and used for reverse transcription-PCR (RT-PCR).
RT-PCR analysis.
RT-PCR was performed using an mRNA Selective RT-PCR kit (Takara Shuzo) to determine the transcriptional organization of the sboR and sboA genes. The following primer sets were designed specifically for the indicated regions: the intergenic region between the two genes, sboRA-F (5′ CTAAAGGCAGGCCGATGTCG 3′) and sboRA-R (5′ CTATAGCCAGACCGATACCG 3′); the sboR region, SboR2-F (5′ CCTCATAACTGGCTGAGTAG 3′) and SboR2-R (5′ GTCACAGGCACGGCTAATAC 3′); and the sboA region, sboAF (5′ TTGTCCTGGTTGCTCGTCAG 3′) and sboAR (5′ CCAGTCGCCGCCATTATCTT 3′). RT was carried out at 40°C for 15 min with 0.1 μg of total RNA and each downstream primer, and subsequently, PCR was performed, consisting of denaturing at 82°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1 min, using two primers for each gene. The PCR products after 30 cycles for each gene were analyzed by 0.9% (wt/vol) agarose gel electrophoresis and stained with ethidium bromide.
Construction of gene disruption mutants from G. frateurii THD32.
To construct an sboA mutant, a 1.4-kb HindIII-SphI fragment was subcloned from a DNA fragment obtained by colony hybridization into pUC119, resulting in pUCSDR. The tetracycline resistance gene (Tcr) obtained from pKRP12 (10) was ligated into pUCSDR digested with PstI, to generate pUCSDRTc. Construction of an sboR-defective mutant was carried out using a 0.75-kb PCR fragment amplified using the primers SldR-F1 (5′ AGCCTAGAGAAGGCCGAAGGAGTC 3′) and SldR-R1 (5′ GTGACCCGTTCTGGCCGCATGAAC 3′) under PCR conditions consisting of 30 cycles of denaturing at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The amplified fragment was subcloned into pUC119, resulting in pUCsboR. The tetracycline resistance gene (Tcr) obtained from pKRP12 was ligated into pUCsboR digested with HincIII; to generate pUCsboRTc. Each plasmid harboring the disrupted gene was introduced into G. frateurii THD32 by electroporation. Transformants harboring the plasmid sequence integrated at the corresponding chromosome locus by homologous recombination were selected on d-sorbitol-glycerol agar containing 25 μg/ml of tetracycline. Disruption of sboA was confirmed by Southern hybridization, whereas disruption of the sboR gene was confirmed by PCR using the primers described above.
Complementation of sboR and sboA in disrupted mutant strains.
The sboR gene was obtained from pUC46 digested with SalI and SmaI and subcloned into pSA19 (24) to obtain plasmid pSAsboR. To construct a complementing sboA plasmid under the control of the lac promoter, the SmaI-EcoRI sboA gene fragment obtained from pBluesboA was subcloned into pSA19, resulting in pSABsboA. The sboA gene transcribed in the opposite direction of the lac promoter of pSA19 was prepared from plasmid pETsboA digested with SmaI and SacI and subcloned into pSA19, resulting in pSAETsboA. The resulting plasmids were incorporated into the gene-disrupted mutant by electroporation and selected on 1% d-sorbitol and glycerol plates containing 500 μg/ml of ampicillin and 25 μg/ml of tetracycline.
Southern hybridization.
Genomic DNA was digested with suitable restriction enzymes, electrophoresed in an agarose gel, and then transferred to a Hybond N+ membrane (Amersham Biosciences) by capillary blotting. DNA bands were then fixed to the membrane by exposure to UV light for 5 min. Hybridization and detection were carried out using the ECL direct nucleotide labeling system (Amersham Biosciences) according to the protocol provided by the supplier.
Colony hybridization.
Colonies of the gene library constructed from the chromosomal DNA of G. frateurii THD32 in pUC119 were grown on an LB agar plate containing 50 μg/ml of ampicillin, transferred to a Hybond N+ membrane, and lysed. Hybridization and detection were performed using the DIG system (Roche Diagnostics) according to the protocol provided by the supplier.
Nucleotide sequence analysis.
Plasmids for sequencing were prepared with a QIAprep Spin miniprep kit (QIAGEN). Sequencing was performed using an ABI PRISM 310 instrument (Applied Biosystems). Sequence data were analyzed using Genetyx-Mac (Software Development) and Clone Manager (Scientific and Educational Software). Homology search analysis and alignment were performed with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Expression of sboA in E. coli.
Expression of sboA was assessed under the control of the T7 promoter. The 1.4-kb DNA fragment harboring sboA was subcloned from pUCSDR into pET28a(+) to generate pETsboA. The resulting plasmid was then transformed into E. coli BL21(DE3) by electroporation. The transformant was cultured at 30°C in 100 ml of LB medium containing 50 μg/ml of kanamycin. After 6 to 8 h, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the culture medium at a final concentration of 0.5 mM and further cultured for 5 h. The cells were then harvested by centrifugation at 6,000 rpm for 10 min, washed with 10 mM KPB (pH 6.0), and disrupted with a French press. The debris was removed by centrifugation at 6,000 rpm for 10 min.
Purification of SboA.
One gram of wet cells was resuspended in 4 ml of 10 mM KPB (pH 6.0) containing 1 mM EDTA and 10% (wt/vol) glycerol (buffer A). The cell extract was obtained by passage through a French press followed by ultracentrifugation at 40,000 rpm for 60 min at 4°C. The soluble fraction was put into DEAE-cellulose which had previously been equilibrated with buffer A. After being washed in the column with the same buffer, the enzyme was eluted using a linear gradient of 0 to 0.2 M KCl and eluted around 0.1 to 0.15 M KCl. The active fraction was pooled and dialyzed overnight against a fivefold volume of 10 mM KPB (pH 7.5) containing 10% (wt/vol) glycerol, with 30% saturation of ammonium sulfate, for 3 h. The dialysate was used for hydrophobic column chromatography with a butyl-Toyopearl column which had previously been equilibrated with the same buffer. After washing of the column with the same buffer, the enzyme was eluted using a linear gradient of ammonium sulfate at 20 to 0% saturation and further washed with a threefold volume of 10 mM KPB (pH 7.5). The enzyme fractions were dialyzed and stored at −80°C.
Nucleotide sequence accession number.
Sequence data obtained for this study are available under accession number AB192961 in the DNA Data Bank of Japan (DDBJ).
RESULTS
Nucleotide sequence upstream of sldSLC.
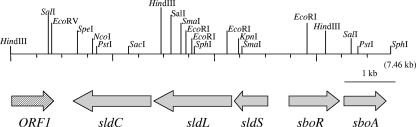
In a previous study, part of a gene showing similarity to the araC transcriptional regulator was found upstream of sldSLC (25). Thus, a new 3.5-kb SphI gene fragment containing a further upstream region was cloned and showed a positive signal with a 0.75-kb PCR fragment containing part of the transcriptional regulator as a probe (Fig. 1). The total amino acid sequence of the transcriptional regulator also showed significant similarity to the AraC/XylE family of transcription activator proteins. We designated the gene sboR, encoding an l-sorbose oxidoreductase regulatory protein, according to several results, as described below. Downstream of sboR, another ORF was found, encoding 263 amino acid residues similar to those of many proteins in the short-chain dehydrogenase/reductase family (30 to 38% identity) in the protein database, especially two putative dehydrogenases predicted from the genome sequences of Rhizobium etli and Azotobacter vinelandii (71 and 69%, respectively). This ORF was designated sboA. The calculated molecular mass and pI of the encoded protein were 28,320 Da and 5.72, respectively. From the genome sequence of G. oxydans ATCC 621H (17), the genes for proteins showing the highest similarity were GOX0646 and GOX2040 (30% identity each). 5-Keto d-gluconate reductase (GOX2187), d-glucose 1-dehydrogenase (GOX2015), and xylitol dehydrogenase (GOX0865) genes also showed identity to sboA (29, 28, and 26%, respectively); however, none showed high identity to sboA, and thus it is difficult to estimate the substrate of the enzyme from this amino acid sequence only.
FIG. 1.
Schematic representation of gene organization, including that of sboRA, in the DNA fragment obtained from G. frateurii THD32. ORFs are represented by arrows indicating their orientation. ORF1 is not complete at its N terminus.
SboA is an NADPH-SR.
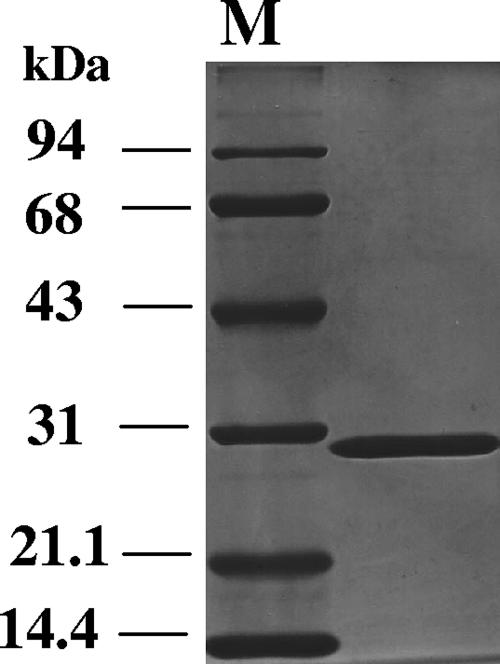
In order to characterize the sboA gene, it was expressed in E. coli BL21(DE3) under the control of the T7 promoter. Enzyme activity measured in the cell extract revealed that SboA showed high specific activity with l-sorbose as a substrate in the presence of NADPH; therefore, purification was then performed by measuring the NADPH-SR activity as described in Materials and Methods. By SDS-PAGE, the purified SboA appeared as an almost homogeneous band at a position corresponding to 29 kDa (Fig. 2). The NH2-terminal amino acid sequence of the purified enzyme was found to be MDMGISGKVA, which is identical to the deduced amino acid sequence obtained from the DNA sequence of G. frateurii THD32. Gel filtration chromatography showed that the molecular mass of the native purified SboA protein was 87 kDa, suggesting that this enzyme might be a trimer; however, this is ambiguous, because other short-chain dehydrogenases appear as dimers.
FIG. 2.
SDS-PAGE analysis of purified SboA. Fifteen micrograms of purified protein was applied to a 12.5% (wt/vol) polyacrylamide gel and stained with Coomassie brilliant blue R-250.
The activities of the purified enzyme were examined with various sugars and sugar alcohols, in the presence of either NAD(H) or NADP(H). It was found that the enzyme utilized NADP, but not NAD, when d-sorbitol was used as a substrate. Other substrates, such as d-mannitol, xylitol, l-arabitol, d-arabitol, meso-erythritol, and glycerol, were inert. For the reduction of l-sorbose, NADPH was utilized, but not NADH. The optimum pH for the oxidation of d-sorbitol was around pH 10, while that for the reduction of l-sorbose was around pH 6 to 7. Under the respective optimum conditions, specific activity for the reduction of l-sorbose (95.70 U/mg) was much higher than that for the oxidation of d-sorbitol (11.38 U/mg); therefore, this enzyme was named NADPH-SR, and the corresponding EC number is EC 1.1.1.289. The Km and Vmax values of the purified enzyme were 167 mM and 250 U/mg, respectively. The addition of MgSO4, ZnSO4, or CaCl2 did not affect the enzyme activity at all, while CuSO4 inhibited up to 20% of the original activity at a final concentration of 1 mM. There was no influence of EDTA at a final concentration of up to 20 mM.
SboA is required for efficient growth on l-sorbose.
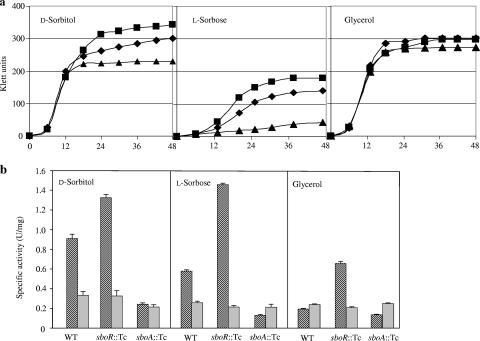
To understand the biological function of sboA existing in this Gluconobacter strain, a mutant defective in sboA was obtained by homologous recombination, with the sboA gene fragment inserted by the tetracycline resistance gene (sboA::Tcr). Growth of the mutant on l-sorbose was severely impaired, and that on d-sorbitol was also impaired to some extent (Fig. 3a). The growth yield of the mutant strain on d-sorbitol was much more dependent on the concentration of d-sorbitol in the culture medium, and at 5% (wt/vol) or higher, its growth was similar to that of the wild type (data not shown). Gluconobacter cells strongly oxidized d-sorbitol to l-sorbose by using two membrane-bound enzymes, PQQ-GLDH and FAD-SLDH (25); therefore, it seemed that to gain efficient growth, mutant cells required a higher concentration of d-sorbitol to utilize this growth substrate before complete conversion to l-sorbose occurred by membrane-bound enzymes. NADPH-SR activity in the cytoplasmic fraction was high for wild-type cells grown on either d-sorbitol or l-sorbose and low for cells grown on glycerol (Fig. 3b), indicating that this activity is inducible. Enzyme activity was not completely diminished in the mutant strain (sboA::Tcr), and residual activity seemed constant, irrespective of the growth substrate. This residual activity was similar to that in wild-type cells grown on glycerol, indicating that another enzyme has NADPH-SR activity but that it is constitutively expressed. For Gluconobacter cells, it is known that there is a NAD(H)-dependent dehydrogenase (NAD-SLDH) catalyzing the oxidation of d-sorbitol to d-fructose (1); therefore, NAD-SLDH activity in the cytosolic fraction was also measured. This enzyme activity changed very little with different carbon sources or with sboA disruption (Fig. 3b).
FIG. 3.
Growth and enzyme properties of wild-type and mutant strains. (a) Growth profiles of wild-type (diamonds), sboR disruptant (squares), and sboA disruptant (triangles) strains grown on different C sources, including 1% d-sorbitol (left), 0.5% l-sorbose (middle), and 1% glycerol (right). (b) Enzyme activities in soluble fractions from each mutant strain grown on different C sources. NADPH-SR activity (hatched bars) was measured in 50 mM KPB (pH 6.0), and NAD-SLDH activity (gray bars) was measured in 100 mM Tris-HCl (pH 9.0). All assays were performed in triplicate, and the means and standard deviations are shown.
Characterization of sboR disruptant.
It was speculated that sboR might be involved in the induction of sldSLC by d-sorbitol or l-sorbose (25). In order to investigate the physiological role of SboR, a mutant strain defective in sboR was constructed by insertion of the tetracycline resistance gene cassette. Unexpectedly, SLDH activity in membrane fractions from the mutant strain was similar to that of the wild-type strain under several conditions tested and was induced normally by d-sorbitol added to the culture medium, indicating that SboR is not involved in the induction of FAD-SLDH activity (data not shown). Interestingly, the sboR mutant grew on d-sorbitol and l-sorbose even better than the wild-type strain did, and higher NADPH-SR activity was detected in cytoplasmic fractions (Fig. 3b), suggesting that SboR acts as a repressor of SboA.
SboR represses SboA activity involved in l-sorbose assimilation.
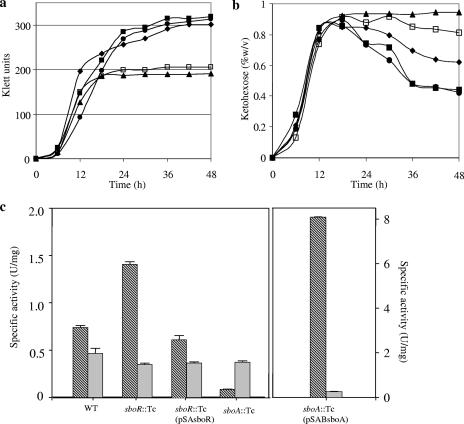
To confirm the repression of SboA by SboR, complementation of the mutant strain defective in the sboR gene was performed. Growth of the complemented strain on d-sorbitol was lower than that of either the sboR mutant or the wild-type strain (Fig. 4a), corresponding to a higher accumulation of l-sorbose (Fig. 4b) and to decreased NADPH-SR activity in cytoplasmic fractions (Fig. 4c). It seems that a higher copy number of sboR in the complemented strain prevented derepression by l-sorbose, even after l-sorbose accumulated in the culture medium. As a result, NADPH-SR activity remained low and l-sorbose assimilation did not occur, leading to its higher accumulation in the stationary phase. Furthermore, we also constructed an sboA-complemented strain. Growth of the complemented strain on d-sorbitol medium was restored to a level comparable to that of wild-type strains, as shown in Fig. 4a. NADPH-SR activity was much higher in the sboA-complemented strain than in other strains, resulting in assimilation of l-sorbose as well as that in the sboR disruptant strain (Fig. 4b). These results clearly indicated that sboA, encoding an NADPH-SR, is responsible for l-sorbose assimilation and is repressed by SboR.
FIG. 4.
Comparison of growth, l-sorbose production, and enzyme activities of wild-type and mutant strains. (a) Wild-type (closed diamonds), sboR mutant (closed squares), sboA mutant (closed triangles), sboR-complemented (closed circles), and sboA-complemented (open squares) strains were cultured on 1% d-sorbitol, and their growth was monitored with a Klett-Summerson photoelectric colorimeter with a red filter. (b) Oxidation products from d-sorbitol in the culture supernatant were measured by the resorcinol test. (c) NADPH-SR activity (hatched bars) and NAD-SLDH activity (gray bars) in the cytoplasmic fractions were measured.
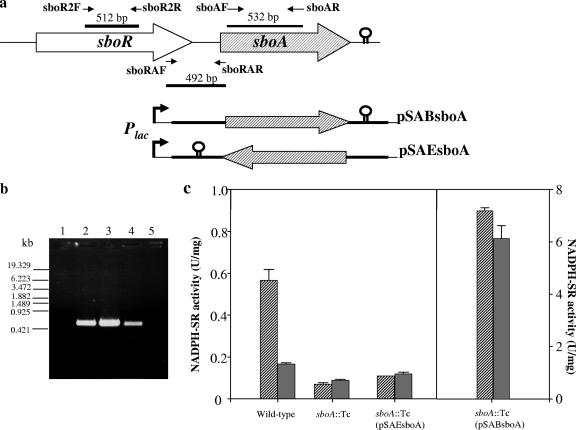
sboR and sboA are in the same transcriptional unit.
Between sboR and sboA, a gap of 132 bp exists, and a terminator-like sequence is found 32 bp downstream of sboA. To confirm whether sboR and sboA are in the same transcriptional unit, RT-PCR was performed, using the RNA fraction isolated from the G. frateurii THD32 wild-type strain as a template and three sets of primers, specific for a part within each gene and an intergenic region between the two genes, as shown in Fig. 5a. RT-PCR amplification products were obtained from all sets of primers (Fig. 5b), indicating that sboR and sboA are cotranscribed from the same upstream promoter. Moreover, complementing plasmids harboring the complete sequence of sboA and the upstream intergenic region were constructed, in which the transcription direction was in either the same or opposite orientation (pSABsboA or pSAEsboA, respectively) to that of the lac promoter in the vector plasmid (Fig. 5a). The strain with plasmid pSABsboA exhibited high NADPH-SR activity, and the enzyme activity was not significantly different between cells grown on d-sorbitol and glycerol (Fig. 5c), indicating that expression occurred constitutively by the lac promoter, not by its own inducible promoter. Furthermore, complementation by plasmid pSAEsboA seemed not to induce sboA, since the enzyme activity of the complemented strain was similar to that of the disruptant (Fig. 5c). These results support the hypothesis that sboA and sboR are organized in an operon under the control of the same promoter.
FIG. 5.
Transcriptional organization of sboRA genes and examination of promoter activity in the intergenic region. (a) Schematic representation of the orientation of genes, indicated by large arrows. Thin arrows show the position and direction of the primers used, and the thick bars indicate the regions amplified by the primer pairs. Also shown is the gene region carried by pSA19 for the complementation of sboA in the same or opposite direction from that of the lac promoter, resulting in plasmid pSABsboA or pSAEsboA, respectively. (b) Agarose gel analysis of RT-PCR amplification products. The primer sets used for RT-PCR were sboR2F and sboR2R (lanes 1 and 2), sboAF and sboAR (lane 3), and sboRAF and sboRAR (lanes 4 and 5). Lanes 1 and 5 contain samples in which reverse transcriptase was omitted. (c) NADPH-SR activity in soluble fractions from wild-type and mutant strains grown on d-sorbitol medium (hatched bars) and glycerol (gray bars).
DISCUSSION
In a previous paper, we found a transcriptional regulator next to sldSLC and assumed that SboR might be an activator of the expression of sldSLC, whose activity was induced according to the increased concentration of d-sorbitol added to the culture medium (25). In this paper, however, we demonstrated that sboR is not involved in the induction of sldSLC because FAD-SLDH activity was unchanged and still induced in the sboR mutant as well as in the wild-type strain (data not shown), and instead, it was shown that sboR is involved in the repression of its own expression and of sboA, which is shown to be in the same transcriptional unit. The deduced amino acid sequence of SboR showed high similarity to proteins in the AraC-XylS family, having two helix-turn-helix DNA binding motifs in the C-terminal domain and a ligand binding site in the N-terminal domain (7). Most proteins in this family are transcriptional activators, whereas SboR is a repressor. l-Sorbose seems to be involved in the derepression of SboR and allows the expression of sboA, since higher NADPH-SR activity was found in cells grown on d-sorbitol or l-sorbose than in those grown on glycerol (Fig. 3). On the other hand, it was also shown that the induction of SboA by d-sorbitol or l-sorbose occurred even in sboR mutant cells, indicating that another transcriptional activator is involved in the activation of sboRA transcription, which might be the same as the activator for sldSLC transcription. We tried to obtained the recombinant SboR protein to examine its binding ability with DNA fragments; however, it was obtained as an inclusion body (data not shown), and thus, DNA binding experiments remain for future work.
Shinjoh et al. reported that an NADPH-SR of 60 kDa from G. suboxydans IFO 3291 was encoded by a gene belonging to the d-mannitol dehydrogenase superfamily and that the enzyme was required for l-sorbose assimilation (21). Similarly, SboA found in G. frateurii THD32 seemed to be essential for efficient l-sorbose assimilation, although SboA has 263 amino acid residues and its molecular size is 28,320 Da, similar to many proteins in the short-chain dehydrogenase/reductase family in the genome database and different from the enzyme of G. suboxydans IFO 3291. The physicochemical and catalytic properties of SboA were compared to those of other NADPH-SRs or d-sorbitol:NADP dehydrogenases reported from Gluconobacter strains, as shown in Table 2. Basically, NADPH-SRs are divided into two types, those of 30 kDa and those of 60 kDa. NADP-SLDH obtained from G. oxydans G624 (19) is similar to the enzymes from G. suboxydans IFO 3291 and G. melanogenus N44-1 (23), and the NADPH-SR from G. melanogenus IFO 3294 (2) shows strong similarity to SboA in its molecular size and catalytic properties; however, there has been no molecular biological study of this enzyme. Since the disruption of sboA did not diminish NADPH-SR activity completely in G. frateurii THD32 and the remaining activity seemed to be constitutive, whereas SboA was induced by l-sorbose, it is supposed that another NADPH-SR exists in this strain, which might be similar to the 60-kDa NADPH-SR. In fact, in the genome sequence of G. oxydans ATCC 621H, two 60-kDa NADPH-SR-like genes, GOX1432 and GOX0849, showing 85 and 44% identities, respectively, to the NADPH-SR gene from G. suboxydans IFO 3291, were found, while homologs of SboA showing only 30% identity or less also existed (see Results). Recently, an NADPH-SR with a molecular mass of 31 kDa was reported for Candida albicans (8); however, its amino acid sequence showed only 27% identity to that of SboA, and it reacted with d-fructose, which was not utilized by SboA at all.
TABLE 2.
Comparison of SboA with d-sorbitol:NADP oxidoreductases (l-sorbose forming) found in Gluconobacter strainsc
| Enyzme | Molecular mass (kDa) | No. of subunits | Substrate specificity (%)b
|
Km (mM)
|
Vmax (U/mg)
|
Optimum pH
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d-Sorbitol | d-Mannitol | d-Arabitol | Xylitol | d-Fructose | l-Sorbose | d-Sorbitol | l-Sorbose | d-Sorbitol | l-Sorbose | d-Sorbitol | l-Sorbose | |||
| SboA in this study | 29 | 3? | 12 | 0 | 0 | 0 | 0 | 100 | 24 | 167 | 14 | 250 | 10 | 6-7 |
| NADPH-SR purified from G. melanogenus IFO 3294 (2) | 30 | 2 | 14 | 0 | 0 | 0 | 0 | 100 | NDc | 35 | 11 | 77 | 7-9 | 5-7 |
| NADPH-SR purified from G. melanogenus N44-1 (23) | 60 | 1 | 134 | 134 | ND | ND | 142 | 100 | 328 | 167 | ND | ND | 10 | 7 |
| NADP-SLDHa | 54 | 1 | 100 | 86 | 87 | ND | ND | ND | 132 | ND | ND | ND | 10 | ND |
Obtained from G. oxydans G624 and expressed in Pseudomonas putida IFO 3738 (19).
Relative to l-sorbose, except for that of NADP-SLDH of G. oxydans G624, which is expressed relative to d-sorbitol.
ND, not determined.
SboA was shown to be an NADPH-SR, whereas SboR is a repressor of the expression of sboRA, and the induction of sboRA occurred by another, unidentified activator with d-sorbitol or l-sorbose. Why is the repressor sboR required to comprise an operon with sboA? It might be a “break” for l-sorbose accumulation by preventing its assimilation or against the expression of sboA after l-sorbose is exhausted. When the strain starts to grow on d-sorbitol, the cells directly incorporate and utilize it in a small part as a carbon source by the pentose phosphate pathway through d-fructose, which is derived from NAD-SLDH and then phosphorylated to d-fructose-6-phosphate. A major part of the substrate is oxidized by membrane-bound PQQ-GLDH and FAD-SLDH, and then l-sorbose accumulates in the culture medium. After l-sorbose accumulates to some extent and d-sorbitol is exhausted, the cells start to utilize l-sorbose, probably similar to the case in C. albicans (8), through fructose 6-phosphate. During the accumulation of l-sorbose, SboR is expressed together with SboA and may compete with the unknown activator by binding to the same operator site for sboRA transcription; however, it probably does not repress its expression because of the presence of a derepressing molecule. In fact, when SboR was expressed under the control of the lac promoter, which works constitutively in Gluconobacter, no consumption of l-sorbose was observed (Fig. 4), indicating that l-sorbose is an inducer that derepresses SboR from the operator site. When l-sorbose is exhausted, the activator without the coactivator no longer works, and then complete repression of sboRA by SboR is accomplished. SboR also works positively for the accumulation of l-sorbose at the beginning of growth on d-sorbitol by preventing the expression of SboA. Gluconobacter strains may quickly oxidize d-sorbitol to an unfavorable substance, l-sorbose, creating acidic conditions to prevent the growth of other bacteria, and after expelling enemies, slowly utilize l-sorbose, similar to the hypothesis for acetic acid use by Acetobacter (16). Actually, compared to l-sorbose, d-sorbitol is a good carbon source for many microorganisms, such as E. coli, which utilizes it via the phosphotransferase system, which transports and phosphorylates the substrate to d-sorbitol-6-phosphate (28).
It has been shown that an NADPH-SR is responsible for the assimilation of l-sorbose in this strain and is required for efficient growth on l-sorbose; however, even though its activity was increased more than five times by introducing the gene via a plasmid, the assimilation rate was not much affected (Fig. 4). In NADPH-SR-overexpressing cells, something other than NADPH-SR activity limits the assimilation of l-sorbose, for example, the transport system of l-sorbose into the cytoplasm or coenzyme NADP regeneration in cells. There is no information about the transport system for l-sorbose. When cells grow on d-sorbitol, a small part of l-sorbose formed during the oxidation reaction can be transported and utilized by coupling with the hydrolysis of ATP (ABC transporter) or with proton motive force; however, it is more plausible that a small part of the substrate, d-sorbitol, is utilized for growth without oxidation and that the oxidized product is accumulated almost quantitatively until the substrate is exhausted (Fig. 4b). l-Sorbose is then gradually consumed with slower growth when the energy source for l-sorbose transport is already lacking, implying that the transporter is a permease requiring no energy source such as ATP or proton motive force. This also suggests that the transport system for l-sorbose is inducible and works only after a high concentration of l-sorbose is accumulated in the culture medium.
This is the first report about the regulation of l-sorbose assimilation in a Gluconobacter strain, although the regulation mechanism is still not clear. To show direct evidence of the actual inducer of SboR, the expression of SboR in E. coli and its use in a gel shift assay are desirable; however, we obtained the recombinant protein as an inclusion body (data not shown). Further analysis to determine the repression mechanism of SboR is required.
Acknowledgments
This work was carried out under the collaboration of the Core University Program between Yamaguchi University and Kasetsart University, supported by the Scientific Cooperation Program of the Japan Society for the Promotion of Science (JSPS) and the National Research Council of Thailand (NRCT). This work was also supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture, Japan (grant 18580078 to H.T.).
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Adachi, O., H. Toyama, G. Theeragool, N. Lotong, and K. Matsushita. 1999. Crystallization and properties of NAD-dependent d-sorbitol dehydrogenase from Gluconobacter suboxydans IFO 3257. Biosci. Biotechnol. Biochem. 63:1589-1595. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, O., H. Toyama, G. Theeragool, N. Lotong, and K. Matsushita. 1999. Crystallization and properties of NADPH-dependent l-sorbose reductase from Gluconobacter melanogenus IFO 3294. Biosci. Biotechnol. Biochem. 63:2137-2143. [DOI] [PubMed] [Google Scholar]
- 3.Adachi, O., D. Moonmungmee, E. Shinagawa, H. Toyama, M. Yamada, and K. Matsushita. 2003. New quinoprotein in oxidative fermentation. Biochim. Biophys. Acta 1647:10-17. [DOI] [PubMed] [Google Scholar]
- 4.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. [PubMed] [Google Scholar]
- 5.Ameyama, M., E. Shinagawa, K. Matsushita, and O. Adachi. 1981. d-Fructose dehydrogenase of Gluconobacter suboxydans: purification, characterization, and application to enzymatic microdetermination of d-fructose. J. Bacteriol. 145:814-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dully, J. R., and P. A. Grieve. 1975. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal. Biochem. 64:136-141. [DOI] [PubMed] [Google Scholar]
- 7.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg, J. R., N. Price, R. P. Oliver, F. Sherman, and E. Rustchenko. 2005. Candida albicans SOUI encodes a sorbose reductase required for l-sorbose utilization. Yeast 22:957-969. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino, T., T. Sugisawa, M. Tazoe, M. Shinjoh, and A. Fujiwara. 1990. Metabolic pathway for 2-keto-l-gulonic acid formation in Gluconobacter melanogenus IFO 3293. Agric. Biol. Chem. 54:1211-1218. [Google Scholar]
- 10.Kimberly, S. R., and J. P. Gregory. 1995. New plasmids carrying antibiotic-resistance cassettes. Gene 165:141-142. [DOI] [PubMed] [Google Scholar]
- 11.Kulka, R. G. 1956. Colorimetric estimation of ketopentoses and ketohexoses. Biochem. J. 63:542-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 14.Matsushita, K., H. Toyama, and O. Adachi. 1994. Respiratory chain and bioenergetics of acetic acid bacteria. Adv. Microb. Physiol. 36:247-301. [DOI] [PubMed] [Google Scholar]
- 15.Matsushita, K., Y. Fujii, Y. Ano, H. Toyama, M. Shinjoh, N. Tomiyama, T. Miyazaki, T. Sugisawa, T. Hoshino, and O. Adachi. 2003. 5-Keto-d-gluconate production is catalyzed by a quinoprotein glycerol dehydrogenase, major polyol dehydrogenase, in Gluconobacter species. Appl. Environ. Microbiol. 69:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushita, K., T. Inoue, G. Theeragool, J. Trcek, H. Toyama, and O. Adachi. 2005. Acetic acid production in acetic acid bacteria leading to their ‘death’ and survival, p. 169-181. In M. Yamada (ed.), Survival and death in bacteria. Research Signpost, Kerala, India.
- 17.Prust, C., M. Hoffmeister, H. Liesegang, A. Wiezer, W. F. Fricke, A. Ehrenreich, G. Gottschalk, and U. Deppenmeier. 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23:195-200. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 19.Shibata, T., C. Ichigawa, M. Matsuura, Y. Noguchi, Y. Saito, and M. Yamashita. 2000. Cloning of a gene for d-sorbitol dehydrogenase from Gluconobacter oxydans G624 and expression of the gene in Pseudomonas putida IFO 3738. J. Biosci. Bioeng. 89:463-468. [DOI] [PubMed] [Google Scholar]
- 20.Shinjoh, M., Y. Setoguchi, T. Hoshino, and A. Fujiwara. 1990. l-Sorbose dissimilation in 2-keto-l-gulonic acid-producing mutant UV10 derived from Gluconobacter melanogenus IFO 3293. Agric. Biol. Chem. 55:2257-2263. [Google Scholar]
- 21.Shinjoh, M., M. Tazoe, and T. Hoshino. 2002. NADPH-dependent l-sorbose reductase is responsible for l-sorbose assimilation in Gluconobacter suboxydans IFO 3291. J. Bacteriol. 184:861-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugisawa, T., T. Hoshino, S. Masuda, S. Nomura, Y. Setoguchi, M. Tazoe, M. Shinjoh, S. Someha, and A. Fujiwara. 1990. Microbial production of 2-keto-l-gulonic acid from l-sorbose and d-sorbitol by Gluconobacter melanogenus. Agric. Biol. Chem. 54:1201-1209. [Google Scholar]
- 23.Sugisawa, T., T. Hoshino, and A. Fujiwara. 1991. Purification and properties of NADPH-linked l-sorbose reductase from Gluconobacter melanogenus N44-1. Agric. Biol. Chem. 55:2043-2049. [Google Scholar]
- 24.Tonouchi, N., T. Tsuchida, F. Yoshinaga, S. Horinouchi, and T. Beppa. 1994. A host-vector system for a cellulose-producing Acetobacter strain. Biosci. Biotechnol. Biochem. 59:1899-1901. [DOI] [PubMed] [Google Scholar]
- 25.Toyama, H., W. Soemphol, D. Moonmangmee, O. Adachi, and K. Matsushita. 2005. Molecular properties of membrane-bound FAD-containing d-sorbitol dehydrogenase from thermotolerant Gluconobacter frateurii isolated from Thailand. Biosci. Biotechnol. Biochem. 69:1120-1129. [DOI] [PubMed] [Google Scholar]
- 26.Wehmeier, U. F., B. Nobelmann, and J. W. Lengeler. 1992. Cloning of the Escherichia coli sor genes for l-sorbose transport and metabolism and physical mapping of the genes near metH and iclR. J. Bacteriol. 174:7784-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wehmeier, U. F., and J. W. Lengeler. 1994. Sequence of the sor-operon for l-sorbose utilization from Klebsiella pneumoniae KAY2029. Biochim. Biophys. Acta 1208:348-351. [DOI] [PubMed] [Google Scholar]
- 28.Yamada, M., and M. H. Saier. 1987. Physical and genetic characterization of the glucitol operon in Escherichia coli. J. Bacteriol. 169:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 30.Yebra, M. J., A. Veyrat, M. A. Santos, and G. Perez-Martinez. 2000. Genetics of l-sorbose transport and metabolism in Lactobacillus casei. J. Bacteriol. 182:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]