Abstract
Magnetotactic bacteria (MTB) are a heterogeneous group of aquatic prokaryotes with a unique intracellular organelle, the magnetosome, which orients the cell along magnetic field lines. Magnetotaxis is a complex phenotype, which depends on the coordinate synthesis of magnetosomes and the ability to swim and orient along the direction caused by the interaction with the Earth's magnetic field. Although a number of putative magnetotaxis genes were recently identified within a conserved genomic magnetosome island (MAI) of several MTB, their functions have remained mostly unknown, and it was speculated that additional genes located outside the MAI might be involved in magnetosome formation and magnetotaxis. In order to identify genes specifically associated with the magnetotactic phenotype, we conducted comparisons between four sequenced magnetotactic Alphaproteobacteria including the nearly complete genome of Magnetospirillum gryphiswaldense strain MSR-1, the complete genome of Magnetospirillum magneticum strain AMB-1, the complete genome of the magnetic coccus MC-1, and the comparative-ready preliminary genome assembly of Magnetospirillum magnetotacticum strain MS-1 against an in-house database comprising 426 complete bacterial and archaeal genome sequences. A magnetobacterial core genome of about 891 genes was found shared by all four MTB. In addition to a set of approximately 152 genus-specific genes shared by the three Magnetospirillum strains, we identified 28 genes as group specific, i.e., which occur in all four analyzed MTB but exhibit no (MTB-specific genes) or only remote (MTB-related genes) similarity to any genes from nonmagnetotactic organisms and which besides various novel genes include nearly all mam and mms genes previously shown to control magnetosome formation. The MTB-specific and MTB-related genes to a large extent display synteny, partially encode previously unrecognized magnetosome membrane proteins, and are either located within (18 genes) or outside (10 genes) the MAI of M. gryphiswaldense. These genes, which represent less than 1% of the 4,268 open reading frames of the MSR-1 genome, as yet are mostly of unknown functions but are likely to be specifically involved in magnetotaxis and, thus, represent prime targets for future experimental analysis.
Magnetotactic bacteria (MTB) represent a diverse group of prokaryotes with respect to morphology and physiology (5) that are capable of magnetic orientation. All known MTB belong to different subgroups of the Proteobacteria and the Nitrospirae phylum, with most known cultivated and uncultivated representatives within the Alphaproteobacteria (2). Magnetotaxis is a complex phenotype, which depends on the synthesis of specific intracellular organelles, the magnetosomes. The synthesis of magnetosomes is a process with genetic control over the species-specific morphology and size of magnetosome crystals and the intracellular assembly of chain-like structures. In addition, magnetotaxis requires active migration along the geomagnetic field, which apparently occurs in a highly coordinated fashion by interaction with other sensory mechanisms including chemo-, aero-, and even phototaxis (11, 12).
Biomineralization of magnetic iron oxide or sulfide crystals proceeds in specific intracellular vesicles formed by the magnetosome membrane (MM). Recently, proteomic analysis of the MM in combination with reverse genetics led to the identification of genes encoding magnetosome membrane proteins (MMPs) (17, 18, 41) in Magnetospirillum gryphiswaldense MSR-1. All MMP-encoding genes were identified within a hypervariable 130-kb genome fragment described as a genomic “magnetosome island” (MAI) (39, 42), which is apparently conserved in other MTB and which may have been transferred horizontally (13, 18, 42).
By genetic analysis the function of several MMPs was recently revealed, including a role in the “activation” of magnetosome vesicles (20) and the assembly and stabilization of magnetosome chains along the cytoskeletal, actin-like magnetosome-filament (21, 32, 37). Although functions of other MMPs have remained obscure, it is clear that the formation of functional magnetosomes also requires control over the intracellular differentiation and formation of MM vesicles, the uptake and transport of supersaturating amounts of iron, its precipitation within the MM vesicle, and the nucleation and growth of the magnetosome crystal. In addition, magnetically directed taxis and motility need to be coordinated and regulated by environmental signals. The first genome-wide studies suggested that MTB genomes contain exceptionally high numbers of chemotaxis proteins. For instance, 65 chemotaxis transducers were identified in Magnetospirillum magnetotacticum strain MS-1 (1), and 62 were found in Magnetospirillum magneticum strain AMB-1 (27).
The abundance of genes of unknown function within the MAI suggested a genetic complexity of magnetotaxis that potentially might go beyond previous expectations. It has been further speculated that additional gene functions essential for magnetotaxis might be located elsewhere in the genome. Therefore, bioinformatic approaches provide an additional step in understanding the unique physiology of magnetotaxis, given the severe difficulties involved in its genetic manipulation and biochemical analysis.
Comparative analysis of a large and growing number of diverse sequenced genomes is revolutionizing the pace of gene discovery (31) and may provide novel functional assignments to genes (14). In addition to organism-specific singleton ORFans (where ORF is open reading frame) that have no similarities in the current databases, a common set of genes without functional assignment encompasses so-called orthologous ORFans (40) or conserved ORFs, which are defined as ORFs with significant similarities to ORFs in the public databases. Within this group, ORFs can be grouped into different subsets, where the definition of subsets is variable and can be based on phylogenetic affiliation, common metabolic traits, or related habitats (metabolism-, phylogeny-, or habitat-specific ORFs). This approach is also often referred to as “phylogenetic profiling” with the underlying assumption that a unique set of genes shared between any two organisms are correlated with a particular phenotype (31). This has led to the identification of, for example, photosynthetic genes in photosynthetic prokaryotes (26, 30, 34) or to the identification of light-inducible proteins which determine the relative fitness of Prochlorococcus strains of the ecotypes (35).
During recent years, genome information from several magnetite-producing, magnetotactic Alphaproteobacteria has gradually become available. In 2000, draft genome assemblies were provided by the Joint Genome Institute (www.jgi.doe.gov) for the marine Magnetococcus strain MC-1 (28, 43) and the magnetotactic spirillum M. magnetotacticum strain MS-1 (7, 18, 38) (the genome of MC-1 was only recently completed in November 2006). A 480-kb genomic fragment including the 130-kb MAI in M. gryphiswaldense strain MSR-1 was analyzed (42), and the first magnetobacterial genome to be completed, that of M. magneticum strain AMB-1, was recently published (27).
In this study, we present the nearly complete draft genome sequence of M. gryphiswaldense strain MSR-1 and extensive comparisons with the three other available magnetobacterial genomes of the Alphaproteobacteria. We analyzed the phylogenetic affiliations of MSR-1 genes and determined the magnetobacterial core genome, i.e., the minimal set of genes shared by all MTB. We further used this set of genes to address the occurrence of group-specific genes. We show that a working set of 28 group-specific genes is exclusively, but universally, associated with the magnetotactic genotype, a number of them encoding previously unrecognized MMPs. The majority of these genes exhibit a conserved organization and are located within the MAI. Our findings suggest that these genes are specifically involved in magnetotaxis and, thus, represent prime targets for future experimental analysis.
MATERIALS AND METHODS
Sequencing and assembly of M. gryphiswaldense strain MSR-1.
Strain MSR-1 was cultivated, and genomic DNA was prepared and used for construction of 1.5-kb and 3.5-kb insert plasmid libraries as previously described (39). Additionally, a cosmid library was constructed according to the manufacturer's instructions (pWEB vector; Epicenter Technologies, Madison, WI). Templates for sequencing were obtained by insert amplification via PCR or by plasmid isolation. End sequences were performed with Big Dye chemistry (ABI) (39), resulting in more than 9.4-fold coverage for the genome (total contig length of 4.5 Mb of draft sequence) and in more than 10-fold coverage for the plasmid (36 kb of finished sequence). Sequences were assembled using Phrap (http://www.genome.bnl.gov/Software/UW/) and the Consed package (version 14.00) (16).
Gene prediction and annotation.
The ORF prediction of strains AMB-1 and MC-1 was used in this study as provided by the NCBI Reference Sequences collection (RefSeq) with the accession number NC_007626 and NC_008576, respectively. On nucleotide sequence scaffolds of strains MSR-1 and MS-1, ORF prediction was carried out by using the in-house gene prediction software MORFind2 (J. Waldmann and H. Teeling, unpublished). This system combines the three gene finders CRITICA (4), GLIMMER3 (8), and ZCURVE (19) and resolves conflicts in an iterative postprocessing algorithm to enhance ORF prediction.
Annotation and data mining were performed using the GenDB, version 2.2 system (29), supplemented by the tool jCOAST, version 1.0 (comparative analysis and search tool; M. Richter, unpublished) developed at MPI Bremen, seeking for each coding region observations from similarity searches against sequence databases (nr, SWISS-PROT, KEGG-Genes, and genomesDB; for an explanation, see “Local genomes database” below) and protein family databases (Pfam, release 20.0; InterProScan 4.2, Interpro release 13.0), and from predictive signal peptide (SignalP, version 3.0 [6]) and transmembrane helix-analysis (TMHMM, version 2.0 [23]). Predicted protein coding sequences were automatically annotated by the in-house software MicHanThi (33). The MicHanThi software predicts gene functions based on similarity searches using the NCBI-nr (including SWISS-PROT) and InterPro database.
Local genome database.
The local genome database (genomesDB) provides a computationally well-defined environment of 426 published whole-genome sequences of bacterial and archaeal origin with all ORFs of each genome carrying a unique identifier. To allow genome comparisons between specific user-defined groups, all ORFs are assigned to the respective organism and metabolic group. In contrast to the general-purpose database NCBI-nr, which contains every sequence ever submitted, the focus of genomesDB is the association of every protein to its phylogenetic affiliation in a defined environment.
Definition and prioritizing of a group.
Group-specific ORFs were extracted, and four gene groups were defined. (i) The MTB-related group of genes with homologs in nonmagnetotactic bacteria are found in M. gryphiswaldense strain MSR-1, M. magneticum strain AMB-1, and M. magnetotacticum strain MS-1. The genes representing this group show moderate sequence similarity in bacteria outside the MTB. (ii) The MTB-related group of genes with occurrence limited to a given genus is found in M. gryphiswaldense strain MSR-1, M. magneticum strain AMB-1, and M. magnetotacticum strain MS-1. Genes within this group are limited in occurrence to these three species of the genus Magnetospirillum. (iii) The MTB-related group of genes contains members with homologs in nonmagnetotactic bacteria. Members of this group are M. gryphiswaldense strain MSR-1, M. magneticum strain AMB-1, M. magnetotacticum strain MS-1, and Magnetococcus strain MC-1. This group of genes shows moderate sequence similarity in bacteria outside the MTB. (iv) The MTB-specific group includes genes whose occurrence is limited to magnetotactic bacteria. Members of this group are M. gryphiswaldense strain MSR-1, M. magneticum strain AMB-1, M. magnetotacticum strain MS-1, and Magnetococcus strain MC-1. Genes included here occur only in these four organisms and are without homologous genes outside this group. It thus seems highly likely that they play a role in the synthesis of magnetosomes. Genes that occur in all four members of this group were classified as having a “high potential” for playing an important role in magnetotaxis or biomineralization of magnetosomes.
Systematic extraction of group-specific genes with the local genomesDB.
We used the following general strategy for the extraction of candidate genes: (i) all predicted ORFs of the MSR-1 genome were initially searched with the BLAST algorithm against the entire set of predicted ORFs of the strain AMB-1, MS-1, and MC-1 genomes, and vice versa (“BLAST all against all”). The results were searched with variable cutoff E values in order to identify sets of genes shared either by all four MTB strains, or at least within the genus Magnetospirillum. (ii) The identified sets of genes were further searched with BLAST against the in-house genomesDB and subsequently extracted with variable cutoff E values. A match was defined as follows: if E values of all BLAST hits of a particular ORF are within the ORF set of one or more group members in the genomesDB and are below the defined cutoff for a given group, there is a match. All extracted group-specific ORFs were manually inspected, and the final set was selected based on high pairwise sequence similarity or the availability of additional information yielded by one of the bioinformatic tools mentioned above.
Phylogenetic affiliation estimation.
To evaluate the phylogenetic assignment of the conserved ORFs in the datasets, all conserved ORFs were tested by BLAST for the phylogenetic distribution of best hits against a local genomes database (genomesDB) comprising a hit with an expectation value of E <1e-05.
Comparison of the shared gene content by reciprocal best matches (RBMs).
To compare the datasets for gene content determination, a BLAST search between all predicted ORFs in the four MTB was performed. Reciprocal best matches were counted by a BLAST result with an expectation value E of <1e-05 each and a subject coverage of over 65%.
Functional classification with COG.
All predicted ORFs were also searched for similarity against the initial cluster of orthologous groups (COG) database (containing 44 organisms; http://www.ncbi.nlm.nih.gov/COG/). A match was counted if the similarity search resulted in an E value below 1e-05.
Phylogenetic trees.
A phylogenetic tree was calculated using maximum-likelihood analysis with the PROML program of the PHYLIP 3.65 package (9). Bootstrap analysis was carried out by generating datasets with the SEQBOOT program (100 replicates), and a majority rule consensus tree was calculated using the CONSENSE program of the same package.
Nucleotide sequence accession numbers.
The M. gryphiswaldense strain MSR-1 draft genome sequence has been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under the accession number CU459003; the plasmid sequence was deposited under accession number CU45900.
RESULTS
General features of the genome sequences.
General features of the four magnetobacterial genome sequences are summarized in Table 1. The genome draft sequence of M. gryphiswaldense strain MSR-1 comprises 4,264,908 bp of chromosomal sequence and a 35,803-bp plasmid sequence. The closely related Magnetospirillum strains MSR-1, AMB-1, and MS-1 have a comparable genome size, GC content, number of rRNA genes, and COGs, whereas differences are found in the absolute number of predicted ORFs. Despite their close phylogenetic relationship, the genomes of all four strains contain a significant proportion (11 to 18%) of organism-specific ORFs, which is within the average range typical for most microbial genomes (44). The more remotely related coccoid strain MC-1 displays similarities in the number of rRNA genes but strongly differs with respect to the GC content and the number of conserved ORFs in the COG database.
TABLE 1.
General genome features of the four magnetotactic bacteria strains analyzed in this study
| Feature | MSR-1 | AMB-1a | MS-1 | MC-1 |
|---|---|---|---|---|
| Nucleotide statistics | ||||
| Genome size (bp) | 4,264,908 | 4,967,148 | 4,503,280 | 4,719,581 |
| No. of nonregular nucleotides | 17,646 | 33 | 1,060 | 0 |
| GC content (%) | 62.8 | 65.1 | 64.0 | 54.8 |
| No. of contigs | 373 | 1 | 316 | 1 |
| Gene predictions | ||||
| No. of predicted ORFs | 4,268 | 4,559 | 4,925 | 3,716 |
| No. of genes with similarity to genomesDBb | 3,726 | 4,044 | 4,011 | 3,074 |
| No. of genes with similarity to COG | 2,797 | 2,937 | 2,851 | 1,486 |
| No. of organism-specific genesc | 542 | 515 | 914 | 642 |
| No. of 16S/23S/5S rRNA genes | 2/2/2 | 2/2/NA | 2/2/2 | 3/3/3 |
| No. of tRNA genes | 47 | 50 | 44 | 45 |
Data are from Matsunaga et al. (27). NA, not annotated in the original publication.
Based on genomesDB and comprising 426 whole-genome sequences.
Based on genomesDB.
A further test revealed that 48 of 49 genes, which typically occur in single copy in microbial genomes, are found only singular in the sequence assembly of MSR-1. In general, it can be concluded from the retrieval of characteristic genome features known from the closely related AMB-1, such as the absolute number of tRNA genes and rRNA genes, the conformance of the ratio of total predicted to conserved ORFs, and the single-copy gene distribution, that the available data for MSR-1 represent the nearly complete genome sequence.
Phylogenetic distribution of the best BLAST hits of all conserved ORFs from MSR-1.
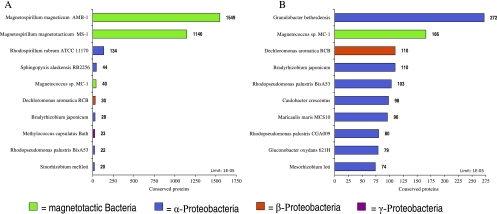
To evaluate the phylogenetic affiliation of all ORFs with known similarities (conserved ORFs) from MSR-1, all ORFs were searched with BLAST for the distribution of the best hit against genomesDB. This revealed that 63% (2,695 ORFs) yielded best hits (lowest BLAST E value) either in AMB-1 (1,549 ORFs) or MS-1 (1,146 ORFs) (Fig. 1A). The remaining 37% are predominantly distributed among different genera of Alphaproteobacteria with most hits to ORFs in the genomes of Rhodospirillum rubrum (Rhodospirillaceae, 134 ORFs) and of Sphingopyxis alaskensis (Sphingomonadaceae, 44 ORFs). Remarkably, a substantial number yielded best hits to ORFs in the betaproteobacterium Dechloromonas aromatica (30 ORFs) and in the gammaproteobacterium Methylococcus capsulatus Bath (23 ORFs).
FIG. 1.
Phylogenetic affiliation of best BLAST hits of all conserved ORFs from MSR-1. Bars represent the top-10 numbers of the best E-value hits from each conserved gene in MSR-1. (A) Distribution with all database species from genomesDB included. (B) Distribution after closest relatives AMB-1, MS-1, and R. rubrum were excluded from analysis.
To eliminate the bias generated by closely related species like AMB-1, MS-1, or R. rubrum, hits to these three organisms were excluded for further analysis (Fig. 1B). This revealed that with only one exception (D. aromatica), all organisms yielding top-10 numbers of hits belong to the Alphaproteobacteria. Interestingly, even after exclusion of hits to the other Magnetospirillum strains, ORFs with best hits to MC-1 represent only the second-largest group after Granulobacter bethesdensis.
Distribution of RBMs between MTB genomes.
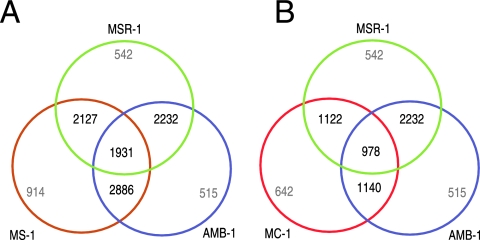
The RBM analysis was performed by searching all predicted MSR-1 ORFs against all ORFs from each individual genome of strains AMB-1, MS-1, and MC-1. This revealed that 2,232 genes are conserved between MSR-1 and AMB-1 (52% of MSR-1). A similar number (2,127 genes) is conserved between MSR-1 and MS-1 (50% of MSR-1), whereas only 1,122 genes (26% of MSR-1) display RBM characteristics between MSR-1 and MC-1.
Comparison of RBMs between the three Magnetospirillum strains revealed that a much higher number of genes are shared between strains AMB-1 and MS-1 (2,886) than between strains MSR-1 and AMB-1 (2,232) and between MSR-1 and MS-1 (2,127). Overall, a conserved core set of 1,931 genes (equivalent to 45% of the MSR-1 genome) is shared between all three Magnetospirillum species (Fig. 2A). As expected, a much smaller proportion of genes are conserved between the magnetospirilla and the magnetic coccus MC-1: if MS-1 is replaced by MC-1 in the three-genome comparison, only 23% (978 genes) of the MSR-1 genes are shared between the three strains (Fig. 2B). Intercomparison of all four MTB identified a magnetobacterial “core genome” that comprises 891 genes, corresponding to approximately 20% of the overall gene content of MSR-1.
FIG. 2.
Comparative gene content analysis of MTB based on reciprocal best matches. The Venn diagrams illustrate the shared gene content between the four genomes. For visualization, individual diagrams for three genomes are shown. The numbers of species-specific genes and shared genes are indicated. (A) Shared gene content between MSR-1, AMB-1, and MS-1. (B) Shared gene content between MSR-1, AMB-1, and strain MC-1.
Identification of group-specific ORFs in MTB.
Magnetotaxis and magnetite biomineralization are unique metabolic traits that require very specific metabolic pathways. We reasoned that putative magnetosome genes should be conserved in most MTB genomes or even be universally shared by all MTB while, on the other hand, they should be either entirely restricted to MTB or exhibit weaker similarity to homologous genes identified in related but nonmagnetotactic organisms. Based on this, each gene was scored for the presence or absence of homologs in nonmagnetotactic bacteria from the genomesDB and classified according to one of the groups described in the following sections.
(i) MTB-specific genes.
MTB-specific genes are a subset of known genes, such as mamC, that are either confined to the three Magnetospirillum strains (“Magnetospirillum specific”) or all four MTB but do not have any obvious homologs in non-MTB (17, 18, 41). To systematically extract members of this class, all genes from MSR-1 with counterparts in strains AMB-1, MS-1, and optionally MC-1 (E value of <1e-05) but no similarity in nonmagnetotactic bacteria (E value of >1e-05) were filtered from the list of BLAST hits. Two groups of MTB-specific genes were distinguished based on their absence or presence in MC-1. A total of 152 genes from MSR-1 are shared only by other species of Magnetospirillum; i.e., they are also absent from strain MC-1 (see Table S1 in the supplemental material for a complete list). It must be noted, however, that the lack of further constraints (e.g., minimum coverage) has the risk of generating false positives, in particular, for short or modular genes. Twenty-six of these 152 genes are located on the previously identified 480-kb genomic fragment which encloses the MAI, and 15 are located immediately within the 135-kb MAI. Six of these 15 MAI genes represent previously known genes located within the mam and mms operons (mamW, mms6, mamG, mamR, mamJ, and mamL), for which an essential role in magnetosome formation has already been confirmed experimentally (3, 17, 37, 39, 42). The other nine Magnetospirillum-specific MAI genes represent previously unrecognized genes (Table 2).
TABLE 2.
Summary of MTB-specific genes and characteristics of their encoded productsa
| Gene | No. of TMH | No. of aa | IEP | Size (kDa) | E value
|
Gene product | Located within the MAI | Conserved neighborhood | Identified as MMP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB-1 | MS-1 | MC-1 | |||||||||
| Magnetospirillum-specific genesb | |||||||||||
| mms6 | 1 | 135 | 9.6 | 12.7 | 2e-14 | 2e-14 | Magnetosome protein Mms6, control of crystal growthe | + | + | +f,g,h | |
| mamGc | 2 | 83 | 9.5 | 7.6 | 7e-22 | 7e-23 | Magnetosome protein MamG | + | + | +g | |
| mamR | 72 | 8.9 | 8.0 | 2e-28 | 7e-29 | Magnetosome protein MamR | + | + | +f | ||
| mamJ | 425 | 3.8 | 44.2 | 2e-44 | 2e-52 | Magnetosome protein MamJ, control of chain assembly | + | + | +f,g | ||
| mamL | 2 | 122 | 11.5 | 13.2 | 2e-29 | 1e-28 | MamL protein | + | + | ||
| mamW | 1 | 137 | 12.8 | 15.0 | 1e-42 | 8e-43 | Magnetosome protein MamW | + | +i | ||
| MGR4045 | 135 | 4.3 | 15.0 | 2e-68 | 3e-66 | Conserved hypothetical protein | + | ||||
| MGR4047 | 102 | 9.0 | 11.2 | 8e-11 | 2e-16 | Conserved hypothetical protein | + | ||||
| MGR4052 | 115 | 5.0 | 12.8 | 2e-48 | 2e-48 | Conserved hypothetical protein | + | ||||
| MGR4063 | 160 | 10.0 | 17.2 | 2e-28 | 4e-29 | Conserved hypothetical protein | + | ||||
| MGR4066 | 1 | 108 | 12.9 | 11.8 | 2e-10 | 2e-10 | Conserved hypothetical protein | + | |||
| MGR4114 | 1 | 68 | 10.0 | 6.8 | 1e-06 | 1e-07 | Conserved hypothetical protein | + | |||
| MGR4115 | 114 | 5.0 | 11.8 | 2e-06 | 3e-07 | Conserved hypothetical protein | + | ||||
| mamY (MGR4150) | 2 | 370 | 4.5 | 40.8 | 2e-137 | 5e-138 | Conserved protein | + | + | +g | |
| MGR4153 | 307 | 10.3 | 33.4 | 2e-139 | 2e-140 | Conserved hypothetical protein | + | ||||
| mmeA (MGR1900)d | 364 | 7.6 | 38.4 | 6e-124 | 1e-124 | Conserved protein | +g | ||||
| MTB specific genes present in MC-1 | |||||||||||
| mamD | 1 | 313 | 10.4 | 12.3 | 8e-87 | 3e-87 | 4e-11 | Magnetosome protein MamD | + | + | +f,g,h |
| mamT | (1) | 173 | 10.4 | 18.8 | 1e-84 | 1e-80 | 1e-24 | Magnetosome protein MamT; conserved cytochrome c heme-binding site | + | + | +g |
| mamS | 1 | 179 | 7.2 | 18.6 | 2e-57 | 1e-57 | 5e-11 | Magnetosome protein MamS | + | + | +f,h |
| mamF | 3 | 110 | 9.7 | 12.2 | 6e-40 | 6e-40 | 5e-15 | Magnetosome protein MamF | + | + | +f,g |
| mamC | 2 | 124 | 4.8 | 12.3 | 3e-18 | 2e-18 | 4e-05 | Magnetosome protein MamC | + | + | +f,g,h |
| mamI | 2 | 76 | 9.2 | 8.2 | 2e-12 | 2e-12 | 1e-02 | MamI protein | + | + | |
| mtxA (MGR0208) | 313 | 6.5 | 33.1 | 4e-84 | 6e-99 | 2e-77 | Conserved hypothetical protein, secreted | + | +f | ||
| MGR2333 | 69 | 11.3 | 7.9 | 2e-29 | 2e-29 | 1e-07 | Conserved hypothetical protein; possible phage related | ||||
| MGR2349 | 118 | 5.5 | 13.0 | 2e-55 | 6e-06 | 9e-07 | Conserved hypothetical protein; possible phage related | ||||
| mmsF (MGR4072) | 3 | 124 | 10.0 | 13.87 | 8e-42 | 1e-42 | 7e-15 | MamF-like, conserved hypothetical protein, membrane | + | + | +g |
| mamX (MGR4149) | (1) | 268 | 5.5 | 26.2 | 2e-106 | 2e-106 | 2e-32 | Weakly similar to MamE and MamS proteins | + | + | +f,h |
TMH, transmembrane helices (numbers in parentheses indicate possible overprediction of the number of transmembrane helices); aa, amino acids; IEP, isoelectric point.
Only genes located within the MAI are shown. For a complete list see the supplemental material.
Not extracted by the described algorithm because of low complexity sequence structure.
Additionally identified as MMP; also conserved in R. rubrum (1e-75).
Arakaki et al. (3).
Tanaka et al. (41).
Grünberg et al. (17).
Matsunaga et al. (27).
Ullrich et al. (42).
Eleven of the extracted MTB-specific genes were found present in all four MTB including MC-1 (Table 2). Six of them encode previously identified MMPs (mamD, mamF, mamC, mamT, mamS, and mamI) or are cotranscribed with other MMP-encoding genes within the mamAB, mamGFDC, and mms6 operons located within the MAI (39). In this study, five new candidate genes, which are not located within the MAI, were identified.
We asked if the overlap in gene content is just a reflection of the close phylogenetic relationship of the four MTB strains within the Alphaproteobacteria or whether it is due to the common trait of magnetotaxis. We tested this hypothesis by tentatively including R. rubrum as a control in our analysis, because it represents the closest nonmagnetic 16S-rRNA relative of Magnetospirillum strains with a complete genome sequence available. This analysis yielded no genes that occurred in all four MTB and in R. rubrum. However, the intercomparison between the three magnetospirilla and R. rubrum yielded 41 conserved genes with no homologs in other organisms. Interestingly, one of them (MGR1900) matches an as yet unassigned peptide sequence (LLTELSDDVNR) obtained by a previous proteomic analysis of the MM (17) and thus represents another MMP, which we designate mmeA (magnetosome membrane associated). MmeA has no known function and contains a predicted signal peptide sequence (Table 2).
(ii) MTB-related genes.
A subset of known genes involved in magnetosome formation (exemplified by mamH, mamK, mamO, mamP, mamQ, mamA, mamB, mamM, and mamE) belongs to gene families that also occur in nonmagnetic bacteria, although exhibiting lower similarity. We reasoned that this class defined as MTB-related genes is extractable by a systematic approach, provided that the genes display a phylogeny consistent with their functional adaptation in MTB, i.e., if the degree of similarity within the magnetotactic members of these families is significantly higher than the similarity to genes in nonmagnetotactic organisms. In order to define cutoff values appropriate to achieve this discrimination within the gene families, we analyzed the set of known magnetosome genes of strain MSR-1 with similarity to genes outside the group of MTB for their degree of conservation within and without the MTB by BLAST analysis.
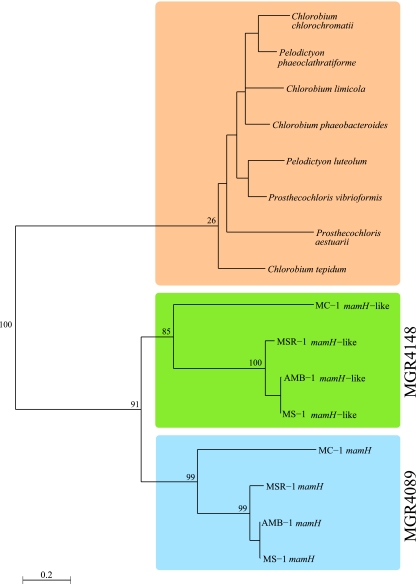
The analysis of other known magnetosome genes shared by all four MTB revealed that, with the exception of mamQ, which has a closer homolog in Thermotoga maritima than in MC-1 based on the BLAST E value, all analyzed genes form coherent groups within their respective gene families and can be discriminated from nonmagnetotactic homologs by their E values. These are consistently between 5e-05 and 1e-48 for the closest homologs in non-MTB, whereas they are generally below 1e-80 for the Magnetospirilla and below 1e-30 for Magnetococcus. A characteristic example for the relationship between these homologous genes is illustrated by the phylogenetic analysis of the MamH protein (Fig. 3). MamH (MGR4089) belongs to the major facilitator superfamily (MFS), a family of transporters capable of transporting small solutes. MamH orthologs from the four MTB form a coherent group compared to the remotely related genes from nonmagnetotactic MTB protein to members of the Chlorobia phylum. Interestingly, an additionally identified MamH-like MTB-related gene (MGR4148 in MSR-1) forms another distinct branch.
FIG. 3.
Phylogenetic tree of MamH (MGR4089) orthologous and paralogous proteins including the MTB-related MGR4148 (maximum-likelihood analysis). MamH represents a typical example for an MTB-related protein defined in this study; i.e., it forms a coherent phylogenetic branch within its family tree. In addition, the newly identified MTB-related MGR4148 gene is related to MamH but forms a distinct group. The three major clusters are indicated by different colors. The numbers indicate the bootstrap support for selected nodes.
Based on the inspection of relationships between known mam genes and nonmagnetobacterial homologs, the following criteria were set up to search for MTB-related genes: (a) best BLAST hit to MC-1 E value of <1e-30, (b) best BLAST hit to MS-1 E value of <1e-80, (c) best BLAST hit to AMB-1 E value of <1e-100, and (d) best BLAST hit to nonmagnetic genomesDB E value of >1e-50.
Seventeen MTB-related genes were identified by these criteria (Table 3, extracted genes), including nine mam genes. In addition, eight novel candidate genes were extracted, which, with the exception of MGR4148, are located outside the MAI (Table 3).
TABLE 3.
Summary of MTB-related genes with homologs in nonmagnetotactic bacteria and characteristics of their productsa
| Geneb | No. of TMH | No. of aa | IEP | Size (kDa) | E value
|
Gene product | Located within the MAI | Conserved neighborhood | Identified as MMP | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB-1 | MS-1 | MC-1 | Hits outside MTB (organism) | |||||||||
| Nonextractable | ||||||||||||
| mamN | 10 | 436 | 6.7 | 46.1 | <1e-200 | <1e-200 | 7e-35 (Carboxydothermus hydrogenoformans) | Magnetosome protein MamN | + | + | +h | |
| mamU | 296 | 9.7 | 31.9 | 2e-111 | 1e-113 | 3e-21 (Mesorhizobium loti) | MamU protein | + | + | |||
| Extracted | ||||||||||||
| mamH | 12 | 427 | 7.4 | 45.6 | <1e-200 | 4e-85 | 1e-114 | 8e-39 (Pelodictyon luteolum) | MamH protein, MFS | + | + | |
| mamK | 359 | 5.2 | 39.0 | <1e-200 | <1e-200 | 4e-97 | 4e-17 (Methanopyrus kandleri) | MamK protein, MreB-actin-like, magnetosome filamente | + | + | +g | |
| mamO | 8 | 631 | 6.5 | 65.3 | <1e-200 | <1e-200 | 2e-76 | 2e-14 (Caulobacter crescentus) | Magnetosome protein MamO; putative serine protease | + | + | +g,h |
| mamP | (1) | 269 | 7.6 | 28.2 | 6e-106 | 4e-106 | 3e-32 | 5e-05 (Legionella pneumophila) | MamP protein; PDZ/DHR/GLGF domains | + | + | |
| mamQ | 1 | 271 | 6.1 | 29.9 | 3e-109 | 3e-109 | 3e-14 | 8e-16/ (Thermotoga maritima) | Magnetosome protein MamQ, LemA family | + | + | +h |
| mamA | 216 | 5.6 | 23.9 | 2e-110 | 2e-110 | 4e-35 | 7e-15 (Methanosarcina mazei) | Magnetosome protein MamA; TPR-like, activation of magnetosome vesiclesf | + | + | +g,h,i | |
| mamB | 3 | 296 | 5.2 | 31.9 | 5e-157 | 5e-157 | 4e-76 | 4e-41 (Hahella chejuensis) | Magnetosome protein MamB, cation efflux protein family, magnetosome-directed iron transport | + | + | +g,h |
| mamM | 3 | 317 | 5.8 | 34.4 | 1e-170 | 2e-170 | 8e-77 | 5e-34 (Geobacter metallireducens) | Magnetosome protein MamM; cation efflux protein family; magnetosome-directed iron transport | + | + | +g,h |
| mamE | (1) | 771 | 8.9 | 77.9 | <1e-200 | <1e-200 | 1e-54 | 1e-37 (Rhodopirellula baltica) | Magnetosome protein MamE; putative trypsin-like serine protease; PDZ domain | + | + | +g,h,i |
| MGR0611c | 872 | 5.2 | 97.7 | <1e-200 | <1e-200 | <1e-200 | 2e-21 (Geobacter sulfureducens) | Chemotaxis protein | ||||
| MGR1882/1883c,d | 871 | 5.1 | 96.6 | <1e-200 | <1e-200 | <1e-200 | 9e-20 (Desulfovibrio desulfuricans) | Chemotaxis protein | ||||
| MGR4148 | 18 | 660 | 9.9 | 71.7 | <1e-200 | <1e-200 | 3e-161 | 2e-27 (Chlorobium tepidum) | Similar to MamH protein; MFS | + | + | |
| MGR3500 | 479 | 6.2 | 52.9 | <1e-200 | <1e-200 | 1e-36 | 1e-48 (Idiomarina loihiensis) | Signal transduction protein containing cAMP-binding and CBS domainsj | + | |||
| MGR0292 | 419 | 6.3 | 46.1 | 1e-140 | 3e-144 | 7e-35 | 2e-38 (Azoarcus strain EbN1) | Type I secretion membrane fusion protein; HlyD | ||||
| MGR0267 | 1 | 362 | 9.5 | 39.5 | 5e-143 | 2e-143 | 3e-33 | 3e-43 (Silicibacter pomeroyi) | ATPase involved in chromosome partitioning | |||
| MGR0626 | 632 | 6.8 | 67.9 | 2e-131 | 9e-131 | 2e-33 | 3e-44 (Rhodospirillum rubrum) | Outer membrane receptor protein; mostly Fe transport | ||||
| MGR1564 | (1) | 494 | 6.6 | 53.5 | 2e-147 | 8e-152 | 7e-31 | 2e-45 (Pseudomonas fluorescens) | GGDEF domain protein | |||
TMH, transmembrane helices (numbers in parentheses indicate possible overprediction of the number of transmembrane helices); aa, amino acids; IEP, isoelectric point.
Nonextractable, Magnetospirillum-specific genes absent from MC-1; extracted, MTB-related genes present in MC-1.
Shows suspicious hit outside in NCBI-nr to “Candidatus Kuenenia stuttgartiensis.”
Separated in two genes in MSR-1, because of a frameshift.
Komeili et al. (21).
Komeili et al. (20).
Tanaka et al. (41).
Grünberg et al. (17).
Matsunaga et al. (27).
cAMP, cyclic AMP; CBS, cystathionine-beta-synthase.
MGR0611 and MGR1882/83 contain a weak conserved metallo-beta-lactamase superfamily domain (Pfam lactamase_B), which can be found apart from beta-lactamases in a couple of other proteins with uncertain functions. In addition, MGR0611 contains a cyclic nucleotide-binding domain (Pfam cNN_binding) and hemerythrin HHE cation binding domain (Pfam hemerythrin). Multiple hemerythrin genes have already been identified within the MAI of MSR-1 that are known to bind and transport oxygen and iron (11), and all four MTB show a significantly higher number of predicted proteins with a hemerythrin domain. For comparison, the closely related R. rubrum contains only 5 genes, whereas AMB-1 contains 30 genes. This might imply an important function in magnetotaxis for these proteins.
The genes MGR0611 and MGR1882/83 represent a special case because they have only weak similarity to hits in the genomesDB but display significant similarities (E value of <1e-200; 49% identity) to genes found in the draft genome of the planctomycete “Candidatus Kuenenia stuttgartiensis.” Since Planctomycetes, in common with the MTB, have intracellular membranes and form unique compartments (pirellulosome) (15), it might be speculated that these shared genes are somehow related to the formation of similar subcellular organelles in both organisms.
If hits to R. rubrum with an E value of <1e-80 are included, only one gene (MGR0464) was identified that meets the required criteria. MGR0464 (in R. rubrum annotated as ArgK protein) is related to genes that encode proteins having ATPase and kinase activity and is possibly involved in lysine-arginine-ornithine/arginine-ornithine transport systems.
Analysis of conserved neighborhoods.
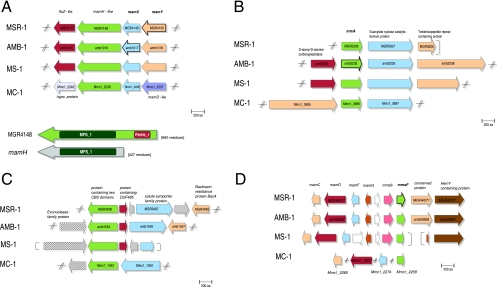
Genes involved in the same pathway tend to cluster in prokaryotic chromosomes. This can be exploited to infer functional coupling between genes (31). All identified MTB-specific and MTB-related genes present in the four MTB strains were manually inspected for conserved content. Three of the group-specific genes display no synteny (MGR0611, MGR1564, and MGR1882/MGR1883). Two genes (MGR2333 and MGR2349) showing a conserved neighborhood were identified as phage related and therefore were not further considered. Four genes display a conserved cluster organization in the three Magnetospirillum strains (MGR0292, MGR0267, MGR0626, and MGR3500), and five display a conserved cluster organization in all four analyzed MTB (MGR0208, MGR3500, MGR4072, MGR4148, and MGR4149). These genes, which are highly conserved with respect to both sequence and organization, are discussed in detail in the following paragraphs (Fig. 4).
FIG. 4.
Gene neighborhood representation of selected group-specific genes. Identical colors indicate homologous genes in the corresponding genomes. Arrows in bold lines indicate identification of the gene product within the magnetosome membrane. (A) mamXY cluster. Conserved gene neighborhood of MGR4148, mamX (MGR4149), and mamY (MGR4150) (top). Schematic representation of the different Pfam domain structure of the MTB-related gene MGR4148 compared to mamH (bottom) (B) Gene neighborhood of mtxA. The corrected annotation for the MGR0208 homolog of AMB-1 is shown. (C) Gene neighborhood of MGR3500. (D) Gene neighborhood of mmsF (MGR4072).
The mamXY cluster encodes several group-specific MMPs.
In all four MTB, MGR4148 and MGR4149 are part of a cluster containing four genes. However, only two of them are identical between the Magnetospirillum strains and MC-1. In the latter strain, one gene encodes a protein that exhibits the highest similarity to the MamD protein, which in MSR-1 was identified as one of the most abundant MMPs containing an LG repeat domain (17).
The highly conserved MTB-related gene MGR4148 (Fig. 4A, top) is located within the MAI and encodes a protein that exhibits a unique two-domain organization exclusively found in MTB (Fig. 4A, bottom): the N-terminal domain is similar to the Pfam MFS (Pfam MFS_1) and is homologous to the MamH protein. The C-terminal domain is similar to a ferric reductase-like transmembrane component (Pfam Ferric_reduct). Interestingly, a mutant lacking the Ferric_reduct domain of frp1 in Schizosaccharomyces pombe is deficient in ferric iron uptake (36).
MGR4148 is preceded in all four MTB by the MTB-specific gene MGR4149, whose product displays a weak and partial similarity to both the magnetosome proteins MamE (N-terminal domain) and MamS (central domain). Since this protein (amb1017) was experimentally identified as MMP in AMB-1 (41), we designate MGR4149 as MamX.
The Magnetospirillum-specific gene MGR4150, which in all magnetospirilla is the first gene of the cluster, encodes a fragment (VLSQEITQELSHIAQSR) that shows a 100% match to a peptide sequence obtained by previous analysis of the magnetosome subproteome but that could not been assigned yet to a known gene sequence (17). We therefore designated MGR4150 as MamY. The last gene of this operon in the magnetospirilla is a ftsZ-like gene fragment consisting of Pfam Tubulin and Tubulin_C domains. These domains are present in prokaryotic homologs of eukaryotic tubulins or bacterial FtsZ-like proteins, which are involved in cell division (25). The colocalization of magnetosome proteins with genes potentially involved in signal transduction and cytoskeletal elements makes it tempting to speculate that they might be involved in environmental regulation of magnetosome biomineralization and perhaps in signal transduction to the magnetosome chain.
The mtx cluster is putatively involved in magnetotaxis.
The MTB-specific protein MGR0208 contains a predicted signal peptide sequence. Its corresponding orthologs in AMB-1, MS-1, and MC-1 exhibit high amino acid identity (60%, 60%, and 50%, respectively) and have identical lengths. In the published original annotation of AMB-1 (27), the corresponding ORF is apparently mispredicted and yields a truncated polypeptide (251 amino acids) lacking the signal peptide. However, manual inspection revealed a start codon in a conserved position yielding a longer sequence, as shown in Fig. 4B. Remarkably, this polypeptide was recently identified as an MMP by a proteomic approach in AMB-1 (41), and accordingly we propose to designate it mtxA (mtx for magnetotaxis).
MtxA is part of a conserved cluster of genes. In all four MTB strains, it is followed by a highly conserved gene with similarities to Pfam's adenylate cyclase family model and CHASE2 domain. This domain organization belongs to a widespread group of transmembrane receptors that function as sensors for monitoring environmental changes and regulatory circuit function in catabolite repression in microorganisms (45).
MGR3500 contains two cystathionine-beta-synthase domains.
A further highly conspicuous candidate protein is encoded by the MTB-related gene MGR3500 (Fig. 4C). This gene is embedded within a gene cluster that displays some variation yet exhibits recognizable conservation between all four MTB. The cluster consists of MGR3500, MGR3497 (a symporter family protein), and MGR3499 (containing the Pfam domain of unknown function, DUF485). However, MGR3499 is missing in MC-1.
The mamGFDC and mms6 operons.
The MTB-specific gene MGR4072 located within the mms6 operon (Fig. 4D) also matches to an unassigned peptide sequence (LPVVSWVADRI) obtained by proteomic analysis of the MM (17) (and thus represents another MMP designated as mmsF). The predicted protein sequence resembles the magnetosome protein MamF with 62% identity. Additionally, the genome of AMB-1 contains three mamF-like genes ranging from 83% to 59% amino acid identity, whereas MC-1 contains two mamF-like genes (with a lower conservation level). MSR-1 and MS-1 contain no further copies of mamF-like genes. Although MamF is one of the most abundant proteins in MM in M. gryphiswaldense, its function is still unknown.
MGR4072 is followed by two conspicuous genes: MGR4071 is an Alphaproteobacteria-specific gene, which narrowly escaped the filter criteria for a Magnetospirillum-specific gene (best hit to non-MTB Sinorhizobium meliloti of 1e-05 with 55% coverage). MGR4070 contains a weak hit to the tetratricopeptide-like helical domain (InterPro IPR011990), which is known from other magnetosome proteins (MamA) and assumed to be involved in protein-protein interactions. Most genes of this region have counterparts also in strain MC-1, in which, however they are not clustered but are scattered around the genome.
DISCUSSION
In this study, we determined the nearly complete genome sequence of M. gryphiswaldense strain MSR-1 and used these data for the first detailed report on the computational identification of magnetosome-specific genes by a straightforward comparative genomics approach. Consistent with the 16S rRNA-based phylogeny and the common morphology and physiology, the MSR-1 genome is similar to the genomes of the two other Magnetospirillum species with respect to size, gene content, and phylogenetic distribution of genes. Analysis at the genome level also confirmed the close relationship of members of the genus Magnetospirillum to the Rhodospirillaceae, in particular R. rubrum. The magnetotactic coccus strain MC-1 appears to be only distantly phylogenetically related to Magnetospirillum, based on 16S rRNA analysis. The genomic comparisons support this conclusion and show that a magnetotactic phenotype is only weakly reflected by the overall gene content of the genome.
Extraction of group-specific signature genes had to be sufficiently stringent in order to exclude false positives but also sensitive enough to not increase the number of false negatives. It might be argued that the selection of our E-value thresholds is somewhat arbitrary. However, the cutoff E values are consistent with the range of similarity found in the previously known MM proteins, and variation of the cutoff E value within a conservative range did not affect the number of identified hits (data not shown).
The first approximation of a magnetobacterial signature set of genes will likely be subject to modification as further data emerge. Apart from a small number of genes that might have escaped detection because of the incompleteness of two of the MTB genomes, the number of MTB-specific genes is likely to shrink further, given the rapid growth of the protein database, which will reveal homologous genes in non-MTB that so far are classified as missing. On the other hand, the future addition of genomes from currently unrepresented, uncultivated branches and phenotypes of MTB will reveal further genes specific for MTB, for example, those associated with the formation of uncommon numbers, shapes (e.g., bullet and needle-like), or arrangements (e.g., bundles of chains) of magnetite crystals dissimilar from the investigated four strains or those controlling the formation of greigite instead of magnetite crystals in some uncultivated MTB. Regardless of uncertainties in precisely defining the signature set, even this first approach returned highly interesting results.
Out of the magnetobacterial core genome of about 891 genes, we identified 28 signature genes as magnetobacterial group specific, i.e., which occur in all four analyzed MTB. Eleven genes, designated as MTB specific, from this set do not have any obvious similarity to genes in nonmagnetotactic organisms. In addition to MTB-specific genes, relaxing our constraints to include genes with similarity in non-MTB increased this set with the risk of potentially increasing the number of false positives. However, only as few as 17 MTB-related genes present in all four genomes were extracted, which exhibit only remote similarity to genes from nonmagnetotactic organisms (MTB-related genes). Another subset of genes is conserved in all Magnetospirillum strains, but they do not have counterparts in MC-1 (Magnetospirillum-specific genes), as exemplified by mamN and mamU (Table 3). We failed to extract further members of this particular group with the algorithm described above because of the general phylogenetic coherency of the genes between the closely related Magnetospirillum strains, which makes it impossible to discriminate between “general” genes and putative magnetosome genes based on simple similarity filtering.
Using a similar approach, Raymond et al. (34) described the detection of photosynthetic-specific and photosynthesis-related genes by finding all genes shared within the subset of photosynthetic genes and then subtracting from this set those genes found in nonphotosynthetic organisms. This revealed a set of 53 photosynthesis-related and photosynthetic-specific candidate proteins. A similar approach was used for the identification of potential genetic determinants of hyperthermophily (24). Fifty core cyanobacteria-specific COGs exclusively shared by Cyanobacteria were extracted by genome analysis of 15 complete cyanobacterial genomes (30). In contrast, only one protein, reverse gyrase, was encoded in the genomes of all hyperthermophiles and not in any other genome, as identified by a further comparative approach by phylogenetic profiling (10). Compared to this latter situation, the set of MTB-specific and -related genes seems to be rather complex and approaches the number of genes implicated in bacterial photosynthesis.
A substantial number of Magnetospirillum-specific genes are currently annotated as hypothetical in MC-1, MS-1, and AMB-1. Since we found homologous genes in at least three of the MTB, it is clear that these are actual genes of unassigned function. In addition, four (mamY [MGR4150], mtxA [MGR0208], mmsF [MGR4072], and mamX [MGR4149]) novel MMPs were identified in MSR-1 by linking identified signature genes to experimental evidence obtained from previously unassigned peptide sequences of purified magnetosomes (17) or proteomic data from AMB-1 (27, 41).
MTB-related and MTB-specific proteins conserved in all MTB are of special interest as they may represent a core set of genes required for the magnetotactic phenotype. However, the largest set of group-specific MSR-1 genes (152 genes) identified in this study is represented by Magnetospirillum-specific genes (see Table S1 in the supplemental material). Remarkably, they include a number of genes encoding MMPs or which have a confirmed role in magnetosome biomineralization. For example, a crucial function in iron nucleation was attributed to the magnetosome-associated Mms6 protein by in vitro crystallization experiments (3). Another Magnetospirillum-specific protein, the acidic protein MamJ, was demonstrated to have an essential role in the proper alignment of magnetosome chains along the cytoskeletal magnetosome filament (37).
A special case is MGR1900, which encodes an MMP and was designated as MmeA. It represents an exception as it does not strictly meet the criteria for MTB-specific genes because its only known nonmagnetic homolog is in the genome of the closely related R. rubrum but is not present in that of the magnetic coccus MC-1. The significance of this finding is not clear at the moment, but one might speculate that its presence in nonmagnetotactic bacteria could be associated with common features of cellular organization shared by MTB and photosynthetic bacteria, such as, for example, the ability to synthesize intracytoplasmatic membrane vesicles invaginating from the cell membrane (21).
The apparent absence of genes in MC-1 could, on the one hand, mean that some of these genes are not essential for magnetotaxis and have entirely unrelated functions or, on the other hand, that they have functions specifically required for magnetotaxis only in the magnetospirilla. For example, the different shape, number, and organization of magnetosomes as well the different mode of magnetotaxis and niche adaptation between the freshwater magnetospirilla and the marine coccus MC-1 might explain the genetic variability between them. Moreover, the physiology of strain MC-1 is very different from magnetospirilla. Another explanation could be that the same essential functions are performed in MC-1 by unrelated or at least nonorthologous proteins, a phenomenon described as nonorthologous gene displacement (22).
The predominant fraction of identified MTB-specific and MTB-related genes displays synteny. Our screen of the signature set for genes of conserved context revealed three experimentally proven and four predicted operons. Several group-specific genes are colocalized with genes implicated in signal transduction, suggesting that these operons might have a function in the control of magnetotactic motility. Interestingly, several of the MTB-specific and -related genes were found in close proximity to another signature gene, which corroborates their suspected functions in magnetotaxis. The cotranscription together with nonsignature genes within the same operon suggests that less specialized gene functions may contribute to the magnetotactic phenotype, perhaps by carrying out supporting or accessory roles, such as coordinating and integrating magnetite formation and magnetotaxis with more general metabolic activities.
With the exception of MTB-specific genes confined to the genus Magnetospirillum, which are likely to include several false positives, the majority of identified candidate genes are located within the MAI in MSR-1. Many group-specific genes are located within the mamAB, mamGFDC, and mms6 operons. In addition, multiple hits were found in another cluster within the MAI, encoding two novel MMPs. This mamXY operon in MSR-1 comprises four genes including one that encodes multidomain MMPs with similarity to the MamE and MamS proteins (MamX), an additional gene encoding a MamH-like MFS paralogous protein with a ferric reductase-like domain, and the Magnetospirillum-specific MMP MamY. Altogether, this intriguing characteristic makes it a prime candidate operon for magnetosome formation within the MAI in addition to the known mamAB, mamGFDC, and mms6 operons.
The fact that at least one MMP and a number of other magnetobacteria-specific genes are located outside the MAI suggests that the MAI encodes many, but apparently not all, gene functions required for magnetotaxis. The scattering of group-specific genes argues against a recent single horizontal gene transfer event. In addition, the phylogeny of all analyzed group-specific genes appears fully consistent with the 16S rRNA-derived phylogeny of the four MTB. Several of the group-specific genes located outside the MAI belong to gene families involved in cell signaling and chemotaxis. For example, a large chemotaxis protein (MGR0611) exhibits striking conservation between all four strains (approximately 50% amino acid identity). Another gene, encoding a previously unrecognized MMP (MGR0208, designated mtxA in this study) is located within a predicted operon outside the MAI, which also contains a gene encoding a putative sensor protein for monitoring environmental changes. Thus, the preliminary experimental analysis and prediction of gene functions might, on the other hand, suggest a scenario in which genes specifically required for the synthesis and biomineralization of magnetosome particles are confined within the MAI, whereas the genomic organization of gene functions required for the “taxis” part of magnetotaxis is less conserved and displays a wider distribution.
In conclusion, we identified a set of 28 group-specific genes likely to be specifically associated with the magnetotactic phenotype in MTB. Fourteen of them were previously unrecognized and represent novel candidate genes for roles in magnetite biomineralization. Although we can only speculate on the function of these unassigned genes at this time, the genes described in this study will allow researchers interested in magnetotaxis and biomineralization to focus efforts to a small subset of genes that are likely to be of key interest which can be tested by experimental techniques.
Supplementary Material
Acknowledgments
We thank T. S. Brettin, H. E. Kiss, D. Martinez, N. Thayer, and G. Xie of the Joint Genome Institute of the U.S. Department of Energy and Los Alamos National Laboratories for sequencing and annotation of the MC-1 genome. R. Kottmann, C. Quast, and E. Prüsse (all MPI Bremen) are acknowledged for helpful suggestions to improve jCOAST, version 1.0. We also thank M. Schüler (MPI Bremen) and C. Jogler (LMU München) for helpful comments and J. Thiel, I. Müller, and A. Beck (all MPI Berlin) for technical and informatics support.
This study was supported by the Max Planck Society and the German BMBF Biofuture program. D.A.B. is supported by U.S. National Science Foundation grant EAR-0311950.
Footnotes
Published ahead of print on 20 April 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexandre, G., S. Greer-Phillips, and I. B. Zhulin. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28:113-126. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., J. Peplies, and D. Schüler. 2006. Diversity and taxonomy of magnetotactic bacteria, pp. 25-36. In D. Schüler (ed.), Magnetoreception and magnetosomes in bacteria. Springer-Verlag, Berlin, Germany.
- 3.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 4.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. [DOI] [PubMed] [Google Scholar]
- 5.Bazylinski, D. A., A. J. Dean, T. J. Williams, L. K. Long, S. L. Middleton, and B. L. Dubbels. 2004. Chemolithoautotrophy in the marine, magnetotactic bacterial strains MV-1 and MV-2. Arch. Microbiol. 182:373-387. [DOI] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Blakemore, R. P., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP, phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 10.Forterre, P. 2002. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 18:236-237. [DOI] [PubMed] [Google Scholar]
- 11.Frankel, R. B. 2006. Aerotaxis, pp. 1-24. In D. Schüler (ed.), Magnetoreception and magnetosomes in bacteria. Springer-Verlag, Berlin, Germany.
- 12.Frankel, R. B., D. A. Bazylinski, M. S. Johnson, and B. L. Taylor. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda, Y., Y. Okamura, H. Takeyama, and T. Matsunaga. 2006. Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB-1 reveals how magnetosome synthesis developed. FEBS Lett. 80:801-812. [DOI] [PubMed] [Google Scholar]
- 14.Galperin, M. Y., and E. V. Koonin. 2004. Conserved hypothetical proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glöckner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 100:8298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Grünberg, K., E. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, F., H. Ou, and C. Zhang. 2003. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 31:1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komeili, A., Z. Li, D. K. Newman, and G. J. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 22.Koonin, E. V., A. R. Mushegian, and P. Bork. 1996. Non-orthologous gene displacement. Trends Genet. 12:334-336. [PubMed] [Google Scholar]
- 23.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 24.Makarova, K. S., Y. I. Wolf, and E. V. Koonin. 2003. Potential genomic determinants of hyperthermophily. Trends Genet. 19:172-176. [DOI] [PubMed] [Google Scholar]
- 25.Margolin, W. 2006. Bacterial division: another way to box in the ring. Curr. Biol. 16:881-884. [DOI] [PubMed] [Google Scholar]
- 26.Martin, K. A., J. L. Siefert, S. Yerrapragada, Y. Lu, T. Z. McNeill, P. A. Moreno, G. M. Weinstock, W. R. Widger, and G. E. Fox. 2003. Cyanobacterial signature genes. Photosynth. Res. 75:211-221. [DOI] [PubMed] [Google Scholar]
- 27.Matsunaga, T., Y. Okamura, Y. Fukuda, A. T. Wahyudi, Y. Murase, and H. Takeyama. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12:157-166. [DOI] [PubMed] [Google Scholar]
- 28.Meldrum, F. C., S. Mann, B. R. Heywood, R. B. Frankel, and D. A. Bazylinski. 1993. Electron microscopy study of magnetosomes in a cultured coccoid magnetotactic bacterium. Proc. Biol. Sci. 251:231-236. [Google Scholar]
- 29.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, J. Clausen, J. Kalinowski, B. Linke, O. Rupp, R. Giegerich, and A. Pühler. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulkidjanian, A. Y., E. V. Koonin, K. S. Makarova, S. L. Mekhedov, A. Sorokin, Y. I. Wolf, A. Dufresne, F. Partensky, H. Burd, D. Kaznadzey, R. Haselkorn, and M. Y. Galperin. 2006. The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. USA 103:13126-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterman, A., and R. Overbeek. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 7:238-251. [DOI] [PubMed] [Google Scholar]
- 32.Pradel, N., C.-L. Santini, A. Bernadac, Y. Fukumori, and L.-F. Wu. 2006. Biogenesis of actin-like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc. Natl. Acad. Sci. USA 103:17485-17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast, C. 2006. MicHanThi-design and implementation of a system for the prediction of gene functions in genome annotation projects. Ph.D. thesis. University of Bremen, Bremen, Germany.
- 34.Raymond, J., O. Zhaxybayeva, J. P. Gogarten, S. Y. Gerdes, and R. E. Blankenship. 2002. Whole genome analysis of photosynthetic prokaryotes. Science 298:1616-1620. [DOI] [PubMed] [Google Scholar]
- 35.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 36.Roman, D. G., A. Dancis, G. J. Anderson, and R. D. Klausner. 1993. The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol. Cell. Biol. 13:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheffel, A., M. Gruska, D. Faivre, A. Linaroudis, P. Graumann, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along filamentous structure in magnetotactic bacteria. Nature 440:110-114. [DOI] [PubMed] [Google Scholar]
- 38.Schleifer, K., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Köhler. 1991. The genus Magnetospirillum gen. nov. description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 39.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siew, N., Y. Azaria, and D. Fischer. 2004. The ORFanage: an ORFan database. Nucleic Acids Res. 32:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, M., Y. Okamura, A. Arakaki, T. Tanaka, H. Takeyama, and T. Matsunaga. 2006. Origin of magnetosome membrane: proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics 6:5234-5247. [DOI] [PubMed] [Google Scholar]
- 42.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams, T. J., C. L. Zhang, J. H. Scott, and D. A. Bazylinski. 2006. Evidence for autotrophy via the reverse tricarboxylic acid cycle in the marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 72:1322-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, G. A., N. Bertrand, Y. Patel, J. B. Hughes, E. J. Feil, and D. Field. 2005. Orphans as taxonomically restricted and ecologically important genes. Microbiology 151:2499-2501. [DOI] [PubMed] [Google Scholar]
- 45.Zhulin, I. B., A. N. Nikolskaya, and M. Y. Galperin. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.