Abstract
Pyruvate-dependent reduction of NADP has been demonstrated in cell extracts of the human gastric pathogen Helicobacter pylori. However, NADP is not a substrate of purified pyruvate:ferredoxin oxidoreductase (PFOR), suggesting that other redox active enzymes mediate this reaction. Here we show that fqrB (HP1164), which is essential and highly conserved among the epsilonproteobacteria, exhibits NADPH oxidoreductase activity. FqrB was purified by nickel interaction chromatography following overexpression in Escherichia coli. The protein contained flavin adenine dinucleotide and exhibited NADPH quinone reductase activity with menadione or benzoquinone and weak activity with cytochrome c, molecular oxygen, and 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB). FqrB exhibited a ping-pong catalytic mechanism, a kcat of 122 s−1, and an apparent Km of 14 μM for menadione and 26 μM for NADPH. FqrB also reduced flavodoxin (FldA), the electron carrier of PFOR. In coupled enzyme assays with purified PFOR and FldA, FqrB reduced NADP in a pyruvate- and reduced coenzyme A (CoA)-dependent manner. Moreover, in the presence of NADPH, CO2, and acetyl-CoA, the PFOR:FldA:FqrB complex generated pyruvate via CO2 fixation. PFOR was the rate-limiting enzyme in the complex, and nitazoxanide, a specific inhibitor of PFOR of H. pylori and Campylobacter jejuni, also inhibited NADP reduction in cell-free lysates. These capnophilic (CO2-requiring) organisms contain gaps in pathways of central metabolism that would benefit substantially from pyruvate formation via CO2 fixation. Thus, FqrB provides a novel function in pyruvate metabolism and, together with production of superoxide anions via quinone reduction under high oxygen tensions, contributes to the unique microaerobic lifestyle that defines the epsilonproteobacterial group.
Much of the world's population is infected with Helicobacter pylori, a genetically diverse microaerophile that establishes lifelong infections of the gastric mucosa and causes chronic gastritis and duodenal and gastric ulcers (3, 12, 13). Infection with H. pylori is also a risk factor in development of mucosa-associated lymphoid tissue lymphoma and adenocarcinoma, one of the most frequently lethal of all cancers (12, 32). The ability to persist for decades in the gastric mucosa is largely dependent on the microbe's ability to promote sufficient inflammation to acquire nutrients from serum leakage (carbon and energy sources and urea for the pH-neutralizing urease) without succumbing to the deleterious reactive oxygen and nitrogen intermediates generated by the innate immune response (41). The related epsilonproteobacterium Campylobacter jejuni establishes lifelong infections of the intestinal tracts of birds, particularly domestic poultry, and is the leading cause of food-borne enteritis in humans (5). In contrast to H. pylori infection, infections with C. jejuni are usually self-limiting and generally do not require antimicrobial intervention.
These microaerophiles efficiently colonize and thrive within the gastrointestinal mucus and tend to concentrate deep in the crypts and near the underlying epithelial cells. Accordingly, the central intermediary metabolism of these organisms is very conserved and optimized for this microaerobic environment. Both C. jejuni and H. pylori utilize organic acids and amino acids as primary carbon and energy sources and rely on gluconeogenic pathways to provide the necessary intermediates for biosynthesis of cell wall material, vitamins, and nucleic acids (1, 11, 17, 19, 27, 39). Other general features of these pathogens include (i) a strictly respiratory form of metabolism; (ii) reliance on molecular hydrogen as a key energy source (14, 17, 18, 20, 28); (iii) a preference for NADPH, rather than NADH, as the primary electron donor; and (iv) pyruvate oxidation via a reversible pyruvate:ferredoxin oxidoreductase (PFOR) (6, 20). While the PFOR of H. pylori is composed of four subunits encoded by the porGDAB operon (20, 23, 37), the PFOR of C. jejuni is the product of a single gene, similar to the PFORs of anaerobic bacteria and amitochondriate protozoan parasites (21). The preference for flavodoxin as an electron carrier distinguishes the epsilonproteobacteria from other PFOR-containing organisms, which utilize low-redox-potential ferredoxins (10). It is the low-redox-potential ferredoxins that reduce the antimicrobial prodrug metronidazole to toxic, DNA-damaging hydroxylamine adducts (38).
The microaerophilic nature of the epsilonproteobacteria has been linked to intrinsic oxygen toxicity resulting from inactivation of PFOR, iron sulfur proteins, and other respiratory components and to the activities of NADPH oxidases, flavoproteins, and nitroreductases (8, 16, 27, 42). Our previous studies established that alkyl hydroperoxide reductase (AhpC) activity of H. pylori was dependent on the thioredoxin/thioredoxin reductase system (HP0824/HP0825) and displayed a preference for NADPH (2). Further inspection of the H. pylori KE26695 genome reveals two thioredoxin reductase genes (trxR1 [HP0825] and trxR2 [HP1164]) (1, 2, 39). However, HP1164 lacks the catalytic CXXC motif required for thioredoxin reductase activity and, unlike trxR1, is essential for viability (2, 6, 9). HP1164 contains motifs that place it in the general class of NADPH oxidase/disulfide reductases of unknown function (2).
Here we show that HP1164, which is uniquely conserved among the epsilonproteobacteria (6), exhibits strong NADPH flavodoxin and quinone reductase activity, and we designate it FqrB (flavodoxin quinone reductase). Our studies show that FqrB is coupled by flavodoxin to the PFOR system and catalyzes pyruvate-dependent production of NADPH. Moreover, the reverse reaction produces pyruvate via CO2 fixation, perhaps the major route of carbon assimilation by capnophilic members of the epsilonproteobacteria. Finally, we show that in the presence of oxygen, FqrB mediates single-electron reduction of quinones, generating stoichiometric concentrations of superoxide anions that may contribute to oxygen toxicity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strain KE26695 was grown under humid microaerobic conditions at 37°C on brucella-based medium supplemented with 7.5% newborn calf serum (Gibco Laboratories) and antimicrobials as previously described (18, 24). Campylobacter jejuni strain H840 (ATCC 29428) was grown in brucella broth as previously described (17). Escherichia coli strains were cultured in Luria-Bertani medium supplemented with appropriate antibiotics.
DNA techniques.
DNA manipulations were carried out by the general techniques described by Sambrook et al. (35). Plasmid DNA was isolated with the QiaPrep Spin minikit (QIAGEN). DNA fragments and amplicons from PCR were purified from agarose gels with the QIAquick gel extraction kit (QIAGEN). Oligonucleotide primers used in this study are listed in Table 1. PCR mixtures generally contained (in 25 μl) 10 ng of H. pylori genomic DNA, 50 pmol of each primer, 200 μM (each) deoxynucleoside triphosphates, and 1 U of Taq DNA polymerase in standard PCR buffer (MBI Fermentas). Thirty amplification cycles were performed (94°C for 30 s, 53°C for 30 s, and 72°C for 1 min, with a final extension for 10 min) in a Perkin-Elmer 2400 thermal cycler.
TABLE 1.
Oligonucleotide primers
| Primer | Sequencea |
|---|---|
| HP1164 P1 | 5′-ACCCCACTCCTTAAAACCCTCTTG |
| HP1164 P1Ext | 5′-AGCCCCACTCCCTACCAT |
| HP1164 P1Int | 5′-ATCATGCTAAATTCGTGGTGGTTG |
| HP1164 P2 | 5′-AACGCCGGCTATTTCGCATTCTAC |
| HP1164 P2Int | 5′-TAGAGCCGCCGATCGCATACAG |
| HP1164 P3 | 5′-TTATTCAAATCAGGGGCGAGCAT |
| HP1164 P4 | 5′-ACAAGGCGTTAAAGAGCAATACC |
| HP1164 P4Ext | 5′-CTGTTAGTGGGGCATTTGATTACG |
| FqrBNdeI | 5′-AAAGGATACATATGAATCAAGAAATTTTAGACGT |
| FqrBXhoI | 5′-GTGCTCGAGGTGCAACCTTTTAGCGATTTCT |
| C1 | 5′-GATATAGATTGAAAAGTGGAT-3′ |
| C2 | 5′-TTATCAGTGCGACAAACTGGG-3′ |
The underlined nucleotide sequences indicate restriction sites.
Cloning of fqrB (HP1164).
The gene encoding HP1164 (fqrB) was amplified by PCR by using a forward primer containing an NdeI site permitting cloning into the ATG codon of pET29b (Novagen) and a reverse primer designed to produce an in-frame hexahistidine C-terminal tag following restriction with XhoI. The amplicon was ligated into a similarly restricted pET29b vector and transformed into E. coli BL21 CodonPlus(DE3)-RIL (Stratagene), and resulting kanamycin-resistant colonies containing the cloned fqrB were confirmed by PCR. One of these clones, pETfqrB, was chosen for further study. Overexpressed FqrB was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis by comparing extracts from IPTG (isopropyl-β-d-thiogalactopyranoside)-induced and noninduced cultures. FqrB was purified by nickel interaction chromatography as previously described (37). The purity and molecular mass of the eluted protein were confirmed by electrophoresis, and protein concentrations were estimated by the Bradford assay (Bio-Rad) with bovine serum albumin as a standard.
Spectral analysis of FqrB and cofactor determination.
Absolute spectra were recorded for the His6-tagged FqrB protein suspended in 1 ml of 100 mM Tris buffer (pH 8.0), and to ensure complete oxidation, 1 mM hydrogen peroxide was added. The protein concentration was 20 μM. To obtain a reduced spectrum, a few crystals of sodium hydrosulfite were added to the sample. The reference cuvette contained buffer. The spectral range scanned was 250 to 700 nm. The flavin cofactor was identified by spectral characteristics following protein denaturation in trichloroacetic acid and comparison with flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and riboflavin standards.
FqrB substrate specificity.
The activity of His6-tagged FqrB was monitored spectrophotometrically in a modified Cary 14 spectrophotometer (OLIS, Bogart, GA) at 23°C, either by following the decrease in absorbance at 340 nm resulting from the oxidation of NADPH or by following the reduction of substrate at a specified wavelength. The reduction of various substrates was monitored at appropriate wavelengths, which include the following: metronidazole, 320 nm (ɛ = 9.0 mM−1 cm−1); nitrofurazone, 400 nm (ɛ = 12.6 mM−1 cm−1); furazolidone, 400 nm (ɛ = 18.8 mM−1 cm−1); horse heart cytochrome c, 550 nm (ɛ = 18.9 mM−1 cm−1); riboflavin or FMN, 450 nm (ɛ450 = 11.4 mM−1 cm−1); 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB), 412 nm (ɛ = 13.6 mM−1 cm−1); and nitazoxanide (NTZ), 418 nm (ɛ = 18.6 mM−1 cm−1). Substrates assayed by following the oxidation of NADPH included menadione, benzoquinone, o-nitrophenol, oxygen, purified thioredoxin (E. coli), thioredoxin (Spirulina), and H. pylori thioredoxins 1 and 2 (a gift from Leslie Poole, Wake Forest University). Specific activity measurements were performed in 1-cm-path-length quartz cuvettes in buffer A (Tris buffer [100 mM; pH 8.0] and NaCl [100 mM]) containing NADPH (300 μM) and appropriate substrate (100 μM). The reactions were initiated by addition of an appropriate dilution of His6-tagged FqrB, to a final reaction volume of 1.0 ml.
Quinone-cytochrome c reduction assay.
NADPH quinone reductase activity was measured with cytochrome c as an electron acceptor at 550 nm under aerobic conditions. The reaction mixture in buffer A contained 200 μM NADPH, 30 μM horse heart cytochrome c, and His6-tagged FqrB (165 μg protein), and the reaction was started by addition of 100 μM menadione. To assess superoxide anion participation in the reduction of cytochrome c, 200 U superoxide dismutase (SOD) was added to the reaction mixture. The difference in the rates of cytochrome c reduction in the presence and absence of SOD was used to compute the rate of superoxide anion generation.
Kinetic analysis of FqrB.
Initial-velocity kinetic assays were performed in triplicate for FqrB with the substrates NADPH, benzoquinone, and menadione by following the oxidation of NADPH at 340 nm under the following conditions: NADPH (5 to 150 μM) at a saturating menadione concentration (150 μM), benzoquinone (10 to 140 μM) at a saturating NADPH concentration (150 μM), and menadione (5 to 100 μM) at a saturating NADPH concentration (300 μM). The reactions were initiated by addition of His6-tagged FqrB to a final concentration ranging from 10 to 125 ng/ml. The values of Km (mM) and Vmax (mM min−1) were determined from plots of the initial velocity versus substrate concentration by nonlinear regression analysis using the program KaleidaGraph v. 3.5 (Synergy Software). Values for kcat (s−1) were determined from Vmax by using the predicted molecular mass of 37,051 Da for the C-terminally His6-tagged FqrB protein. The reported error is the standard deviation. All enzyme activities reported are within 5% error, and this includes variation from batch to batch purified by these methods.
Substrate inhibition assays with menadione and NADPH were performed in 1-cm-path-length quartz cuvettes in buffer A and were initiated by addition of His6-tagged FqrB to a final concentration of 50 ng/ml. Kinetic constants were determined at eight different concentrations of menadione (1,2, 5, 10, 20, 30, 40, and 60 μM) and at NADPH concentrations of 5, 10, 20, 40, and 60 μM. Apparent values of Vmax (Vmaxapp) were determined by nonlinear regression analysis, and 1/Vmaxapp was replotted against the reciprocal substrate inhibitor concentration (1/[NADPH] or 1/[menadione]) to determine values for Km,A and Km,B, respectively. The replots were derived from the following equation describing the forward velocity for a ping-pong bi-bi system in the absence of products, where NADPH is substrate A and oxidized menadione is substrate B: V = (VmaxAB)/(Km,AB + Km,BA + AB).
Coupled PFOR:FldA:FqrB assays.
Cloning of the porGDAB operon, expression, and purification from E. coli have been previously described (20, 37). H. pylori flavodoxin (FldA) was purified from E. coli as previously described (10). PFOR (EC 1.2.7.1) was assayed at 23°C under anaerobic conditions with 100 mM potassium phosphate (pH 7.0), 10 mM sodium pyruvate, 5 mM benzyl viologen (BV) (ɛ = 9.2 mM−1 cm−1 at 546 nm), 0.18 mM coenzyme A (CoA), and 1 mM MgCl2 as previously described (20, 37). Mixtures for coupled enzyme reactions contained PFOR (0.1 mg/ml), FldA (10 μM), and FqrB (2 μg/ml), and the reaction was started by adding 3 mM NADP to the assay mixture (to get a final concentration of 300 μM). Enzymatic assays were conducted under strict anaerobic conditions using a specially designed anaerobic cuvette that is coupled to an anaerobiosis device. Several cycles of vacuum and argon flow ensured the total absence of oxygen in the cuvette. Under these conditions, the reaction was initiated by introducing CoA, initially located in a separate compartment of the cuvette, to the PFOR assay mixture. The course of the reaction was followed in a diode array spectrophotometer (Agilent/HP 8453) with the cuvette closed to maintain strict anaerobiosis.
The mixture for the reverse reaction (pyruvate forming) contained 300 μM NADPH, 0.1 mM acetyl-CoA, and 5 mM bicarbonate. Alternatively, CO2 was bubbled into the cuvette prior to addition of NADPH to initiate the reaction. The dependence on each enzyme and each substrate in the forward and reverse reactions was determined in independent experiments. The reverse reaction was also monitored by A232 by following the appearance of reduced CoA (ɛ = 4.5 mM−1 cm−1). Enzymatic activities are reported as units/mg of protein (nmol or μmol min−1) for each of the enzymes added to the reaction mix. All assays were performed in triplicate, and means and standard deviations were computed. Variation in enzyme activity from batch to batch was also examined in triplicate in bacterial extracts prepared on different days.
NADP reduction in cell extracts.
Crude cell extracts of C. jejuni and H. pylori were prepared from liquid culture, and PFOR activities were assessed by rapid BV assay as described previously (20, 23, 37). Briefly, ca. 2 g (wet weight) bacterial pellet was suspended, disrupted by sonication, and subjected to low-speed centrifugation to remove cellular debris and unbroken cells. High-speed supernatant was prepared by ultracentrifugation at 100,000 × g for 1 h. Pyruvate-dependent PFOR activity was assayed by following the reduction of BV or NADP under anaerobic conditions. To confirm that NADPH production resulted from the activity of the PFOR-FldA-FqrB complex, the PFOR inhibitor NTZ (5 and 10 μM) was added to extracts. The endogenous NADP reductase activity of the extract was determined in the absence of pyruvate by following the reduction of NADP at 340 nm. Similarly, NADPH oxidase activity was measured in extracts in the absence of added pyruvate.
Bioinformatic analyses.
Whole genome sequences for H. pylori KE26695, H. pylori J99, Helicobacter hepaticus ATCC 51449, C. jejuni NCTC 11168, C. jejuni RM1221, and Wolinella succinogenes DSM 1740 were screened for common genes (e−15) by using the lineage-specific algorithm of Emu Web Services (http://emu.imb.uq.edu.au/index.php). From this list of common genes were subtracted all genes in common at a cutoff of e−25 with matches with any genes of E. coli K-12, Staphylococcus aureus MW2, or Pseudomonas aeruginosa. The short list (∼300 genes), including fqrB (HP1164), represents highly conserved genes unique to this group of microaerophiles. Motif searches utilized SMART software (36), homology searches used BLAST of microbial genomes (www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), and alignments of closely related sequences were done with CLUSTAL-W (www.ebi.ac.uk/clustalw/). Protein-protein interactions were evaluated using the PimRider (Hybrigenics), and gene organization was examined on the Pedant web site (http://pedant.gsf.de/).
RESULTS
Whole-genome analyses revealed that H. pylori expresses multiple thioredoxin reductases (HP0824 and HP1164) as well as two thioredoxins (HP0825 and HP1553) (1, 2, 39). Based on alignments between the two deduced thioredoxin reductase protein sequences and the absence of the characteristic CXXC motif required for thioredoxin reductase function, we concluded that HP1164 was not a thioredoxin reductase and therefore was incorrectly annotated. SMART analysis revealed that HP1164 is a member of a general class of NADPH-specific thiol reductases or NADPH oxidase proteins. To investigate the function of HP1164, an essential gene (2, 6), we purified the protein following overexpression and designed enzyme assays to generally test for NADPH oxidase activity using substrates such as cytochrome c, nitroblue tetrazolium, and molecular oxygen. These preliminary assays (NADPH oxidase activity) confirmed HP1164 as a member of this general class of NADPH oxidoreductases but of unknown biological function.
Phylogenetic analysis.
Concordance analyses of whole genomes of epsilonproteobacteria revealed that HP1164 (FqrB) was among some 300 uniquely conserved genes that are most likely associated with a microaerobic lifestyle. Figure 1 presents an alignment of the FqrB orthologs, highlighting regions that are highly conserved and pointing out putative functional domains. The group shared identities in amino acid conservation of 45 to 51% and similarities ranging from 66 to 69%. The closest matches outside this group were with members of the genus Bacillus, with best identities of 24% and similarities of 43%. FqrB showed no similarity with nitroreductases or other quinone reductases encoded within the H. pylori genome. Similarities with TrxR1 (HP0825) suggest that FqrB has diverged from thioredoxin reductases. The annotation error relates to a conserved region of HP1164 that contains the sequence INDC in H. pylori and VSDC in W. succinogenes (Fig. 1), which are similar to the CXXC motif found in thioredoxins. In Campylobacter species and other species of Helicobacter, this motif lacks the one cysteine and these sequences no longer match thioredoxin reductases by BLAST search.
FIG. 1.
Clustal W alignment of FqrB in epsilonproteobacteria. The protein sequences were obtained from the Pedant website. The boldface region contains a single cysteine in H. pylori (INDC) and W. succinogenes FqrB orthologs, a region in thioredoxin reductase proteins that contains a CXXC motif. In the C-terminal region there is a highly conserved cysteine (bold) that is likely involved in catalysis.
Purification and spectral properties.
His6-tagged FqrB was purified from E. coli BL21(DE3) by Ni2+ interaction chromatography as a pale yellow, ∼36-kDa protein. To confirm FqrB as a flavoprotein, absolute spectra were collected using the scanning mode of the Cary 14 spectrophotometer. As can be seen in Fig. 2, a typical flavin spectrum was obtained with protein treated with 1 mM H2O2 to completely oxidize the flavin. The flavin spectrum was bleached by addition of several grains of sodium hydrosulfite (spectrum not presented). The spectral properties of His6-tagged FqrB were very similar to those previously described for FldA (10). However, spectral analysis of the cofactor, released from the protein, showed an absorption maximum at 450 nm rather than 445 nm (typical of FMN), suggesting that the cofactor is FAD (data not presented).
FIG. 2.
Optical spectrum of FqrB showing the flavin. Absolute spectra were collected under oxidized conditions and a protein concentration of 1.75 mg/ml. Major peaks were detected at 378 nm and 462 nm, and a shoulder was detected at 465 nm. Comparison of the spectrum of the released cofactor with those of FAD, FMN, and riboflavin suggests that the cofactor is FAD.
Substrate specificity and kinetic constants.
Based on cytochrome c reductase activity, FqrB was much more active with NADPH (267 nmol min−1 mg protein−1) than with NADH (∼20 nmol min−1 mg protein−1). To determine whether FqrB was a thioredoxin reductase, the purified enzyme was monitored for NADPH thioredoxin reductase activity in the presence of several thioredoxins. FqrB did not reduce commercially available thioredoxins from E. coli and Spirulina, nor was it able to reduce Trx2 from H. pylori. The enzyme demonstrated weak activity with Trx1 from H. pylori (∼10 nmol min−1 mg protein−1), well below that which would be expected of a true substrate (2).
Several electron acceptors were examined as potential substrates in the presence of NADPH. A low enzyme-catalyzed basal rate of NADPH oxidase activity (∼5 nmol min−1 mg protein−1), in the absence of any added electron acceptors, implies that oxygen serves as a poor terminal electron acceptor. Low oxygen-specific activity is typical of most flavoproteins. The basal rate in the presence of oxygen was eliminated by purging the system with nitrogen or argon. Both cytochrome c and DTNB served as weak electron receptors for FqrB, with specific activities of 0.3 and 0.7 μmol min−1 mg protein−1, respectively. The purified enzyme exhibited no measurable activity with nitrofurans, o-nitrophenol, or metronidazole. As a control for these reactions, we examined the substrate preferences for the His6-tagged NfsB, the nitroreductase of E. coli, which exhibited nitroreduction of nitrofurans and o-nitrophenol but, like FqrB, exhibited no activity with metronidazole (data not presented). FqrB displayed the highest specific activity with purified flavodoxin (FldA) from H. pylori (126 μmol min−1 mg protein−1), suggesting a possible role as a flavodoxin reductase. However, riboflavin and FMN were not substrates of FqrB.
FqrB also displayed a high specific activity with the quinone substrates menadione and benzoquinone (112.5 and 124 μmol min−1 mg protein−1, respectively). These substrates gave specific activities nearly 3 orders of magnitude greater than those observed for the other examined substrates, implying that FqrB may also function as a quinone-specific oxidoreductase. NfsB also exhibited quinone reductase activity, as previously reported (43), and a specific activity nearly double that of FqrB (249 μmol min−1 mg protein−1). In the presence of menadione, NADPH quinone reduction could also be followed by monitoring cytochrome c reduction by His6-tagged FqrB under aerobic conditions. Since oxidation of semiquinones often generates superoxide anions by single-electron transfers, SOD was added to the reaction mixture to test this possibility. SOD inhibited cytochrome c reduction by 90% (from 58.5 to 7.6 μmol min−1 mg protein−1), confirming single-electron transfer to oxygen and raising the possibility that substantial superoxide (∼50 μmol min−1 mg protein−1) could be generated in whole cells during periods of oxidative stress.
The kinetic constants for menadione, benzoquinone, and NADPH in the presence of a saturating second substrate are reported in Table 2. Assuming that Km is representative of the binding affinity, at saturating NADPH, FqrB binds menadione with sixfold-greater affinity than it does benzoquinone, while it turns over benzoquinone at a rate approximately fourfold greater than it does for menadione. The catalytic efficiencies (kcat/Km) are approximately equal for benzoquinone and menadione. The enzyme displays a relatively high affinity for all three substrates, with Km values on the micromolar scale. Such high binding affinities for the two quinone substrates, combined with catalytic efficiencies of 106 M−1 s−1, are further evidence favoring the quinone-based substrates as the true substrates for FqrB. Kinetic analysis of FqrB reduction of flavodoxin (FldA) also indicated reduction of the FMN of FldA to the semiquinone and quinone in a two-step reaction under anaerobic conditions. Given the two possible functions for HP1164, we named the protein FqrB to reflect flavodoxin and quinone reductase activities.
TABLE 2.
Kinetic constants for menadione, benzoquinone, and NADPH in the presence of a saturating second substratea
| Substrate | Saturating second substrate | Km (μM) | kcat (s−1) | kcat/Km(M−1 s−1) |
|---|---|---|---|---|
| Menadione | NADPH | 14.4 ± 2.4 | 122 ± 12 | 8.4 (±1.6) × 106 |
| Benzoquinone | NADPH | 82.7 ± 18.6 | 474 ± 53 | 5.7 (±1.4) × 106 |
| NADPH | Menadione | 26.3 ± 3.5 | 115 ± 12 | 4.4 (±0.7) × 106 |
Reaction mechanism.
Both menadione and NADPH displayed parallel double-reciprocal plots (Fig. 3 and 4, respectively) characteristic of a ping-pong mechanism for FqrB. Replotting the reciprocal of the apparent Vmax values (Fig. 3 and 4, insets) for both substrates yielded the specific Km values, Km,A = 29.5 μM and Km,B = 10 0.2 μM, for NADPH and menadione, respectively. These values are in excellent agreement with the Km values determined for NADPH and menadione by using a saturating second substrate.
FIG. 3.
Initial velocity of FqrB at fixed [NADPH]. Menadione was the variable substrate, and NADPH was the fixed variable substrate at 5 μM (○), 10 μM (⧫), 20 μM (▴), 40 μM (▪), and 60 μM (•). Inset, replot of the reciprocal of Vmaxapp (obtained from nonlinear regression fit of the initial-velocity data) versus 1/[NADPH], yielding a value of 29.5 μM for the reciprocal of the x intercept.
FIG. 4.
Initial velocity of FqrB at fixed [menadione]. NADPH was the variable substrate, and menadione was the fixed variable substrate at 1 μM (⋄), 2 μM (□), 5 μM (▵), 10 μM (○), 20 μM (⧫), 30 μM (▴), 40 μM (▪), and 60 μM (•). Inset, replot of the reciprocal of Vmaxapp (obtained from nonlinear regression fit of the velocity-velocity data) versus 1/[menadione], yielding a value of 10.2 μM for the reciprocal of the x intercept.
Biological function of FqrB.
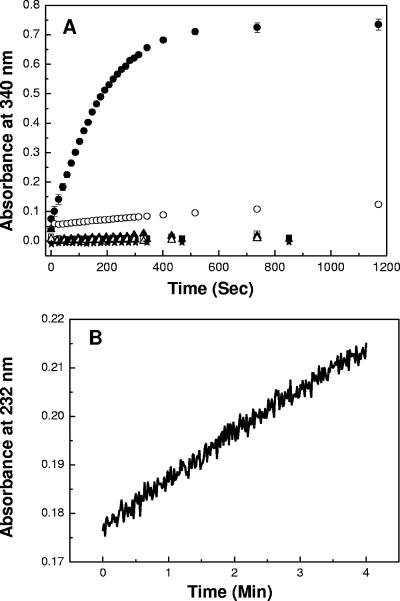
Since flavodoxin (FldA) was the most efficient substrate reduced by FqrB and since FldA was previously established as the electron carrier for PFOR (10, 22), we considered the possibility that FqrB was associated with the oxidative decarboxylation of pyruvate. To test this possibility, we purified PFOR, FldA, and FqrB and included them in assays of pyruvate-dependent NADPH formation under anaerobic conditions. Pyruvate-dependent NADP reduction was demonstrated, and removal of any of the enzymes or substrates resulted in loss of enzyme activity (Table 3 and Fig. 5A). The maximum activity recorded (49 to 52 nmol min−1) was under the optimal conditions of 5 μM FldA, 90 μg/ml PFOR, 38 μg/ml FqrB, and all substrates at saturation. PFOR was the rate-limiting enzyme (0.57 μmol min−1 mg PFOR protein−1) in the reaction, with a Km for pyruvate of ∼300 μM (20), and when the reaction was optimized for FqrB, the specific activity was 24.5 μmol min−1 mg FqrB protein−1. FqrB showed some residual activity which might be attributable to weak nonspecific reduction of FldA or weak interaction of reduced PFOR directly with FqrB. The specific activity of purified PFOR with artificial electron acceptors (BV), when corrected for single-electron transfer, is equivalent to the activities determined for the complex. As previously established, there is a low rate of NADP reductase activity associated with FqrB that is evident in the absence of PFOR and FldA (Table 3). We also noted in these studies that PFOR becomes inactivated in the presence of molecular oxygen, as reflected in assays that did not strictly exclude oxygen from the reaction (Table 3). We had recently shown that the antiparasitic/antibacterial drug NTZ is a noncompetitive inhibitor of PFOR (20). These studies were performed with the artificial electron acceptor BV. To determine if the coupled reaction and NADPH production were also inhibited by NTZ, concentration-dependent inhibition was determined by following the appearance of NADPH with time. At a concentration of 5 μM, NTZ inhibited the coupled reaction by nearly 50% (Table 3), indicating that PFOR activity was necessary for FqrB function.
TABLE 3.
Pyruvate-dependent NADP reduction
| Reaction componentsa | NADP reduction (nmol/min)b | Sp act (U/mg protein)c
|
|
|---|---|---|---|
| PFOR | FqrB | ||
| Complete (NADP) | 27 ± 2 | 0.269 ± 0.02 | 13.5 ± 1 |
| Complete (NAD) | 1.4 ± 0.2 | 0.014 ± 0.002 | 0.7 ± 0.1 |
| −PFOR | 0.03 | 0 | 0.015 |
| −FldA | 0.2 | 0.002 | 0.10 |
| −FqrB | 0.3 | 0.003 | 0 |
| −Pyruvate | 0.1 | 0.001 | 0.05 |
| −CoA | 0.2 | 0.002 | 0.10 |
| +NTZ (5 μM) | 16 ± 2 | 0.160 | 8 |
| 2× PFOR | 49 | 0.233 | 24.5 |
| 6× FqrB | 49 | 0.544 | 3.8 |
| 12× FqrB | 52 | 0.578 | 2.1 |
| 24× FqrB | 50 | 0.556 | 1.3 |
| Pseudoanaerobiosisd, complete (NADP) | 12 | 0.08 | 2.4 |
Reaction components included flavodoxin (FldA), PFOR, and FqrB, and activities are computed in the absence (−) or presence (+) of components. The protein concentrations used were as follows: FldA, 9.5 μM; PFOR, 0.1 mg/ml; and FqrB, 0.002 mg/ml. The effect of different protein concentrations on enzymatic activity was determined at 2×, 6×, 12×, and 24× the indicated protein concentration.
Relative activity of the complex. Values are means and standard deviations.
The specific activity (units) for each protein (PFOR or FqrB) is in μmol min−1 mg protein−1. Values are means and standard deviations.
Pseudoanaerobiosis indicates that no effort was made to sparge the cuvette of oxygen.
FIG. 5.
(A) Initial velocity of PFOR:FldA:FqrB. The reduction of NADP was followed at 340 nm in the presence of pyruvate, reduced CoA, and NADP in saturation as described in the text. Data are shown for the complete reaction (•), the complete reaction with NAD substituted for NADP (○), the reaction without PFOR (▪), the reaction without flavodoxin (□), the reaction without FqrB (▴), the reaction without pyruvate (▵), and the reaction without reduced CoA (*). The reactions were run under strict anaerobic conditions in a cuvette flushed with argon gas. (B) Acetyl-CoA consumption in reverse PFOR complete reaction. The consumption of acetyl-CoA was monitored at 232 nm under anaerobic conditions in 100 mM potassium phosphate (pH 7), 1 mM MgCl2, 0.1 mM acetyl-CoA, 5 mM bicarbonate, 0.3 mM NADPH, 0.3 mg/ml of PFOR, 0.9 μM flavodoxin, 0.15 μM FqrB, and CO2 bubbled into the cuvette.
CO2 fixation and NADPH-driven pyruvate synthesis.
Since flavodoxin was the most efficiently reduced substrate of FqrB, we considered the possibility that the PFOR-coupled reaction also functioned in reverse to produce pyruvate in an NADPH-dependent manner. Unlike pyruvate dehydrogenases, PFORs are reversible (20). As seen in Table 4, the reverse reaction was dependent on NADPH, CO2, and acetyl-CoA for full activity. The specific activity for the reverse reaction was ca. 75% of that for the forward reaction (9 versus 12 nmol min−1, corrected for pseudoanaerobic assay conditions). In the absence of NADPH or FqrB, there was no measurable activity. However, in the absence of PFOR or FldA, the partial remaining activity measured might be attributable to both proteins serving as substrates for FqrB. If FldA were present in excess, the initial velocity for NADPH oxidation by FqrB would be unchanged. In the case of PFOR (absence of FldA) the remaining activity likely represents direct transfer of reducing equivalents from FqrB or NADPH oxidation by residual oxygen in the cuvette. We directly measured the disappearance of acetyl-CoA (formation of reduced CoA) in the reverse reaction to verify that substrates were converted to pyruvate in the reaction (Fig. 5B). The specific activity based on acetyl-CoA consumption was 294 nmol min−1 mg of PFOR−1. In the absence of acetyl-CoA, CO2, or NADPH, this reaction had no measurable activity. Similarly, leaving out PFOR, FldA, or FqrB also resulted in no enzymatic activity (data not presented). While not depicted, NTZ also inhibited the reverse reaction. Taken together, these results show that the PFOR:FldA:FqrB complex of H. pylori is bidirectional and might represent an important pathway for incorporation of CO2 into biomass.
TABLE 4.
Pyruvate synthesis (reverse reaction)
| Reaction componentsa | NADPH oxidation (nmol/min) | Sp act (units/mg protein)b
|
|
|---|---|---|---|
| PFOR | FqrB | ||
| Complete | 15.4 ± 2 | 0.055 ± 0.007 | 3 ± 0.5 |
| −NADPH | 0 | 0 | 0 |
| −FqrB | 0 | 0 | 0 |
| −PFOR | 5.9 | 0.021 | 1.2 |
| −FldA | 4.7 | 0.017 | 0.9 |
| −Acetyl-CoA | 6.4 | 0.023 | 1.3 |
| −CO2 | 6.2 | 0.022 | 1.2 |
| Corrected valuec | 9 | 0.032 | 1.8 |
Complete reverse reaction under pseudoanaerobic conditions was monitored following the oxidation of NADPH in presence of 1 μM Fld, 0.3 mg/ml PFOR, and 5 μg/ml Fqr. Formation of reduced CoA from acetyl-CoA was demonstrated during the reaction (Fig. 5). The activities were computed in the absence of one component of the reaction (−). Some residual activity remains as long as FqrB, NADPH, and some electron acceptor remain in the reaction. This electron acceptor likely could be residual oxygen in the sample.
The specific activity (Units) for each protein (PFOR or FqrB) is in μmol min−1 mg protein−1.
Initial velocities were corrected for residual oxidase activity.
Pyruvate-dependent NADP reduction in cell extracts.
Since C. jejuni also contains orthologs of FqrB, FldA, and PFOR, we examined extracts from this organism as well as H. pylori for pyruvate-coupled NADPH formation. In high-speed supernatants from C. jejuni, the coupled reaction rate was 6.43 nmol min−1 mg protein−1. For H. pylori, the rate was 9 nmol min−1 mg protein−1, compared with 35 nmol min−1 mg protein−1 with BV as the electron acceptor. In both cases, the PFOR inhibitor NTZ inhibited the pyruvate-dependent reduction of NADP in both organisms (data not presented). The low activities measured in cell extracts are consistent with activities previously published for these organisms (18, 19, 22, 23) and reflect the low metabolic activity that is typical for these slow-growing bacteria.
DISCUSSION
The genome of H. pylori encodes a variety of redox-active enzymes, many of which were identified through functional assays or through mutation-based strategies (2, 9, 15, 25, 30, 31, 42). Since a large percentage of genes in H. pylori are essential for viability (6), it is likely that some genes associated with redox activities, such as fqrB (HP1164), would not be identified in genetic screens. Phylogenetic studies show that FqrB is highly conserved among members of the epsilonproteobacteria and has functionally diverged from the thioredoxin reductase family. FqrB lacks the CXXC catalytic domain (Fig. 1) that is required for thioredoxin reductase activity, and direct enzyme assay confirmed that thioredoxin substrates are not reduced by FqrB. Our studies show that FqrB is a member of the NADPH oxidase class of flavoprotein dehydrogenases, displaying a strong substrate affinity for quinones and flavodoxin and a preference for NADPH. Previous studies had demonstrated pyruvate-dependent reduction of NADP in extracts from H. pylori (23), but the protein responsible for this activity was never identified. Here we report that FqrB (HP1164) is a very good candidate for the NADPH oxidoreductase enzyme predicted by Hughes et al. (23) that couples pyruvate oxidation by PFOR to the production of NADPH.
Since fqrB is both unique to the epsilonproteobacteria and essential in H. pylori (2, 6), the biological role for this protein must be highly specialized. This distinction is important, since FqrB also displays NADPH quinone reductase activity and may also participate in electron transport reactions. In this regard, H. pylori expresses many redox-active enzymes and quinone reductases, many of which are not essential (37, 41). In contrast, porGDAB and fldA are also essential genes and together with FqrB appear to form a novel pyruvate-metabolizing pathway. We provide the following evidence to suggest that these three enzymes represent unique adaptations in the epsilonproteobacteria: (i) PFORs of this group utilize FldA (rather than ferredoxin) as an electron carrier, (ii) phylogenetic studies indicate that FldA and FqrB are highly conserved, and (iii) pyruvate formation through CO2 fixation is consistent with both the capnophilic nature of these organisms and their unique requirement for pyruvate to satisfy gluconeogenic demands (ribose and hexose sugars).
Our studies also indicate that FqrB might inadvertently contribute to the microaerophilic nature of the epsilonproteobacteria by its ability to reduce quinones during periods of oxidative stress. The subsequent single-electron oxidation of semiquinones has the potential to generate mM levels of superoxide anions, as demonstrated in assays of SOD inhibition of cytochrome c reduction. Quinones play a central role in respiratory metabolism by cycling electron transfers between the dehydrogenases and the redox centers and cytochromes of the electron transport system (7, 16, 17, 29). Redox cycling by FqrB might also deplete pools of NADPH during periods of oxidative stress. FqrB exhibits no similarity with MdaB (HP0630), a previously described NADPH-specific quinone reductase that is nonessential and appears to function in protection from oxidative stress (42). Further studies will be required to fully assess the role (if any) of FqrB in oxygen toxicity and oxidative stress.
The annotation error in the two genome sequences of H. pylori arises from a conserved region of FqrB, which in thioredoxin reductases contains a catalytic CXXC motif (2). In H. pylori and W. succinogenes, the motifs INDC and VSDC, respectively (Fig. 1) have amino acid I or V in place of C within a conserved 60-amino-acid stretch, which accounts for the low-probability match of e−15 (1, 2, 39). In species of Campylobacter, there are no cysteine residues in this region and similarities are even lower. FqrB activity is robust with quinone substrates (nearly double that of MdaB) and together with a near absence of thioredoxin reductase activity with authentic thioredoxins from H. pylori strongly supports annotation as either a quinone or flavodoxin reductase. The enzyme did exhibit weak activity with DTNB that was above that in the background nonenzymatic reaction with thiol residues of the protein. Often DTNB reduction is attributed to thioredoxin reductases (33, 34). In contrast to NfsB, FqrB exhibited no nitroreductase activity with the substrates tested. Kinetic studies support a ping-pong catalytic mechanism based on parallel double-reciprocal plots with quinone substrates and NADPH. This mechanism is common to flavoproteins and nitroreductases (26, 34) and for FqrB likely involves redox-active FAD and a conserved cysteine located in the C-terminal region of the protein (Fig. 1). The ping-pong mechanism must hold true for flavodoxin as both electron donor and receptor. Further studies are required to distinguish functions in pyruvate metabolism from functions in quinone reduction and electron transport.
Several studies have examined CO2/bicarbonate fixation in H. pylori. Acetyl-CoA carboxylase activity was studied by Burns et al. (4) and confirmed by Hughes et al. (22). The product of the carboxylase is malonyl-CoA, which is speculated to participate in fatty acid biosynthesis (4). Hughes et al. (22) also examined H14CO3 incorporation by cell extracts and concluded that a pyruvate-dependent isotope exchange was catalyzed by PFOR. In reviewing the data, it is evident that the incorporation of radiolabel into pyruvate resulted from the bidirectional nature of PFOR and the fact that the incorporation was dependent on acetyl-CoA, NADP, and pyruvate but not on malate dehydrogenase or NADH (22). Pyruvate oxidation would be expected to generate NADPH by FqrB, and at some point, the reverse reaction (pyruvate synthesis) would have catalyzed incorporation of labeled bicarbonate. Taken together, our studies support the conclusions of Hughes et al. that PFOR plays a major role in CO2 fixation in H. pylori. While our studies did not specifically address the possibility of NADPH production from oxoglutarate:ferredoxin oxidoreductase, which is also highly conserved between Helicobacter and Campylobacter species, previous studies of this enzyme would suggest that FldA and FqrB reversibly participate in this reaction. In general the metabolism of CO2 and molecular hydrogen has been understudied, but both gases are abundant in the gastrointestinal tract, and it is highly likely that these organisms have evolved the ability to capitalize on them (14, 22, 28).
There has been considerable interest in the intermediary metabolism of small-genome organisms such as H. pylori, where few redundancies or backup pathways are available to circumvent metabolic pathway deficiencies (1, 6, 39). For example, H. pylori lacks the genes encoding phosphofructokinase and pyruvate kinase, which are involved in key steps in the forward direction of the glycolytic pathway (the reverse gluconeogenic pathway is complete), and it appears to compensate by routing glucose catabolism through the Entner Doudorof pathway (1, 6, 11, 18, 39). Ironically, glucose is sparingly utilized by H. pylori, whereas amino acids and organic acids serve as the major carbon and energy sources (1, 6, 11, 18, 39). H. pylori also lacks anaplerotic enzymes that typically connect the Krebs cycle with gluconeogenesis (e.g., oxaloacetate ←→ phosphoenolpyruvate [PEP]) and must rely on phosphoenolpyruvate synthase (pyruvate → PEP) to satisfy gluconeogenic demands, as depicted in Fig. 6. Our studies suggest that CO2 fixation by the PFOR:FldA:FqrB pathway must play a central role in this process and may partly explain the capnophilic nature of this pathogen. In the case of asaccharolytic C. jejuni, which lacks phosphofructokinase and phosphoenolpyruvate synthase, organic acids enter the gluconeogenic pathway via PEP carboxykinase (oxaloacetate ←→ PEP) (18, 19, 40). As shown in Fig. 6, PEP synthesis from pyruvate is possible through the oxaloacetate intermediate via pyruvate carboxylase and PEP carboxykinase (40). The fate of PEP (catabolic versus gluconeogenic) appears to be regulated through allosteric effectors that modulate pyruvate kinase activity (40). In both organisms, the demand for acetyl-CoA for CO2 fixation can be satisfied through both fatty acid catabolism and synthesis of acetyl-CoA from acetate (acetate kinase and acetyl-CoA synthetase) (1, 11, 19, 39). For systems biologists who have modeled the metabolic activities of H. pylori, it will be of interest to see how pyruvate synthesis (CO2 fixation) by PFOR alters carbon flow predictions. Thus, FqrB is a reminder that in silico modeling is only as good as the accuracy and completeness of existing functional genomic information and underscores the need for rigorous biochemical analysis of genes of unknown function.
FIG. 6.
Reversible PFOR:NADPH reductase complex. All reactions are shown as reversible, with FldA shuttling reducing equivalents between PFOR and FqrB. FldARd indicates the reduced form, and FldAOx indicates the oxidized form. Pyruvate formation (reverse reaction) may be favored due to the unique gluconeogenic nature of central metabolism in microaerophiles. The dashed lines indicate pyruvate carboxylation (pyruvate carboxylase [PYC]) and oxaloacetate (OAA) decarboxylation (phosphoenolpyruvate carboxykinase [PCK]) enzymes found in C. jejuni and phosphoenolpyruvate synthase (PPS) found only in H. pylori. C. jejuni also expresses pyruvate kinase (not depicted). Sources of acetyl-CoA include fatty acid catabolism and free acetate and the associated enzymes.
In summary, we have determined that the trxR2 gene (HP1164), previously annotated as encoding a thioredoxin reductase, encodes an NADPH oxidoreductase that is part of the PFOR complex in the epsilonproteobacteria. We named the enzyme FqrB to reflect activities associated with both pyruvate metabolism and quinone reduction, with the latter potentially contributing to oxidative stress through the production of superoxide anions. Finally, perhaps the most important role for FqrB in H. pylori may be in driving CO2 fixation (PFOR:FldA:FqrB pathway), which is so necessary for replenishing pyruvate levels consumed by gluconeogenesis.
Acknowledgments
We thank Leslie Poole for kindly providing purified H. pylori thioredoxins used in this study.
This work was supported by grants from the Canadian Institutes for Health Research and the National Institutes of Health (DK073823) to P.S.H. and by grants BFU2004-01411 (Spanish Government) and PM076/2006 (Aragonese Government) to J.S. N.C. was supported by an FPU fellowship (Spain).
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Alm, R. A., L. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:186-190. [DOI] [PubMed] [Google Scholar]
- 2.Baker, L. M. S., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaser, M. J., and D. E. Berg. 2001. Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Investig. 107:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, B. P., S. L. Hazell, and G. L. Mendz. 1995. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology 141:3113-3118. [DOI] [PubMed] [Google Scholar]
- 5.Butzler, J.-P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868-876. [DOI] [PubMed] [Google Scholar]
- 6.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, H. T., S. W. Marcelli, A. A. Davison, P. A. Chalk, R. K. Poole, and R. J. Miles. 1995. Kinetics of substrate oxidation by whole cells and cell membranes of Helicobacter pylori. FEMS Microbiol. Lett. 129:33-38. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M., L. P. Anderson, L. Zhai, and A. Kharazmi. 1999. Characterization of the respiratory chain of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 24:169-174. [DOI] [PubMed] [Google Scholar]
- 9.Comtois, S. L., M. D. Gidley, and D. J. Kelly. 2003. Role of the thioredoxin system and the thiol-peroxidase Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149:121-129. [DOI] [PubMed] [Google Scholar]
- 10.Cremades, N., M. Bueno, M. Toja, and J. J. Sancho. 2005. Towards a new therapeutic target: Helicobacter pylori flavodoxin. Biophys. Chem. 115:267-276. [DOI] [PubMed] [Google Scholar]
- 11.Doig, P., B. L. de Jounge, R. A. Alm, E. D. Brown, M. Uria-Nickelson, B. Noonan, S. D. Mills, P. Tummino, G. Carmel, B. C. Guild, D. T. Moir, G. F. Vovis, and T. J. Trust. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev. 63:675-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 13.Glupczynski, Y. 1998. Antimicrobial resistance in Helicobacter pylori: a global overview. Acta Gastroenterol. Belgium 61:357-366. [PubMed] [Google Scholar]
- 14.Goodman, T. G., and P. S. Hoffman. 1983. Hydrogenase activity in catalase-positive strains of Campylobacter spp. J. Clin. Microbiol. 18:825-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin, A., D. Kersulyte, G. Sisson, S. J. O. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 16.Hazell, S. L., A. G. Harris, and M. A. Trend. 2001. Evasion of the toxic effects of oxygen, p. 167-175. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed]
- 17.Hoffman, P. S., and T. G. Goodman. 1982. Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J. Bacteriol. 150:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman, P. S., A. Goodwin, J. Johnsen, K. Magee, and S. J. O. Veldhuyzen van Zanten. 1996. Metabolic activities of metronidazole-sensitive and -resistant strains of Helicobacter pylori: repression of pyruvate oxidoreductase and expression of isocitrate lyase activity correlate with resistance. J. Bacteriol. 178:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman, P. S., N. R. Krieg, and R. M. Smibert. 1979. Studies of the microaerophilic nature of Campylobacter fetus subsp. jejuni. I. Physiological aspects of enhanced aerotolerance. Can. J. Microbiol. 25:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, P. S., G. Sission, M. A. Croxen, K. Welch, W. D. Harman, N. Cremades, and M. G. Morash. 2007. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob. Agents Chemother. 51:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horner, D. S., R. P. Hirt, and T. M. Embley. 1999. A single eubacterial origin of eukaryotic pyruvate:ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 16:1280-1291. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, N. J., P. A. Chalk, C. L. Clayton, and D. J. Kelly. 1995. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J. Bacteriol. 177:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong, J.-Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatiño, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Y. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H.-K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen, M. A., M. A. Trend, S. L. Hazell, and G. L. Mendz. 2001. Potential involvement of several nitroreductases in metronidazole resistance in Helicobacter pylori. Arch. Biochem. Biophys. 392:180-191. [DOI] [PubMed] [Google Scholar]
- 26.Koder, R. L., and A. F. Miller. 1998. Steady state kinetic mechanism, stereospecificity, substrate and inhibitor specificity of Enterobacter cloacae nitroreductase. Biochim. Biophys. Acta 1387:395-405. [DOI] [PubMed] [Google Scholar]
- 27.Krieg, N. R., and P. S. Hoffman. 1986. Microaerophily and oxygen toxicity. Annu. Rev. Microbiol. 40:107-130. [DOI] [PubMed] [Google Scholar]
- 28.Maier, R. J. 2005. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochem. Soc. Trans. 33:83-85. [DOI] [PubMed] [Google Scholar]
- 29.Marcelli, S. W., H. T. Chang, T. Chapman, P. A. Chalk, R. J. Miles, and R. K. Poole. 1996. The respiratory chain of Helicobacter pylori: identification of cytochromes and the effects of oxygen on cytochrome and menaquinone levels. FEMS Microbiol. Lett. 138:59-64. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay, A. K., J. Y. Jeong, D. Dailidiene, P. S. Hoffman, and D. E. Berg. 2003. The fdxA ferredoxin gene can down regulate frxA nitroreductase gene expression and is essential in many strains of Helicobacter pylori. J. Bacteriol. 185:2927-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olczak, A. A., J. W. Olson, and R. J. Maier. 2002. Oxidative-stress resistance mutants of Helicobacter pylori. J. Bacteriol. 184:3186-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 33.Poole, L. B., C. M. Reynolds, Z. A. Wood, P. A. Karplus, H. R. Ellis, and M. L. Calzi. 2000. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267:6126-6133. [DOI] [PubMed] [Google Scholar]
- 34.Race, P. R., A. L. Lovering, R. M. Green, A. Ossor, S. A. White, P. F. Searle, C. J. Wrighton, and E. I. Hyde. 2005. Structural and mechanistic studies of the Escherichia coli nitroreductase with the antibiotic nitrofurazole. J. Biol. Chem. 280:13256-13264. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sisson, G., A. Goodwin, A. Raudonikiene, N. J. Birks, H. Han, A. Mukhopadhyay, D. E. Berg, and P. S. Hoffman. 2002. Enzymes associated with the reductive activation and action of nitazoxanide, nitrofurans and metronidazole in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sisson, G., J.-Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomb, J. F., O, White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. Ketchum, H. Klenk, S. Gill, B. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. Khalak, A. Glodek, K. McKenney, L. Fitzegerald, N. Lee, M. Adams, E. Hickey, D. Berg, J. Gocayne, T. Utterback, J. Peterson, J. Kelley, M. Cotton, J. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. Hayes, M. Borodovsky, P. Karp, H. Smith, C. Fraser, and J. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 40.Velayudhan, J., and D. J. Kelly. 2002. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148:685-694. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G., P. Alamuri, and R. J. Maier. 2006. The diverse antioxidant systems of Helicobacter pylori. Mol. Microbiol. 61:847-860. [DOI] [PubMed] [Google Scholar]
- 42.Wang, G., and R. J. Maier. 2004. An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect. Immun. 72:1391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to Frase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736-744. [DOI] [PubMed] [Google Scholar]