Abstract
The fungal pathogen Cryptococcus neoformans is killed by the bacterium Staphylococcus aureus, and the killing is inhibited by soluble capsular polysaccharides. To investigate the mechanism of killing, cells in coculture were examined by scanning and transmission electron microscopy. S. aureus attached to the capsule of C. neoformans, and the ultrastructure of the attached C. neoformans cells was characteristic of dead cells. To identify the molecules that contributed to the fungal-bacterial interaction, we treated each with NaIO4 or protease. Treatment of C. neoformans with NaIO4 promoted adherence. It was inferred that cleavage of xylose and glucuronic acid side chains of glucuronoxylomannan (GXM) allowed S. aureus to recognize mannose residues in the backbone, which resisted periodate oxidation. On the other hand, treatment of S. aureus with protease decreased adherence, suggesting that protein contributed to attachment in S. aureus. In confirmation, side chain-cleaved polysaccharide or defined α-(1→3)-mannan inhibited the killing at lower concentrations than native GXM did. Also, these polysaccharides reduced the adherence of the two species and induced clumping of pure S. aureus cells. α-(1→3)-Mannooligosaccharides with a degree of polymerization (DP) of ≥3 induced cluster formation of S. aureus in a dose-dependent manner. Surface plasmon resonance analyses showed interaction of GXM and surface protein from S. aureus; the interaction was inhibited by oligosaccharides with a DP of ≥3. Conformations of α-(1→3) oligosaccharides were predicted. The three-dimensional structures of mannooligosaccharides larger than triose appeared curved and could be imagined to be recognized by a hypothetical staphylococcal lectin. Native polyacrylamide gel electrophoresis of staphylococcal protein followed by electroblotting, enzyme-linked immunolectin assay, protein staining, and N-terminal amino acid sequencing suggested that the candidate protein was triosephosphate isomerase (TPI). The enzymatic activities were confirmed by using whole cells of S. aureus. TPI point mutants of S. aureus decreased the ability to interact with C. neoformans. Thus, TPI on S. aureus adheres to the capsule of C. neoformans by recognizing the structure of mannotriose units in the backbone of GXM; we suggest that this contact is required for killing of C. neoformans.
Cryptococcus neoformans is an encapsulated yeast that causes fatal meningitis. This fungus is predominantly found in pigeon excreta, and the route of entry into the human body is through the respiratory system (9, 32). Nasal colonization of C. neoformans was experimentally shown in the mouse model (2). In that report, intranasally inoculated C. neoformans cells colonized the nose for 14 to 28 days before dissemination. Many cases of cryptococcal sinusitis have been reported in animals (14, 40). In humans, the nose is not considered the site of cryptococcal colonization, rather, in patients receiving care in hospitals, the organ that C. neoformans colonizes is the lung (1). Apparently, the pathogen is somehow excluded from the upper respiratory tract in most immunocompetent human hosts. However, specific evidence has not been shown.
Interaction between microbes in mucosal colonization is known to occur. In the oral cavity, a large number of species compete for space (29). In dental biofilm, there is a subtle balance between Streptococcus mutans and Streptococcus sanguinis depending on environmental conditions, cell density, nutrition, pH, etc. (31). Within the gastrointestinal tract, lactic acid bacteria reduce colonization by harmful bacteria (34). Such “probiotics” appear to prevent pathogenicity of Helicobacter pylori or enterobacteria by induction of cytokines and by interruption of adhesion to target cells (5, 56, 69). Collectively, microbes communicate sometimes exclusively and sometimes synergistically in an animal or in the environment by multiple mechanisms, including static or cidal effects of antibiotics, signal transduction systems, collaboration with host cells, and others. Quorum-sensing and two-component signal transduction systems are possible mechanisms (16, 65). Molecules related to inter- or intraspecies communications were characterized as “autoinducers.”
Recently, we discovered that the bacterium Staphylococcus aureus, which is found in the nasopharynx of a large proportion of healthy human subjects (28, 57), killed C. neoformans and fragmented its DNA, suggesting that S. aureus in human nasal cavities may prevent the entry of C. neoformans (61). Since the killing required contact of the cells, it seemed unlikely that soluble substances, such as bacteriocin (21, 60) or killer toxin (53), were involved. We also showed that soluble capsular polysaccharides from C. neoformans inhibited killing. In contrast, the presence of S. aureus did not affect the growth of Candida albicans, a well-known endogenous opportunistic pathogen. One could argue that this resistance helps to explain why C. albicans is a more successful colonizer than C. neoformans.
The surface of C. neoformans appears to be organized and defined by the extracellular capsule (9, 52). The main component of the capsule is the polysaccharide glucuronoxylomannan (GXM). It seemed reasonable to imagine that S. aureus might recognize moieties in this polysaccharide. We had already suggested that soluble capsular polysaccharides from C. neoformans bound to cells of S. aureus (61). In the present study, we examine the molecules contributing to protein-carbohydrate interactions in attachment of C. neoformans and S. aureus. We have also studied the ultrastructure of interacting S. aureus and C. neoformans.
MATERIALS AND METHODS
Strains and plasmids used.
The strains and sources of C. neoformans are listed in Table 1. C. neoformans strains were cultivated at 37°C for 48 h in Sabouraud dextrose broth (SB) containing 2% glucose, 0.5% yeast extract, and 1% polypeptone. S. aureus ATCC 6538P and Staphylococcus epidermidis NBRC 12993 grown at 37°C for 48 h in nutrient broth (NB; Eiken, Tokyo, Japan) were used to investigate interactions with C. neoformans unless indicated otherwise. S. aureus tpi mutants TS1505 and TS3048 harboring plasmids or not and their parent strain RN4220 (47) were cultured at 30°C for 48 h. The tpi mutants were isolated by mutagenesis of RN4220 with ethylmethanesulfate (Sigma) as described previously (35). Mutations in the tpi gene have been identified by complementation analysis for their temperature-sensitive cell growth by plasmid pStpi. pStpi carried the chromosomal tpi gene region of 2612 bp at the SmaI site in vector pKE515 (35), which corresponded to 834446 to 837057 on the S. aureus N315 strain genome registered in the genome information broker program in the DNA Data Bank of Japan. TS1505 and TS3048 had single amino acid substitutions of E106K and E79K in TPI protein, respectively.
TABLE 1.
C. neoformans strains used in this study
| Strain | Serotype | Source |
|---|---|---|
| B-3501 | D | Single-basidiospore isolate from a cross of ATCC 28957 × ATCC 28958 |
| M9106 | A | Cerebrospinal fluid |
| M9172 | A | Cerebrospinal fluid |
| M9174 | A | Cerebrospinal fluid |
| M9116 | D | Cerebrospinal fluid |
| M9117 | D | Skin |
| M9254 | A | Bird dropping (67) |
| M9255 | A | Bird dropping (67) |
| M9256 | A | Bird dropping (67) |
| M9257 | A | Bird dropping (67) |
| M9258 | A | Bird dropping (67) |
Coculture of C. neoformans and S. aureus.
Cultures of 0.3 ml each of C. neoformans and S. aureus or S. epidermidis were mixed, and SB was added to adjust the total volume to 3 ml. The mixture was incubated at 37°C while being shaken. After 1, 2, 3, 4, and 5 days, CFU were determined on Sabouraud dextrose agar (SA) containing streptomycin (100 μg/ml). The SA plates were incubated at 27°C for 48 h. Each experiment was performed two or more times.
Scanning electron microscopy.
Samples were prepared as described previously (63) and observed under an S-800 scanning electron microscope (Hitachi, Tokyo, Japan) at 3 kV.
Transmission electron microscopy.
Cells were collected by centrifugation, sandwiched between two 3-mm-diameter copper discs, and rapidly frozen by plunging into propane slush kept in liquid nitrogen (70). The discs were transferred to liquid nitrogen and separated to expose the cells. Specimens were freeze-substituted in acetone containing 2% osmium tetroxide at −85°C for 2 to 3 days. Samples were then gradually warmed to room temperature, rinsed with acetone, and embedded in epoxy resin. Ultrathin sections were cut to a thickness of 70 to 80 nm, mounted on grids with plasma-polymerized naphthalene support film (72), stained with uranyl acetate and lead citrate (71), and observed under a JEM 1200EX transmission electron microscope (JEOL, Tokyo, Japan).
Sensitivity of adherence to protein and carbohydrate disruptions.
To determine the molecules contributing to the adherence of S. aureus to C. neoformans, cells were pretreated with protease K or sodium metaperiodate (37). Since heat-killed cells of C. neoformans (100°C, 20 min) and S. aureus (80°C, 30 min) were found to retain cell-cell interactions, the heated cells were used for adherence assays to avoid any effects, aside from pretreatment, that could stem from the growth of the cells. After the cells were washed, they were suspended in water to 1/10 volume of the original culture. The cell suspension was mixed with an equal volume of protease K (1 mg/ml in Tris-HCl buffer, 100 mM [pH 8.0]) and incubated at 37°C for 4 h; 1/10 volume of 100 mM phenylmethylsulfonyl fluoride was added. The cells were then washed with sterilized water.
For treatment with NaIO4, 100 μl of the heat-killed cell suspension was mixed with 100 μl of 0.08 M NaIO4 and incubated at 4°C overnight in the dark. To arrest oxidation, we added 13.2 μl ethylene glycol. The cells were then washed with sterilized water.
An adherence assay was performed using live cells of C. neoformans or S. aureus and heat-treated cells treated with protease or NaIO4. After incubation at 37°C for 3 h, the percent adherence was determined as follows: percent adherence = (number of C. neoformans cells to which S. aureus attached × 100)/(total number of C. neoformans observed microscopically) (three experiments).
Preparation of chemically modified capsular polysaccharides.
To obtain oxidation of polysaccharides, capsular polysaccharides were oxidized with 0.02 M NaIO4 at 4°C for 120 h in the dark, treated with ethylene glycol to stop oxidation, and dialyzed against water. The oxidized polysaccharides were reduced with NaBH4 for 20 h at room temperature. Following dialysis, the polysaccharides were hydrolyzed with 0.5 M HCl at room temperature for 12 h, neutralized with NaOH, dialyzed against water, and evaporated to dryness. A 1H nuclear magnetic resonance (NMR) spectrum of the product was recorded using a GSX-400 NMR spectrometer (JEOL) in 99.9% D2O at 90°C; sodium 3-(trimethylsilyl)-1-propanesulfonate (DSS) was used as the external standard (26, 27).
Inhibition of killing by chemically modified capsular polysaccharides.
The modified polysaccharide was added to the coculture of C. neoformans B-3501 and S. aureus at a final concentration of 1 mg/ml.
Preparation of oligosaccharides.
The controlled Smith degradation product was hydrolyzed with 0.4 M H2SO4 at 100°C for 1 h (46). After neutralization with BaCO3, the product was applied to a column of Bio-Gel P-2 (2.5-cm diameter, 120-cm length; Bio-Rad Laboratories, Hercules, CA) and eluted with water (10 ml/h, 2 ml/tube). The amounts of carbohydrate were determined by the phenol sulfuric acid method. NMR spectra of the oligosaccharide fractions were recorded as described above for identification.
S. aureus clumping in the presence of oligosaccharides from C. neoformans.
As the coagulation of S. aureus with α-(1→3)-mannan was observed in culture, the degree of mannan polymerization required for the clumping was determined. The suspension of heat-killed cells of S. aureus (a McFarland standard of about 10) was incubated with 100 μM or 500 μM of the oligosaccharides prepared from mannan degraded by controlled Smith degradation. After incubation overnight at 37°C, the number of clumped cells (larger than about 1/16 mm) was determined with a microscope using a hemocytometer.
Preparation of surface protein from S. aureus.
The surface proteins from S. aureus were prepared by the method of Komatsuzawa et al. (30). S. aureus cells cultured in Trypticase soy broth (TSB; Becton Dickinson Company, Sparks, MD) for 6 h were suspended in 3 M lithium chloride. After incubation for 15 min on ice, the cell extract was dialyzed against 0.1 M phosphate buffer, pH 6.8. After addition of sodium chloride at a concentration of 2.5 M, the sample was applied to a column of phenyl-Sepharose CL-4B using about 0.5 ml of the gel. The column was eluted with phosphate buffer containing a decreasing concentration (2.5, 2.0, 1.5, 1.0, 0.5, and 0 M) of NaCl.
Interaction of cryptococcal GXM and protein from S. aureus by SPR.
The interaction of GXM with protein fraction obtained from S. aureus was analyzed by surface plasmon resonance (SPR) by using a Biacore 3000 (Biacore, Uppsala, Sweden). For immobilization of GXM on CM5 sensor chips, the acidic GXM was reduced to neutral polysaccharide by the method described previously (26, 27), because native GXM was not immobilized perhaps due to its charge. Reduced GXM (500 μg) was dissolved in 500 μl of 0.1 M acetate buffer (pH 5.5), and 10 μl of 50 mM NaIO4 in 100 mM acetate buffer (pH 5.5) was added. After incubation for 20 min on ice, the solution was applied to a column of NAP-5 (Amersham Biosciences, Uppsala, Sweden) to remove reagents and to change buffer (Amersham Biosciences, Uppsala, Sweden) using 0.1 M acetate buffer, pH 4.0. The fraction containing carbohydrate (0.5 to 1.0 ml) was diluted to a concentration of 100 μg/ml with 10 mM acetate buffer, pH 4.0. The polysaccharide was immobilized by the method of aldehyde coupling using the reagent in an amine coupling kit by the method described by the manufacturer. For the binding experiments, analyte protein fractions from S. aureus were diluted with 10 mM HEPES-buffered saline containing 3 mM EDTA and 0.005% surfactant P20 and injected into a flow cell at a flow rate of 20 μl/min.
Characterization of adhesion molecules on S. aureus.
An extract of S. aureus into 3 M LiCl was separated by a Diaflo ultrafiltration membrane XM50 (Amicon, Beverly, MA). As the interaction with GXM was found in the upper fraction, this was further fractionated by phenyl-Sepharose CL-4B as described above. The fraction eluted by 2.5 M NaCl was dialyzed against 0.01 M phosphate buffer (pH 7.4), concentrated by lyophilization, and applied to three lanes of 10% polyacrylamide gel without sodium dodecyl sulfate, and the gel was run at a constant current of 20 mA. After electrophoresis, proteins in the gel were electroblotted onto a polyvinylidene difluoride (PVDF) membrane (60 V, 90 min). The protein involved in interaction with cryptococcal GXM was visualized by an enzyme-linked immunosorbent assay for detection of lectin as follows. A portion of the PVDF membrane corresponding to one lane was washed with blocking buffer (10 mM Tris-HCl containing 0.15 M NaCl and 0.05% Tween 20 [pH 7.4]), allowed to react with GXM solution (10 μg/ml) for 1 h, and stained with anti-C. neoformans serotype D rabbit immunoglobulin (1 μg/ml) for 30 min, diluted (1:250) alkaline phosphatase-labeled anti-rabbit immunoglobulin G (Santa Cruz Biotechnology Inc., Santa Cruz, CA) for 30 min, substrate solution (15 ml of 100 mM Tris-HCl [pH 9.5] containing 100 mM NaCl, 5 mM MgCl2,100 μl of NBT solution [0.017% Nitro Blue Tetrazolium, 0.23% N,N-dimethylformamide], and 50 μl of BCIP solution [5% 5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt in N,N-dimethylformamide]). Washing with blocking buffer was performed after each step. The part of the membrane corresponding to another lane was stained with 0.02% of Coomassie brilliant blue (CBB) for 2 min and washed with 50% methanol for 15 min.
TPI activity in intact cells.
The assay for TPI was carried out by the method of Rozacky et al. (58) with minor modifications. To determine whether the enzyme was located on the surface of S. aureus, intact cells were used. The cells grown in TSB at 37°C for 6 h were harvested and washed with assay buffer (30 mM triethanolamine-HCl buffer [pH 7.8] containing 1 mM EDTA). Different amounts of cells were incubated with 1.5 mM of dl-glyceraldehyde 3-phosphate (Sigma), 0.1 mg of NADH, and 10 μg of glycerol-3-phosphate dehydrogenase (Wako, Osaka, Japan) in a final volume of 1 ml. After incubation at 25°C for 5 min, cells were removed by centrifugation at 5,000 rpm for 5 min at 4°C. The absorbance of the supernatant at 340 nm was measured. TPI activities were expressed as the decrease of absorbance at 340 nm by coupling the reaction to oxidation of NADH by glycerol-3-phosphate dehydrogenase.
Adherence of TPI mutants of S. aureus to C. neoformans.
A stationary-phase culture of 0.3 ml of S. aureus at 30°C was mixed with heat-killed cells of C. neoformans B-3501 (about 3 × 107/ml) in SB, and the total volume was adjusted to 3 ml. The mixture was incubated at 30°C while being shaken. After 6 h, the percent adherence was calculated. To suppress growth of the mutants, a suspension of S. aureus cells in PBS was adjusted to twice the stationary phase and used for an adherence experiment.
Statistics.
The percent adherence data were determined as the mean values of at least three replicates ± standard deviations. Statistical significance was determined by Student's t test.
RESULTS
Scanning and transmission electron microscopy.
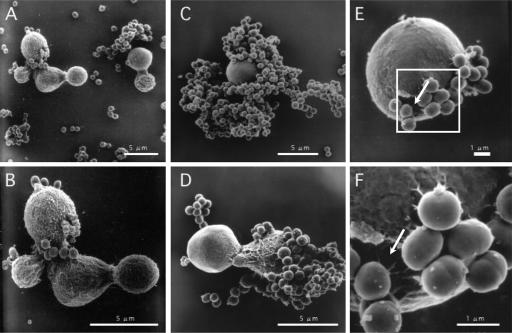
Previously, we used light microscopy to demonstrate that C. neoformans cells were surrounded by S. aureus (61). To confirm and to investigate the mechanism of binding between the fungi and bacteria, cells in coculture were observed by scanning electron microscopy. S. aureus attached to the surface of C. neoformans (Fig. 1C and D). Some fungal cells were entirely surrounded by S. aureus, while other cells had several bacteria adhering to them. After 2 days of culture, it was difficult to find C. neoformans cells, as most of the yeast cells were covered by bacteria. We observed that C. neoformans was damaged near the position of attachment of S. aureus (Fig. 1E and F). The surface of the cryptococcal cells to which S. aureus adhered appeared smooth compared to the surface before coculture (Fig. 1A and B). The surface structure may have been changed by the adherence of S. aureus, although the reason is unknown.
FIG. 1.
Scanning electron micrographs of cells in cocultures of C. neoformans B-3501 and S. aureus. (A and B) C. neoformans B-3501 and S. aureus cells before incubation, after mixing; (C, E, and F) 1-day cultures; (D) 2-day cultures. S. aureus attached to the surface of C. neoformans as seen in panels C and D. Damage to C. neoformans can be seen in panels E and F (see arrows). (F) Enlargement of the white square in panel E.
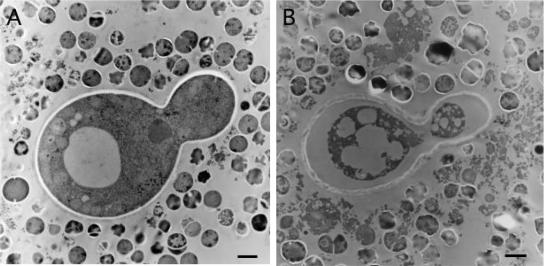
Figure 2 shows ultrathin sections of C. neoformans and S. aureus. By transmission electron microscopy, the ultrastructure of most of the C. neoformans cells remaining after 1 day showed characteristics of dead cells (Fig. 2B). The remaining cytoplasm stained more intensely, and the organelles appeared to be destroyed, whereas before the mixture was incubated, the ultrastructure of C. neoformans cells showed characteristics of live cells (Fig. 2A). However, the mechanism of death could not be elucidated from the electron micrographs.
FIG. 2.
Transmission electron micrographs of cells in cocultures of C. neoformans B-3501 and S. aureus. (A) C. neoformans B-3501 and S. aureus before incubation, after mixing. (B) One-day culture showing darkened cells and cell organelles that appear to be destroyed. Bars, 1 μm.
Coculture of C. neoformans isolates and S. aureus.
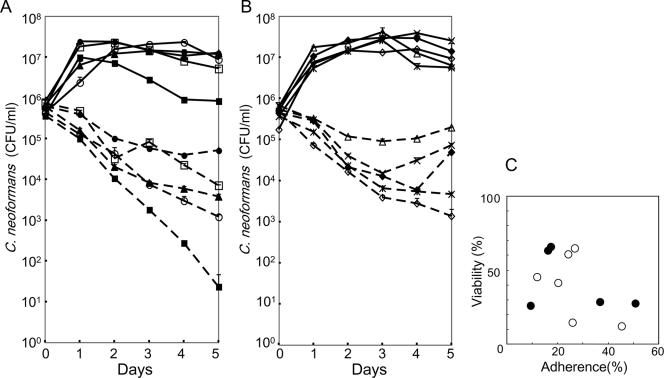
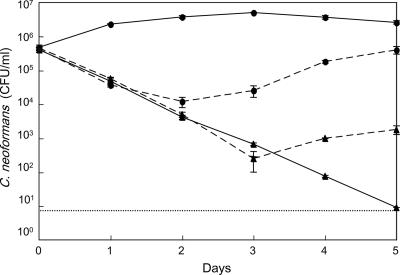
Populations of viable cells of C. neoformans in coculture with S. aureus were followed for 5 days. As shown in Fig. 3, fungal populations decreased for 2 days. After 3 days, responses varied. C. neoformans populations decreased in strains M9116, M9174, and M9255 in a time-dependent manner, the rate of dying of strains M9106 and M9258 slowed, and strains M9172, M9256, M9257, and M9254 began to multiply. Clinical and environmental isolates behaved alike. We examined the correlation between adherence and death using data from 11 isolates, including B-3501. One day after coculture, the percent adherence was calculated using the formula given in Materials and Methods. Viability was determined on day 1 as follows: (CFU of C. neoformans in coculture on day 1 × 100)/(CFU of C. neoformans on day 0) (Table 2). Viabilities were lower when the bacterial burdens were larger. The proportion of burdened yeast tended to correlate negatively with viability (Fig. 3C). The correlation coefficient between the proportion of burdened yeast and viability on day 1 was −0.472.
FIG. 3.
Coculture of C. neoformans strains with S. aureus. (A) CFU of C. neoformans isolates in coculture with S. aureus. The isolates were from clinical specimens. C. neoformans strains M9106 (▴), M9116 (▪), M9117 (□), M9172 (•), and M9174 (○) are shown. (B) CFU of C. neoformans isolates from pigeon droppings. C. neoformans strains M9254 (♦), M9255 (⋄), M9256 (▵), M9257 (×), and M9258 ( ) are shown. Solid line, pure culture of C. neoformans; dotted line, coculture of C. neoformans and S. aureus. The values are means ± standard deviations for three replicate samples from one representative experiment. (C) Correlation between adherence and mortality. Viability equals (CFU in coculture on day 1 × 100)/(CFU in coculture on day 0). Percent adherence was determined after 1day of cultivation. Closed circles, clinical isolates; open circles, environmental isolates.
) are shown. Solid line, pure culture of C. neoformans; dotted line, coculture of C. neoformans and S. aureus. The values are means ± standard deviations for three replicate samples from one representative experiment. (C) Correlation between adherence and mortality. Viability equals (CFU in coculture on day 1 × 100)/(CFU in coculture on day 0). Percent adherence was determined after 1day of cultivation. Closed circles, clinical isolates; open circles, environmental isolates.
TABLE 2.
Percent adherence and CFU of 11 C. neoformans strains in coculture with S. aureus for 1 day
| Strain | Adherence (%) (mean ± SD)a | CFU/mlb (mean ± SD) after coculture for:
|
% CFUc | |
|---|---|---|---|---|
| 0 day | 1 day | |||
| B-3501 | 45 ± 3 | (4.0 ± 0.7) × 105 | (4.8 ± 1.1) × 104 | 11.9 |
| M9106 | 9 ± 3 | (6.2 ± 0.6) × 105 | (1.6 ± 0.2) × 105 | 25.8 |
| M9116 | 51 ± 7 | (3.7 ± 0.3) × 105 | (1.0 ± 0.2) × 105 | 27.2 |
| M9117 | 17 ± 7 | (7.3 ± 0.4) × 105 | (4.8 ± 0.2) × 105 | 65.5 |
| M9172 | 16 ± 6 | (6.2 ± 0.5) × 105 | (4.0 ± 0.9) × 105 | 63 |
| M9174 | 37 ± 6 | (4.5 ± 0.8) × 105 | (1.3 ± 0.1) × 105 | 28.6 |
| M9254 | 27 ± 1 | (4.4 ± 0.3) × 105 | (2.9 ± 0.1) × 105 | 64.8 |
| M9255 | 26 ± 7 | (5.0 ± 1.4) × 105 | (7.3 ± 0.8) × 104 | 14.6 |
| M9256 | 24 ± 3 | (5.3 ± 0.6) × 105 | (3.2 ± 0.2) × 105 | 60.6 |
| M9257 | 12 ± 6 | (6.8 ± 1.3) × 105 | (3.1 ± 0.2) × 105 | 45.2 |
| M9258 | 20 ± 3 | (3.7 ± 0.2) × 105 | (1.5 ± 0.1) × 105 | 41.1 |
Percent adherence = (number of C. neoformans cells to which S. aureus attached × 100)/(total number of C. neoformans observed) (n = 3).
CFU were determined on Sabouraud dextrose agar (SA) containing streptomycin (100 μg/ml). The SA plates were incubated at 27°C for 48 h (n = 3).
Percent CFU = (CFU on day 1 × 100)/(CFU on day 0).
Coculture of C. neoformans and S. epidermidis.
To examine whether killing was specific to S. aureus, a coculture was performed using C. neoformans B-3501 and S. epidermidis. The population of C. neoformans fell drastically over time compared to the pure culture. Adherence of S. epidermidis to C. neoformans was also observed by light microscopy. Most fungal cells (67% ± 2%) had bacterial burdens after 1 day of cocultivation (three experiments).
Pretreatment of cells with protease or sodium metaperiodate.
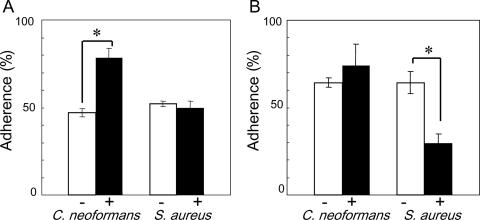
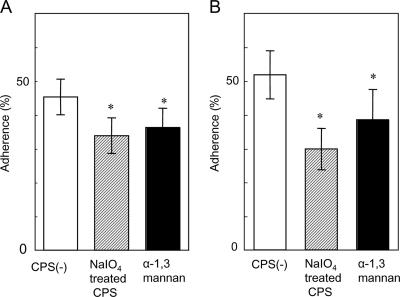
To determine which molecules contribute to cell-cell interactions, cells were treated with protease or NaIO4 prior to cocultivation. Pretreatment of C. neoformans with NaIO4 increased the proportion of burdened cells by about 30% (Fig. 4A), whereas pretreatment of S. aureus with protease decreased it by approximately 35% (Fig. 4B). In general, after treatment with NaIO4, the percent adherence decreased when carbohydrate was involved, but in this case, it increased. However, major changes suggest that carbohydrate and protein may participate in the adherence of C. neoformans and S. aureus, respectively. To examine the effect of treatment of C. neoformans on capsule size, we observed the cells before and after NaIO4 treatment using India ink preparations, measuring the diameter of whole cells and the diameter of cells without capsule. A difference between treated and untreated cells was not observed. The ratios of the diameter of whole cells/diameter of cells without capsule for treated and untreated cells were 1.375 ± 0.141 and 1.375 ± 0.116, respectively. We obtained almost the same results as shown in Fig. 4A when live cells of C. neoformans were used for NaIO4 treatment.
FIG. 4.
Effect of pretreatment with sodium metaperiodate (A) or protease K (B) on the adherence of cells. C. neoformans B-3501 was used. −, before treatment; +, after treatment. Each bar represents the mean percentage of adherence ± standard deviation (error bar) for three experiments. Student's t test was used to compare values between groups. Values that were significantly different (P value of <0.05) are indicated (*) (n = 3).
Inhibition of killing with chemically modified capsular polysaccharides.
The pretreatment of C. neoformans with NaIO4 increased adherence, suggesting that mannose residues resistant to NaIO4 in the backbone of GXM (6, 12) may be the recognition site for S. aureus cells. Polysaccharide fractions were prepared in which xylose and glucuronic acid side chains were cleaved chemically. The controlled Smith degradation product was α-(1→3)-mannan based on a characteristic single signal at 5.115 ppm in the 1H NMR spectrum. We tested whether this mannan protects the fungus. In the presence of 1 mg/ml of oxidized polysaccharides with NaIO4 or mannan, C. neoformans B-3501 CFU increased in coculture with S. aureus after day 3 (Fig. 5). The amount of modified polysaccharides required for the inhibition of killing was 1/10 that of native polysaccharides (61).
FIG. 5.
CFU of C. neoformans B-3501 in coculture with S. aureus in the presence of modified polysaccharides. Circles with solid line, pure culture; triangles with solid line, coculture of C. neoformans and S. aureus; circles with broken line, NaIO4-treated polysaccharide added (1 mg/ml) in coculture; triangles with broken line, α-(1→3)-mannan added (1 mg/ml) in coculture. The values are means ± standard deviations (error bars) for three replicate samples of one representative experiment. The horizontal dotted line represents the lower limit of detection.
We also examined the effects of modified polysaccharides on adherence. As shown in Fig. 6, after 1 and 2 days, the percent adherence significantly decreased in the presence of α-(1→3)-mannan (about 10%) or oxidized polysaccharides (about 10 to 20%). Furthermore, α-(1→3)-mannan and NaIO4-oxidized polysaccharides agglutinated heat-killed cells (80°C, 30 min) of S. aureus. These results suggest that S. aureus may have an affinity for backbone mannose residues in GXM. The reason why CFU increased after 3 days, as shown in Fig. 5, is not clear. One possible explanation is that S. aureus cells aggregated over time into large clusters in the presence of the chemically modified polysaccharides, allowing escaped C. neoformans cells to grow.
FIG. 6.
Adherence of S. aureus to C. neoformans B-3501 heat-killed cells in the presence of modified polysaccharides. (A) One-day cultures; (B) two-day cultures. The values are means ± standard deviations for three replicate samples from one representative experiment. Values that are significantly different (P < 0.05) from the value for CPS are indicated (*). CPS(−), capsular polysaccharide not treated with NalO4.
Cluster formation of S. aureus by mannooligosaccharides.
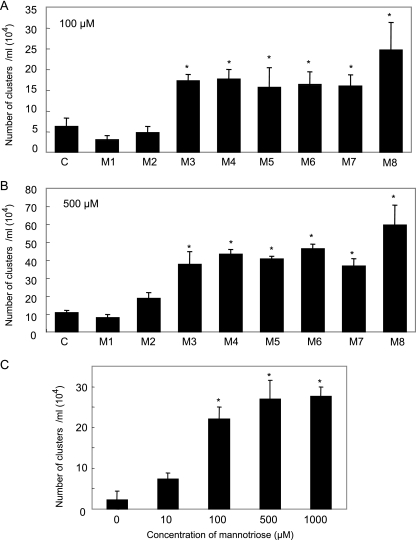
The series of mannooligosaccharides, mannose to suspected mannooctaose, were obtained by mild acid hydrolysis and gel filtration, and the structures were confirmed as α-(1→3)-mannan by 1H NMR spectra. The cells of S. aureus were clustered in the presence of the oligosaccharides. Quantitative analyses were performed, and the number of very large clusters (diameter of >1/16 mm) increased significantly in the presence of oligosaccharides with a DP of ≥3 residues as shown in Fig. 7A and B. The dose dependency was shown using trisaccharide (Fig. 7C). These data suggest the presence of a protein on S. aureus that recognized the α-(1→3)-mannotriose moiety of the mannan backbone of GXM.
FIG. 7.
Quantitative coagulation assay of S. aureus in the presence of oligosaccharides from GXM of C. neoformans. (A and B) Cells of S. aureus were incubated with 100 μM (A) or 500 μM (B) of the oligosaccharides. C, control (without saccharide); M1, mannose; M2, mannobiose. (C) Cells of S. aureus were incubated with 0, 10, 100, 500, and 1,000 μM of trisaccharide. After incubation overnight at 37°C, the number of clumps per milliliter that were larger than about 1/16 mm was determined with a microscope. Values that are significantly different (P < 0.05) from the control value (A and B) or the no-mannotriose value (C) are indicated (*).
SPR analysis of interaction of molecules on C. neoformans and S. aureus.
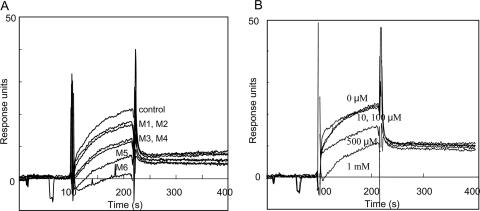
As shown in Fig. 8A, SPR experiments showed that protein from S. aureus interacted with reduced GXM from C. neoformans. In the inhibition assay, oligosaccharides larger than mannotriose showed inhibitory activities. Furthermore, a dose dependency was observed in the experiment using mannotriose (Fig. 8B).
FIG. 8.
Sensorgrams showing the interaction of immobilized reduced GXM and protein fraction from S. aureus by SPR. (A) Oligosaccharides were added to the protein fraction at a concentration of 500 μM for an inhibition assay. M1 to M6, mannose to mannohexaose. (B) Mannotriose was added to the protein fraction at a concentration of 10, 100, 500, and 1000 μM for inhibition assay.
3D structures of oligosaccharides.
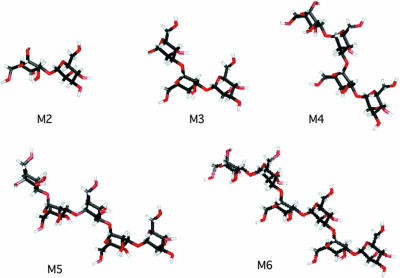
In a further step, three-dimensional (3D) structures of oligosaccharides were investigated. The PDB file was obtained from the SWEET website (http://www.dkfz-heidelberg.de/spec/) (7), and the 3D structures were constructed with MolFeat version 2.1 (Hulinks) as shown in Fig. 9. Curved structures were predicted in oligomannans larger than mannotriose. The presence of a protein on S. aureus which recognizes this conformation should be considered.
FIG. 9.
3D structure of mannooligosaccharides. The PDB file was obtained from the SWEET website (http://www.dkfz-heidelberg.de/spec/), and the 3D structures were constructed with MolFeat version 2.1 (Hulinks). M2 to M6, mannose to mannohexaose.
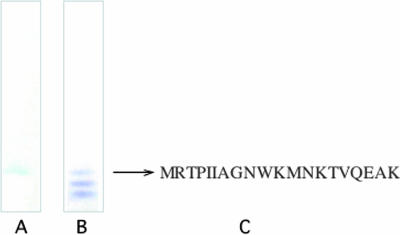
Identification of protein interacting with GXM on S. aureus.
As shown in Fig. 10, a protein band at the same position as immunolectin was observed on the PVDF membrane after CBB staining. Amino acids at the N-terminal region were determined to be MRTPIIAGNWKMNKTVQEAK by Procise 492 (Applied Biosystems). The sequence was identified by homology search as TPI of S. aureus and S. epidermidis.
FIG. 10.
Native polyacrylamide gel electrophoresis of protein fraction from S. aureus followed by electroblotting to a PVDF membrane. (A) Interaction of protein fraction from S. aureus with cryptococcal GXM; (B) CBB-stained membrane; (C) N-terminal amino acid sequence.
TPI activity.
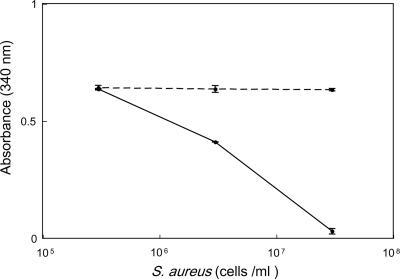
To determine whether TPI was present on the surface of S. aureus, the enzymatic assay was performed using whole cells. TPI activity was shown by the decrease of absorbance at 340 nm due to consumption by the coupling enzyme, glycerol-3-phosphate dehydrogenase. As shown in Fig. 11, TPI activity was demonstrated in a dose-dependent manner. The average values of absorbance without cells at 0 min and 5 min were 0.649 and 0.645, respectively. On the other hand, when the substrate (dl-glyceraldehyde-3-phosphate) was absent, the concentration of NADH was constant. Furthermore, the fraction applied to native polyacrylamide gel electrophoresis showed TPI activity. When a sample of 1 μl was incubated instead of a cell suspension, absorbance at 340 nm decreased to 0.027.
FIG. 11.
Assay of TPI using whole cells of S. aureus. Decrease of absorbance at 340 nm for 5 min revealed TPI activity. Whole cells were incubated with substrate dl-glyceraldehyde 3-phosphate (solid line) or incubated without substrate (broken line).
Adherence of TPI mutant strains to C. neoformans.
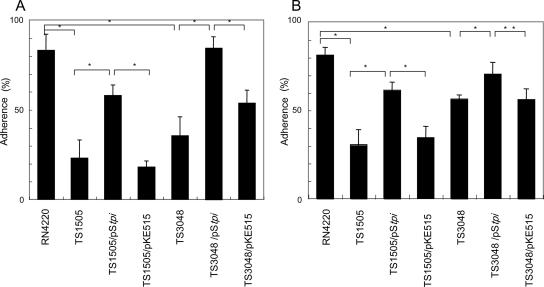
To ascertain the contribution of TPI to adherence, C. neoformans cells were incubated with TPI mutants of S. aureus. As shown in Fig. 12, percentages of adherence were decreased in strains TS1505 and TS3048 and were restored by complementation of the gene.
FIG. 12.
Adherence of TPI mutants of S. aureus to C. neoformans. S. aureus strains were incubated at 30°C with heat-killed cells of C. neoformans B-3501 (about 3 × 107/ml) for 6 h, and the percent adherence was calculated. Each bar represents the percentage of adherence ± standard deviation (error bar) for three experiments. Values that were significantly different are indicated by asterisks as follows: *, P < 0.01; **, P < 0.05. (A) S. aureus in SB was incubated with C. neoformans. (B) Twice the stationary-phase concentration of S. aureus was incubated with C. neoformans cells.
DISCUSSION
The most striking feature of the external cryptococcal surface is the large external gel formed by capsular polysaccharide. It is easy to assume that competing microorganisms must deal with this material, just as hosts with cryptococcosis must. Indeed, we had previously shown that dissolved GXM protects C. neoformans from contact-mediated killing by S. aureus, although the concentration required for inhibition was high (10 mg/ml). Thus, we wondered whether dissolved cryptococcal capsular polysaccharide competed for binding sites on a hypothetical staphylococcal binding protein. In the present study, pretreatment of C. neoformans cells with NaIO4 increased adherence of S. aureus, and the concentration of dissolved, side chain-cleaved polysaccharides required for the inhibition of killing was only 1/10 that of native polysaccharides. We inferred that S. aureus recognized mannose residues in the backbone of GXM and that the side chains of xylose and glucuronic acid sterically hinder binding to C. neoformans.
The contribution of proteins on the surface of S. aureus was suggested by the result of pretreatment by protease. To investigate the interactions of GXM from C. neoformans and proteins from S. aureus, SPR analysis was used. SPR analysis has been used to investigate interactions of various molecules, including proteins, ligand and receptor, antigen and antibody, lectin and carbohydrate, and so on (8, 15, 51). The existence of a surface molecule interacting with GXM on S. aureus was demonstrated objectively in the present SPR analyses.
Binding of a hypothetical protein on S. aureus cells to mannooligosaccharide with a DP of ≥3 should depend on their tertiary structures. To investigate the possibilities for binding, 3D structures of cryptococcal α-(1→3)-mannooligosaccharides were obtained. Mannooligosaccharides with a DP of ≥3 showed a curved conformation. The anomeric bonds rotate 180 degrees in successive residues. In tetra-, penta-, and hexasaccharides, the curved shape repeats itself, so that the polymer has a repeating wave-like shape (Fig. 9). The overall form of a larger α-(1→3)-mannan approached linearity. In contrast, α-(1→6)-mannan, the backbone in the Candida or Saccharomyces cell wall, was predicted to appear helical by the same system used. We speculate that a peptide on S. aureus recognizes the characteristic structure seen in C. neoformans.
Antagonistic interferences between fungus and bacterium or between fungi were reported in the literature. It is interesting that these interactions sometimes result in fungal apoptosis. The number of live cells of C. neoformans is decreased in the presence of Pseudomonas aeruginosa or Bacillus subtilis isolated from pigeon droppings, a common source of C. neoformans (59). Hogan et al. showed an interesting and complicated interaction between C. albicans and P. aeruginosa (23, 24). The filamentous form of the C. albicans yeast is attached and killed by P. aeruginosa. C. albicans reduces filamentation in response to N-(dodecanoyl)-1-l-3-oxo homoserine lactone (3OC12HSL) a cell-cell signaling molecule produced by P. aeruginosa, and grows in yeast form. Farnesol, a quorum-sensing molecule in C. albicans (25), also inhibits filamentation. Both 3OC12HSL and farnesol have a C12 backbone in the structure. Farnesol from C. albicans has another activity in interaction with Aspergillus nidulans: it induces apoptosis in A. nidulans (62). Recently we reported the killing of C. neoformans by a bacterium, S. aureus, with fragmentation of C. neoformans DNA, a characteristic of apoptosis (61). We speculate that effectors from S. aureus could damage the pathogenic yeast by stimulating a signal transduction pathway leading to death.
Apoptosis is known as a programmed process of cell death in multicellular organisms. Madeo et al. (38), Laun et al. (33), and Yamaki et al. (73) described cell death with predominantly apoptotic features in the eukaryotic microorganism Saccharomyces cerevisiae. In dead cells of this yeast, fragmentation of condensed chromatin and fragmentation of the nuclei resembling apoptosis in multicellular organisms were observed by transmission electron microscopy (39, 73). We considered fungal apoptosis in the staphylococcal-cryptococcal interaction. In cocultured C. neoformans and S. aureus, the positive results of the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assay suggested that the death of C. neoformans was accompanied by fragmentation of C. neoformans DNA, a characteristic of apoptosis. By transmission electron microscopy, the ultrastructure of C. neoformans surrounded by S. aureus indicated cell death. We looked for the structural features of nuclear fragmentation observed in S. cerevisiae; however, we were unable to find them in our sections of cryptococcal cells so far. Thus, it remains unknown whether apoptosis was the predominant mechanism of the death of C. neoformans.
Apoptosis-like cell death in pathogenic fungi has been described (13, 54, 55, 62). The contributions of caspase, Ras, or reactive oxygen species to apoptosis were shown, as described for multicellular organisms (38, 39, 54). The utility of the apoptosis-like cell death of fungi or other unicellular eukaryotes is not clear. Perhaps the population could escape from serious mutations by excluding damaged cells, although individuals are lost by programmed cell death. Induction of apoptosis-like cell death could be a future strategy to control fungal growth.
We wondered whether the adherence and killing were specific to S. aureus. S. epidermidis, a more commonly isolated species in the nasal cavities of healthy humans (36), killed C. neoformans as well. Streptococcus pyogenes neither adhered to nor killed C. neoformans, and the yeast grew well in its presence. However, when C. neoformans was cocultured with Escherichia coli, CFU of C. neoformans decreased, although adherence was not observed. The mechanism of injury by E. coli might be different from the contact-mediated killing by S. aureus.
Mannose-containing molecules of C. neoformans are recognized by mannose-binding receptors on mammalian mononuclear cells, macrophages, and dendritic cells, which serve for defense against infections (10, 11, 41). In the contact-mediated killing of C. neoformans by S. aureus, mannose-containing carbohydrate perhaps is recognized by mannose-binding lectin on the bacterial cell surface. Many proteins have been identified on the surface of S. aureus cells; these function in growth, division, attachment, and infection (44). While mannose-binding lectin in serum is known to bind Staphylococcus (45), a lectin on Staphylococcus that binds mannooligosaccharide has not been characterized so far. However, various bacterial lectins have been reported, and the structures of carbohydrate on host target cells have been identified in relation to infection by Escherichia coli, Klebsiella pneumoniae, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Streptococcus pneumoniae, etc. (64), and Streptococcus macacae (48). Of these species, E. coli, K. pneumoniae, and Salmonella bind mannose residues in glycoproteins. E. coli type 1 was shown to recognize the mannotriose, Man-α-(1→3)-Man-α-(1→6)-Man. The proteins on mammalian cells and on bacteria presumably are not homologous molecules. However, it is interesting that mannose residues on C. neoformans are recognized by such different cells as mammalian defensive cells and a bacterium, S. aureus. Furthermore, these phenomena may be related to the fate of C. neoformans in immunocompetent hosts. Interestingly, stress by a mannose-binding antibiotic induced apoptosis-like cell death in Saccharomyces cerevisiae (22). Thus, mannose residues may have an important biological role in receiving signals for proceeding to live or to die in yeasts, both Cryptococcus and Saccharomyces.
TPI of S. aureus, a glycolytic enzyme, was found to be a candidate protein for the decreased ability of S. aureus to interact with C. neoformans. A single amino acid substitution (E79K, E106K) appeared to affect the interaction. We hope to find out whether this lectin site is also the catalytic site of TPI. We would expect that decreased binding to the yeast by the TPI mutants should result in increased yeast viability. Unfortunately, killing of yeast is temperature dependent, occurring only at 37°C; therefore, such direct experiments are not feasible, due to the temperature sensitivity of the staphylococcal mutants.
Glycolytic enzymes are known to act in the cytoplasm. However, very interestingly, glycolytic enzymes located in cell walls of microorganisms have been reported; these had roles besides metabolism. Cell surface-associated glyceraldehyde-3-phosphate dehydrogenases (GAPDHs) of S. epidermidis, S. pyogenes, S. pneumoniae, and Candida albicans have been reported (4, 17, 20, 49). On S. epidermidis, GAPDH was found to act as a transferrin-binding protein on the cell surface (42, 43). In S. pyogenes, GAPDH on the cell surface binds fibronectin, lysozyme, and the cytoskeletal proteins myosin and actin (49). A role for GAPDH in prokaryotic-eukaryotic cell-cell communication is suggested by the finding that GAPDH of S. pyogenes interacts with pharyngeal cells of the host and affects signal transduction (50). GAPDH has been described as a multifunctional protein that helps S. pyogenes escape from the host immune system (66). GAPDH was reported to have a role in recognizing the C5a component of complement. Concerning TPI in Lactobacillus delbrueckii subsp. lactis, it was reported that a cell wall-associated protein homologous to TPI enhanced synthesis of a bacteriocin, lactacin B (68). In S. aureus, TPI has been included in a list of cell envelope proteins (18). The TPI gene was up-regulated in biofilm (3). The GAPDH and TPI genes were located in an operon with genes encoding other glycolytic enzymes. The TPI of S. epidermidis was found by a computer homology search to have the same number of amino acid residues (253 residues) as that of S. aureus. The amino acid sequences of the TPI from the two species were 87.7% identical (19), consistent with our finding of a similar phenomenon in S. epidermidis. Further studies on this staphylococcal cell surface glycolytic enzyme-lectin should contribute to the subject of prokaryote-eukaryote communication.
In conclusion, adherence of S. aureus to mannotriose units in the backbone of the cryptococcal capsule leads to death of C. neoformans. The mannotriose units had a characteristic curved tertiary structure. The staphylococcal protein which interacts with the fungal carbohydrate was identified as TPI.
Acknowledgments
We thank E. S. Jacobson for his valuable suggestions and a critical reading of the manuscript. We also thank T. Morita and Y. Yamazaki for their help with SPR analyses.
This study was partly supported by Cooperative Research Program of Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University (2004-26 and 2005-18), Shionogi Co., Ltd., Genome Pharmaceuticals Co., Ltd., and by the Industrial Technology Research Grant Program in 2004 from the New Energy and Industrial Technology Development Organization of Japan.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Aberg, J. A., L. M. Mundy, and W. G. Powderly. 1999. Pulmonary cryptococcosis in patients without HIV infection. Chest 115:734-740. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. A., and H. M. Sagha. 1988. Persistence of infection in mice inoculated intranasally with Cryptococcus neoformans. Mycopathologia 104:163-169. [DOI] [PubMed] [Google Scholar]
- 3.Becker, P., W. Hufnagle, G. Peters, and M. Herrmann. 2001. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using microrepresentational-difference analysis. Appl. Environ. Microbiol. 67:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, and S. Hammerschmidt. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72:2416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergonzelli, G. E., D. Granato, R. D. Pridmore, L. F. Marvin-Guy, D. Donnicola, and I. E. Corthesy-Theulaz. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee, A. K., J. E. Bennett, and C. P. Glaudemans. 1984. Capsular polysaccharides of Cryptococcus neoformans. Rev. Infect. Dis. 6:619-624. [DOI] [PubMed] [Google Scholar]
- 7.Bohne, A., E. Lang, and C. W. von der Lieth. 1999. SWEET—WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics 15:767-768. [DOI] [PubMed] [Google Scholar]
- 8.Bourne, Y., C. H. Astoul, V. Zamboni, W. J. Peumans, L. Menu-Bouaouiche, E. J. Van Damme, A. Barre, and P. Rouge. 2002. Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose. Biochem. J. 364:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, DC.
- 10.Chaka, W., A. F. Verheul, V. V. Vaishnav, R. Cherniak, J. Scharringa, J. Verhoef, H. Snippe, and A. I. Hoepelman. 1997. Induction of TNF-alpha in human peripheral blood mononuclear cells by the mannoprotein of Cryptococcus neoformans involves human mannose binding protein. J. Immunol. 159:2979-2985. [PubMed] [Google Scholar]
- 11.Chang, Z. L., D. Netski, P. Thorkildson, and T. R. Kozel. 2006. Binding and internalization of glucuronoxylomannan, the major capsular polysaccharide of Cryptococcus neoformans, by murine peritoneal macrophages. Infect. Immun. 74:144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherniak, R., and J. B. Sundstrom. 1994. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 62:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dadachova, E., R. W. Howell, R. A. Bryan, A. Frenkel, J. D. Nosanchuk, and A. Casadevall. 2004. Susceptibility of the human pathogenic fungi Cryptococcus neoformans and Histoplasma capsulatum to gamma-radiation versus radioimmunotherapy with alpha- and beta-emitting radioisotopes. J. Nucl. Med. 45:313-320. [PubMed] [Google Scholar]
- 14.Duncan, C., C. Stephen, S. Lester, and K. H. Bartlett. 2005. Follow-up study of dogs and cats with asymptomatic Cryptococcus gattii infection or nasal colonization. Med. Mycol. 43:663-666. [DOI] [PubMed] [Google Scholar]
- 15.Duverger, E., N. Frison, A. C. Roche, and M. Monsigny. 2003. Carbohydrate-lectin interactions assessed by surface plasmon resonance. Biochimie 85:167-179. [DOI] [PubMed] [Google Scholar]
- 16.Federle, M. J., and B. L. Bassler. 2003. Interspecies communication in bacteria. J. Clin. Investig. 112:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischetti, V. A. 2006. Surface proteins on gram-positive bacteria, p. 12-25. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 18.Gatlin, C. L., R. Pieper, S. T. Huang, E. Mongodin, E. Gebregeorgis, P. P. Parmar, D. J. Clark, H. Alami, L. Papazisi, R. D. Fleischmann, S. R. Gill, and S. N. Peterson. 2006. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics 6:1530-1549. [DOI] [PubMed] [Google Scholar]
- 19.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil-Navarro, I., M. L. Gil, M. Casanova, J. E. O'Connor, J. P. Martínez, and D. Gozalbo. 1997. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 179:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gründling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiramoto, F., N. Nomura, T. Furumai, T. Oki, and Y. Igarashi. 2003. Apoptosis-like cell death of Saccharomyces cerevisiae induced by a mannose-binding antifungal antibiotic, pradimicin. J. Antibiot. (Tokyo) 56:768-772. [DOI] [PubMed] [Google Scholar]
- 23.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, D. A., A. Vik, and R. Kolter. 2004. Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54:1212-1223. [DOI] [PubMed] [Google Scholar]
- 25.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichikawa, T., A. Nishikawa, H. Wada, R. Ikeda, and T. Shinoda. 2001. Structural studies of the antigen III cell wall polysaccharide of Trichosporon domesticum. Carbohydr. Res. 330:495-503. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda, R., and T. Maeda. 2004. Structural studies of the capsular polysaccharide of a non-neoformans Cryptococcus species identified as C. laurentii, which was reclassified as Cryptococcus flavescens, from a patient with AIDS. Carbohydr. Res. 339:503-509. [DOI] [PubMed] [Google Scholar]
- 28.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komatsuzawa, H., M. Sugai, S. Nakashima, S. Yamada, A. Matsumoto, T. Oshida, and H. Suginaka. 1997. Subcellular localization of the major autolysin, ATL and its processed proteins in Staphylococcus aureus. Microbiol. Immunol. 41:469-479. [DOI] [PubMed] [Google Scholar]
- 31.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon-Chung, K. J., and J. E. Bennett. 1992. Cryptococcosis, p. 397-446. In Medical mycology. Lea & Febiger, Philadelphia, PA.
- 33.Laun, P., A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K. U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166-1173. [PubMed] [Google Scholar]
- 34.Lee, Y. K., C. Y. Lim, W. L. Teng, A. C. Ouwehand, E. M. Tuomola, and S. Salminen. 2000. Quantitative approach in the study of adhesion of lactic acid bacteria to intestinal cells and their competition with enterobacteria. Appl. Environ. Microbiol. 66:3692-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Y., K. Kurokawa, M. Matsuo, N. Fukuhara, K. Murakami, and K. Sekimizu. 2004. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol. Genet. Genomics 271:447-457. [DOI] [PubMed] [Google Scholar]
- 36.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madeo, F., E. Frohlich, M. Ligr, M. Grey, S. J. Sigrist, D. H. Wolf, and K. U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S. J. Sigrist, S. Wesselborg, and K. U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911-917. [DOI] [PubMed] [Google Scholar]
- 40.Malik, R., D. I. Wigney, D. B. Muir, D. J. Gregory, and D. N. Love. 1992. Cryptococcosis in cats: clinical and mycological assessment of 29 cases and evaluation of treatment using orally administered fluconazole. J. Med. Vet. Mycol. 30:133-144. [DOI] [PubMed] [Google Scholar]
- 41.Mansour, M. K., E. Latz, and S. M. Levitz. 2006. Cryptococcus neoformans glycoantigens are captured by multiple lectin receptors and presented by dendritic cells. J. Immunol. 176:3053-3061. [DOI] [PubMed] [Google Scholar]
- 42.Modun, B., J. Morrissey, and P. Williams. 2000. The staphylococcal transferrin receptor: a glycolytic enzyme with novel functions. Trends Microbiol. 8:231-237. [DOI] [PubMed] [Google Scholar]
- 43.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neth, O., D. L. Jack, M. Johnson, N. J. Klein, and M. W. Turner. 2002. Enhancement of complement activation and opsonophagocytosis by complexes of mannose-binding lectin with mannose-binding lectin-associated serine protease after binding to Staphylococcus aureus. J. Immunol. 169:4430-4436. [DOI] [PubMed] [Google Scholar]
- 46.Nishikawa, A., T. Sekine, R. Ikeda, T. Shinoda, and Y. Fukazawa. 1990. Reassessment of antigenic determinant of Saccharomyces cerevisiae serotype Ia. Microbiol. Immunol. 34:825-840. [DOI] [PubMed] [Google Scholar]
- 47.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto-Shibayama, K., Y. Sato, Y. Yamamoto, K. Ohta, and H. Kizaki. 2006. Identification of a glucan-binding protein C gene homologue in Streptococcus macacae. Oral Microbiol. Immunol. 21:32-41. [DOI] [PubMed] [Google Scholar]
- 49.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pancholi, V., and V. A. Fischetti. 1997. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J. Exp. Med. 186:1633-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattnaik, P. 2005. Surface plasmon resonance: applications in understanding receptor-ligand interaction. Appl. Biochem. Biotechnol. 126:79-92. [DOI] [PubMed] [Google Scholar]
- 52.Perfect, J. R. 2006. Cryptococcus neoformans: a sugar-coated killer, p. 281-301. In J. Heitman, S. G. Filler, J. E. Edwards, Jr., and A. P. Mitchell (ed.), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC.
- 53.Pfeiffer, I., W. I. Golubev, Z. Farkas, J. Kucsera, and N. Golubev. 2004. Mycocin production in Cryptococcus aquaticus. Antonie Leeuwenhoek 86:369-375. [DOI] [PubMed] [Google Scholar]
- 54.Phillips, A. J., J. D. Crowe, and M. Ramsdale. 2006. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 103:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips, A. J., I. Sudbery, and M. Ramsdale. 2003. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 100:14327-14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rastall, R. A., G. R. Gibson, H. S. Gill, F. Guarner, T. R. Klaenhammer, B. Pot, G. Reid, I. R. Rowland, and M. E. Sanders. 2005. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol. Ecol. 52:145-152. [DOI] [PubMed] [Google Scholar]
- 57.Roche, F. M., M. Meehan, and T. J. Foster. 2003. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology 149:2759-2767. [DOI] [PubMed] [Google Scholar]
- 58.Rozacky, E. E., T. H. Sawyer, R. A. Barton, and R. W. Gracy. 1971. Studies on human triosephosphate isomerase. I. Isolation and properties of the enzyme from erythrocytes. Arch. Biochem. Biophys. 146:312-320. [DOI] [PubMed] [Google Scholar]
- 59.Ruiz, A., J. B. Neilson, and G. S. Bulmer. 1982. Control of Cryptococcus neoformans in nature by biotic factors. Sabouraudia 20:21-29. [PubMed] [Google Scholar]
- 60.Saeed, S., S. Ahmad, and S. A. Rasool. 2004. Antimicrobial spectrum, production and mode of action of staphylococcin 188 produced by Staphylococcus aureus 188. Pak. J. Pharm. Sci. 17:1-8. [PubMed] [Google Scholar]
- 61.Saito, F., and R. Ikeda. 2005. Killing of Cryptococcus neoformans by Staphylococcus aureus: role of cryptococcal capsular polysaccharide in the fungal-bacteria interaction. Med. Mycol. 43:603-612. [DOI] [PubMed] [Google Scholar]
- 62.Semighini, C. P., J. M. Hornby, R. Dumitru, K. W. Nickerson, and S. D. Harris. 2006. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 59:753-764. [DOI] [PubMed] [Google Scholar]
- 63.Sharmin, S., A. Ohori, A. Sano, K. Kamei, M. Yamaguchi, K. Takeo, J. Uno, K. Nishimura, and M. Miyaji. 2003. Histoplasma capsulatum variety duboisii isolated in Japan from an HIV-infected Ugandan patient. Jpn. J. Med. Mycol. 44:299-306. [DOI] [PubMed] [Google Scholar]
- 64.Sharon, N. 2006. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim. Biophys. Acta 1760:527-537. [DOI] [PubMed] [Google Scholar]
- 65.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100(Suppl. 2):14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terao, Y., M. Yamaguchi, S. Hamada, and S. Kawabata. 2006. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J. Biol. Chem. 281:14215-14223. [DOI] [PubMed] [Google Scholar]
- 67.Toyazaki. N. 1989. Studies on Cryptococcus neoformans, a human pathogenic fungus. 1. Ecological aspect of C. neoformans in natural environments and the mycoflora of its substrates. Trans. Mycol. Soc. Jpn. 30:149-160. [Google Scholar]
- 68.Ulrich, R. L., and T. A. Hughes. 2001. Cloning and expression analysis of the 28 kDa protein from Lactobacillus delbrueckii subsp. lactis ATCC 4797 hypothesized to influence lactacin B production. J. Appl. Microbiol. 91:1067-1073. [DOI] [PubMed] [Google Scholar]
- 69.Ushiyama, A., K. Tanaka, Y. Aiba, T. Shiba, A. Takagi, T. Mine, and Y. Koga. 2003. Lactobacillus gasseri OLL2716 as a probiotic in clarithromycin-resistant Helicobacter pylori infection. J. Gastroenterol. Hepatol. 18:986-991. [DOI] [PubMed] [Google Scholar]
- 70.Yamaguchi, M., S. K. Biswas, Y. Suzuki, H. Furukawa, and K. Takeo. 2003. Three-dimensional reconstruction of a pathogenic yeast Exophiala dermatitidis cell by freeze-substitution and serial sectioning electron microscopy. FEMS Microbiol. Lett. 219:17-21. [DOI] [PubMed] [Google Scholar]
- 71.Yamaguchi, M., M. Shimizu, T. Yamaguchi, M. Ohkusu, and S. Kawamoto. 2005. Repeated use of uranyl acetate solution in section staining in transmission electron microscopy. Plant Morphol. 17:57-59. [Google Scholar]
- 72.Yamaguchi, M., A. Tanaka, and T. Suzuki. 1992. A support film of plasma-polymerized naphthalene for electron microscopy: method of preparation and application. J. Electron Microsc. 41:7-13. [PubMed] [Google Scholar]
- 73.Yamaki, M., T. Umehara, T. Chimura, and M. Horikoshi. 2001. Cell death with predominant apoptotic features in Saccharomyces cerevisiae mediated by deletion of the histone chaperone ASF1/CIA1. Genes Cells 6:1043-1054. [DOI] [PubMed] [Google Scholar]