Abstract
Ruminococcus flavefaciens produces a cellulosomal enzyme complex, based on the structural proteins ScaA, -B, and -C, that was recently shown to attach to the bacterial cell surface via the wall-anchored protein ScaE. ScaA, -B, -C, and -E are all cohesin-bearing proteins encoded by linked genes in the sca cluster. The product of an unknown open reading frame within the sca cluster, herein designated CttA, is similar in sequence at its C terminus to the corresponding region of ScaB, which contains an X module together with a dockerin sequence. The ScaB-XDoc dyad was shown previously to interact tenaciously with the cohesin of ScaE. Likewise, avid binding was confirmed between purified recombinant fragments of the CttA-XDoc dyad and the ScaE cohesin. In addition, the N-terminal regions of CttA were shown to bind to cellulose, thus suggesting that CttA is a cell wall-anchored, cellulose-binding protein. Proteomic analysis showed that the native CttA protein (∼130 kDa) corresponds to one of the three most abundant polypeptides binding tightly to insoluble cellulose in cellulose-grown R. flavefaciens 17 cultures. Interestingly, this protein was also detected among cellulose-bound proteins in the related strain R. flavefaciens 007C but not in a mutant derivative, 007S, that was previously shown to have lost the ability to grow on dewaxed cotton fibers. In R. flavefaciens, the presence of CttA on the cell surface is likely to provide an important mechanism for substrate binding, perhaps compensating for the absence of an identified cellulose-binding module in the major cellulosomal scaffolding proteins of this species.
Ruminococcus flavefaciens is an important anaerobic, cellulolytic bacterium found in the rumen and hindgut of domestic and wild mammals (13, 17, 19, 22-24, 34, 35). Our understanding of the enzyme systems responsible for degradation of plant cell wall polymers by R. flavefaciens has advanced recently with the identification of cellulosome-like high-molecular-weight enzyme complexes (12, 25, 26, 27). In R. flavefaciens 17, the structural protein ScaA is known to interact via cohesin- dockerin pairings with a range of catalytic polypeptides possessing xylanase, endoglucanase, and esterase activities (2, 18, 26, 28). An additional structural protein, ScaC, acts as an adaptor scaffoldin, binding via its own dockerin to ScaA but also, via an unusual cohesin, to a range of other, so far unidentified proteins (27). ScaA, in turn, connects to a large structural protein, ScaB, which was recently shown to mediate attachment of the cellulosome to the bacterial cell surface through its interaction with the cell wall-anchored protein ScaE (25).
It has long been recognized that noncatalytic carbohydrate-binding modules (CBMs) play an important role in the initial stages of degradation of crystalline cellulose (8, 29). CBMs enhance the hydrolysis of cellulose by targeting and increasing the effective concentration of either individual glycoside hydrolases or multienzyme cellulosome complexes on the surfaces of carbohydrate substrates (4, 6, 33). Thus, adhesion to cellulose is considered a key step in the degradation of plant cell wall polysaccharides by R. flavefaciens. Previous studies have demonstrated that R. flavefaciens binds avidly to cellulosic substrates and that bacterial attachment is necessary for the digestion of highly ordered crystalline cellulose (20). No evidence has been found, however, that either ScaA or the cohesin-containing proteins ScaB and ScaC with which it interacts possess a CBM. This is puzzling, since in other species cellulosomal scaffolding proteins have invariably been found to include a CBM (4, 5). Although at least one of the enzymes that binds to ScaA, the cellulase Cel44A (formerly known as EndB [28]), possesses its own CBM (28), it seems unlikely that this would be the only mechanism in this bacterium for binding of cells and enzymes to cellulosic substrates. It is therefore important to discover what other molecular mechanisms could account for the observed binding of R. flavefaciens to cellulosic substrates.
In earlier work, cultures of another R. flavefaciens strain, 007, were found to have lost the ability to degrade cotton cellulose following serial subcultivation with cellobiose as the energy source (30, 31). The cotton adhesion-deficient strain 007S showed little change in the ability to degrade other forms of cellulose (i.e., Avicel). These changes were tentatively attributed to regulatory mutations affecting attachment to cotton cellulose, but despite morphological differences in the cell surface between the two strains, observable by electron microscopy (EM), the underlying changes at the molecular level remained unresolved (30).
For this paper, we set out to identify cellulose-bound proteins (CBPs) of R. flavefaciens, using a proteomics-based approach. This work reveals that one abundant cellulose-binding protein, CttA, is encoded by the sca gene cluster and that the open reading frame is positioned between the genes that encode ScaB and ScaE. This fact alone underscores the likely importance that CttA would have on the lifestyle of this bacterium. Indeed, CttA appears to be bound to the cell surface by the cell surface-anchored protein ScaE, which is also responsible for binding the cellulosomal structural protein ScaB. Moreover, CttA was absent in the cotton adhesion-deficient mutant R. flavefaciens 007S.
MATERIALS AND METHODS
Strains and growth conditions.
R. flavefaciens strains 17 (14), 007S, and 007C (31) were grown anaerobically in Hungate-Stack medium (15) that contained 1% microcrystalline cellulose (Avicel PH105; Honeywill & Stein, London, United Kingdom) as the sole energy and carbon source. Escherichia coli Solopack Gold XL10 and BL21(DE3) (Stratagene, La Jolla, CA) were used as hosts for transformation and protein expression, respectively. The plasmid pET30Ek/LIC (Novagen, Madison, WI) was used for the cloning and expression of coding regions of the cttA gene. E. coli strains were routinely grown on Luria-Bertani medium with appropriate antibiotic selection. All chemicals were purchased from Sigma-Aldrich unless otherwise stipulated.
Sequence of cttA.
The cttA coding sequence was obtained by PCR extension of the adjacent scaB gene by a combination of genome walking and PCR extension. The genome walking procedure was performed using a GenomeWalker kit (Clontech, BD Bioscience, Oxford, United Kingdom) following the manufacturer's guidelines. Alternatively, combinations of a vector-specific (M13) primer and primers designed to recognize sca sequences were used to amplify libraries of R. flavefaciens 17 chromosomal DNA ligated into pUC13 or λZAPII (26).
Preparation of native proteins bound to residual crystalline cellulose.
R. flavefaciens strains 17, 007C, and 007S were grown in 800 ml of modified Hungate-Stack medium that contained 1% (wt/vol) microcrystalline cellulose (Avicel) as the sole source of carbon. The cells were grown statically for 10 days at 37°C. Residual cellulose was collected after carefully aspirating the supernatant, using a vacuum pump. The residual cellulose was washed vigorously five times with TBS-Ca-T buffer (25 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM CaCl2, and 0.05% [vol/vol] Tween 20). CBPs were then recovered by incubating the residual cellulose with 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} and heating it at 70°C for 1 h. The suspension was centrifuged at 5,000 × g for 5 min, and the supernatant was centrifuged at 140,000 × g for 1 h at 4°C (Optima L-100 XP ultracentrifuge with a type 90 Ti rotor; Beckman-Coulter, Fullerton, CA). The excess CHAPS was removed using an ultrafiltration device (Amicon 10; Millipore, Watford, United Kingdom). The retentate, containing high-molecular-weight proteins, was diluted in deionized water, and the ultrafiltration step was repeated three times to ensure removal of most of the CHAPS. Concentrated CBPs were stored at −20°C until further use.
Two-dimensional gel electrophoresis.
Procedures for sample preparation, isoelectric focusing, two-dimensional gel electrophoresis, and protein detection by Coomassie blue staining were all performed as reported previously (26).
Spot cutting and MALDI spectrum acquisition.
Spots of interest were excised from the gel manually and subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis on an Applied Biosystems Voyager DE instrument, as described previously (26). Electrospray tandem mass spectrometry (ES-MS/MS) analysis was performed using the Ultimate system from LC Packings/Dionex (Camberly, Surrey, United Kingdom), and peptides were separated on a C18 PepMap 100 nanocolumn with a water-acetonitrile gradient (5% acetonitrile to 80% acetonitrile over 42 min). MS was performed using a Q-Trap (Applied Biosystems/MDS Sciex, Warrington, United Kingdom) triple-quadrupole mass spectrometer fitted with a nanospray ion source, where Q3 was operated as a linear ion trap.
Cloning and expression of His6-tagged expressed proteins.
The recombinant protein His6-CttA (46.1 kDa) was constructed by PCR amplification of the coding region for the N-terminal segment of CttA without the signal peptide (amino acids [aa] 30 to 414 [VSTDFV→ILVTDTT]). PCR amplification was conducted using primers designed to contain 14 or 15 bp of sequence specific for the ligation-independent cloning site of the pET30Ek/LIC expression vector (Novagen) (Table 1). Because the protein product of this plasmid was observed to be cleaved by E. coli during expression, a second plasmid, termed 2X-His6-CttA, was generated in the same way, except that the reverse primers were designed to introduce an additional His6 tag, resulting in a His6 tag at each terminus. Two more recombinant protein products, CttAα1 (32.8 kDa) and CttAα2 (24.7 kDa), were also expressed by cloning cttA sequences coding for aa 30 to 289 (VSTDF→GWLKA) and 230 to 414 (EVAAD→VTDTT), respectively, into pET30Ek/LIC. In each case, amplified DNA was initially purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions. The DNA fragment was then treated with 5 U of T4 DNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany) in a 70-μl (total volume) mixture containing 2.5 mM dATP, 1 mM dithiothreitol, 50 mM Tris-HCl (pH 8.8), 15 mM (NH4)SO4, 7 mM MgCl2, 0.1 mM EDTA, 10 mM 2-mercaptoethanol, and 20 μg/ml bovine serum albumin. The reaction mixture was incubated at 37°C for 1 h and heat inactivated at 70°C for 15 min. The treated DNA fragment was again purified using a QIAquick PCR kit and, after concentration under a vacuum, was ligated with pET30Ek/LIC vector that had been linearized and treated with T4 DNA polymerase, as recommended by the manufacturer. One unit of T4 DNA ligase (Roche) was added to the ligation mix. The resulting recombinant plasmids were transformed into chemically competent E. coli SoloPack Gold XL-10 cells (Stratagene, La Jolla, CA) and plated onto Luria-Bertani agar supplemented with 30 μg of kanamycin ml−1.
TABLE 1.
Primers used for CttA expression
| Primer | Sequencea | Purpose |
|---|---|---|
| His6-CttA forward | GACGACGACAAGATGGCTGCTGGTCAGGCTTATG | Cloning of His6-CttA |
| His6-CttA reverse | GAGGAGAAGCCCGGTTTAAGATGTAGTACTCTCAACCTG | Cloning of His6-CttA |
| His6-CttA forward | GACGACGACAAGATGGCTGCTGGTCA GGCTTATG | Cloning of 2X-His6-CttA |
| His6-CttA reverse | GAGGAGAAGCCCGGTTTACACCACCACCACCACCACAGATGTAGTACTCTCAACCTG | Cloning of 2X-His6-CttA |
| CttAα1 forward | GACGACGACAAGATGGCTGCTGGTCAGGCTTATG | Cloning of CttAα1 |
| CttAα1 reverse | GAGGAGAAGCCCGGTTTAGCCTTGAGCCAGCCATTTG | Cloning of CttAα1 |
| CttAα2 forward | GACGACGACAAGATGGAAGTTGCCGCTGATAAGGTT | Cloning of CttAα2 |
| CttAα2 reverse | GAGGAGAAGCCCGGTTTAAGATGTAGTACTCTCAACCTG | Cloning of CttAα2 |
| XynDoc forward | CGCTGGTACCTGCTAACTACGATCACTCCTAC | Cloning of XynT6-XDoc fusion protein |
| XynDoc reverse | GGACAGATCTTATTTACCGAATCTTGCGTC | Cloning of XynT6-XDoc fusion protein |
| CBM-Coh forward | GCTAGGATCCGGTCAGGCTTATGATGC | Cloning of CBM3-Coh fusion protein |
| CBM-Coh reverse | GGCCGCTCGAGTTAAGATGTAGTACTCTC | Cloning of CBM3-Coh fusion protein |
Underlined sections are specific for the LIC site in the vector, the stop codon is highlighted in bold, and nucleotides coding for a second His6 tag are shown in italics.
His6-tagged proteins from pET30Ek/LIC constructs were overexpressed following transformation into E. coli BL21(DE3). Cells were recovered and lysed by three passes in a French press, using a 40K cell type set to work at 2,000 lb/in2 (Thermo Electron Corporation, Basingstoke, United Kingdom), after growth at 37°C in 1 liter of LB supplemented with 1% (vol/vol) glycerol, 1% (wt/vol) glucose, 10 mM dl-threonine, and 30 μg kanamycin ml−1. Induction was carried out by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density at 600 nm of 0.8 to 1.0, and the culture was incubated for another 4 h at 37°C. Recombinant proteins were purified by nickel-affinity chromatography as described previously (27).
Affinity analysis of the cohesin-dockerin interaction.
The cohesin-dockerin interaction was assessed in microtiter plates by affinity analysis using matching fusion protein systems (3). For this purpose, XynDoc fusion proteins, consisting of a His6-tagged Geobacillus stearothermophilus T-6 xylanase cloned upstream of the desired dockerin module, i.e., that derived from R. flavefaciens Cel44A (previously known as EndB) (28), that of Ctta, or the C-terminal module of ScaB (12), were produced using the appropriate plasmid cassette (pETXynDoc). The complementary CBM-Coh fusion protein, comprising the family 3 CBM from the Clostridium thermocellum scaffoldin CipA upstream of the ScaE and ScaA cohesins (25), was prepared in a similar manner, using pETCBMCoh. The various proteins were expressed in an E. coli host cell system. The His6-tagged dockerin-borne constructs were purified using a Ni-nitrilotriacetic acid column, and the CBM-tagged cohesin-containing constructs were purified using a cellulose resin. Wells of microplates were coated with 0.3 μM of the desired CBM-Coh construct, and incremental concentrations of the desired XynDoc constructs were added. The washed plates were incubated sequentially with primary anti-xylanase T-6 antibody and horseradish peroxidase (HRP)-labeled anti-rabbit antibody. TMB+ substrate-chromogen (Dako Corp., Carpinteria, CA) was added, and the absorbance at 450 nm was measured using a microplate reader.
Protein binding assay with insoluble carbohydrate.
Recombinant proteins expressed from cttA (30 μg) were incubated with shaking for 1 h at 4°C in 500 μl of TBS-Ca buffer (25 mM Tris-HCl, pH 7.0, 150 mM NaCl, and 1 mM CaCl2) that contained 10 mg of Avicel PH105, oat-spelt xylan, or prewashed pulverized barley straw. After the binding step, the insoluble substrate was centrifuged for 2 min at 2,500 × g and washed three times with TBS-Ca buffer. Substrate-bound proteins were eluted in 50 μl of Laemmli buffer (21) and heated at 100°C for 3 min. Eluted proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto a polyvinylidene difluoride membrane as indicated earlier (25). His6-tagged proteins were revealed using an enhanced chemiluminescence approach (Supersignal West Pico chemiluminescence substrate; Pierce, Rockford, IL) after incubating the membrane with nickel- chelated peroxidase (HisProbe-HRP; Pierce), which specifically recognizes His6 tags. An inhibition assay was conducted in parallel in which 0.1% hydroxyl-ethyl cellulose (HEC) was added to the solution prior to the addition of His6-tagged proteins.
Phylogenetic analysis.
Phylogenetic trees and sequence alignments were generated using the ClustalX program (32) and manipulated using TreeViewPPC, version 1.5.3 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). The abbreviations and sources for protein sequences used for the analysis are given in Table 2. Terminology for the modules of undefined function (X modules) was adapted from the CAZyMODo website (Bernard Henrissat, personal communication). The various X modules and dockerin sequences were obtained from the Swiss-Prot website (http://ca.expasy.org/uniprot) or via the Carbohydrate-Active Enzymes server (http://www.cazy.org/), designed by Coutinho and Henrissat (9).
TABLE 2.
Sources of sequences for phylogenetic analysis
| Abbreviation in phylogenetic tree | Organism | Protein | GenBank, EMBL, or Swiss-Prot accession code |
|---|---|---|---|
| Acece-ScaA | Acetivibrio cellulolyticus | Scaffoldin A | AF155197 |
| Acece-ScaB | Acetivibrio cellulolyticus | Scaffoldin B | AY221112 |
| Acece-GH9B | Acetivibrio cellulolyticus | Cellulase 9B | AJ969241 |
| Aerhy-ChiA | Aeromonas hydrophila | Chitinase A | Q9L5D5 |
| Aerpu-ChiA | Aeromonas punctata | Chitinase | Q43919 |
| Altsp-ChiC | Alteromonas sp. | Chitinase A | AB004557 |
| Bacce-ScaA | Bacteroides cellulosolvens | Scaffoldin A | AF224509 |
| Bacce-Cel48A | Bacteroides cellulosolvens | Cellulase 48A | AY374129 |
| Bacsp-CelB | Bacillus sp. | Endoglucanase B | AJ133614 |
| Bommo-ChiA | Bombyx mori | Chitinase A | O92482 |
| Celfi-Cel9A | Cellulomonas fimi | Endoglucanase B | P26225 |
| Celfi-Cel48A | Cellulomonas fimi | Cellobiohydrolase B | P50899 |
| Clotm-CbhA | Clostridium thermocellum | Cellobiohydrolase A | X80993 |
| Clotm-Cel48S | Clostridium thermocellum | Cellobiohydrolase S | L06942 |
| Clotm-Cel9F | Clostridium thermocellum | Cellulase F | X60545 |
| Clotm-CelH | Clostridium thermocellum | Lic26A/Cel5E | M31903 |
| Clotm-CelP | Clostridium thermocellum | CelP | AM262818 |
| Clotm-Xyn11A | Clostridium thermocellum | Xylanase A | AF047761 |
| Cloce-CelA | Clostridium cellulolyticum | Endo-1,4-glucanase A | M32362 |
| Cloce-Cel8C | Clostridium cellulolyticum | Endo-1,4-glucanase C | M87018 |
| Cloce-Cel9E | Clostridium cellulolyticum | Cellulase E | M87018 |
| Cloce-Cel48F | Clostridium cellulolyticum | Processive endocellulase F | M87018 |
| Cloce-Cel9G | Clostridium cellulolyticum | Endo-1,4-glucanase G | M87018 |
| Cloce-Cel9H | Clostridium cellulolyticum | Cellulase H | AF316823 |
| Cloce-Ce9lM | Clostridium cellulolyticum | Cellulase M | AF316823 |
| Clotm-CipA | Clostridium thermocellum | Cellulosome-integrating protein A | L08665 |
| Hypcu-ChiA | Hyphantria cunea | Chitinase A | Q9WGG2 |
| Helze-ChiA | Helicoverpa zea | Chitinase A | O10621 |
| Rumal-Cel5G | Ruminococcus albus | Endoglucanase G | AY422810 |
| Rumal-Cel9B | Ruminococcus albus | Endoglucanase B | AY422810 |
| Rumal-Cel9C | Ruminococcus albus | Endoglucanase C | AY632899 |
| Rumal-Cel48A | Ruminococcus albus | Cellulase | AY422811 |
| Rumal-EgV | Ruminococcus albus | Endo-1,4-glucanase V | AB028320 |
| Rumal-EgVI | Ruminococcus albus | Cellulase VI (Cel9A) | AB028321 |
| Rumal-EgVII | Ruminococcus albus | 1,4-Endoglucanase VII | AB028321 |
| Rumal-Xyn11A | Ruminococcus albus | Xylanase A | U43089 |
| Rumal-XynB | Ruminococcus albus | Xylanase B | AB057588 |
| Rumal-XynC | Ruminococcus albus | Xylanase C | AB057589 |
| Rumfl-CttA | Ruminococcus flavefaciens | Cotton-binding protein A | AJ278969 |
| Rumfl-Ce3B | Ruminococcus flavefaciens | Carbohydrate esterase A | AJ238716 |
| Rumfl-Cel44A | Ruminococcus flavefaciens | Endoglucanase A | Z83304 |
| Rumfl-EndA | Ruminococcus flavefaciens | Endoglucanase A | Z83304 |
| Rumfl-ScaB | Ruminococcus flavefaciens | Scaffoldin B | AJ810899 |
| Rumfl-ScaC | Ruminococcus flavefaciens | Scaffoldin C | AJ585075 |
| Rumsp-Xyn11A | Ruminococcus sp. | Xylanase A | Z11127 |
| Rumfl-Xyn11B | Ruminococcus flavefaciens | Xylanase B | Z35226 |
| Rumfl-Xyn11D | Ruminococcus flavefaciens | Xylanase D | S61204 |
| Rumfl-Xyn11E | Ruminococcus flavefaciens | Xylanase E | AJ272430 |
| Rumfl-XynX | Ruminococcus flavefaciens | Xylanase X | AJ586790 |
| Serma-ChiA | Serratia marcescens | Chitinase A | P07254 |
| Stema-ChiA | Stenotrophomonas maltophilia | Chitinase A | AF014950 |
| Strli-ChiA | Streptomyces lividans | Chitinase A | Q59924 |
| Strli-ChiC | Streptomyces lividans | Chitinase C | P36909 |
| Thefu-Cel9A | Thermobifida fusca | Endoglucanase (E4) | L20093 |
| Thefu-Cel9B | Thermobifida fusca | Endoglucanase (E1) | L20094 |
| Thefu-Cel48A | Thermobifida fusca | Exocellulase (E6) | AF144563 |
| Vibfu-Cdx | Vibrio furnissii | Chitodextrinase | VFU41418 |
| Xansp-ChiA | Xanthomonas sp. | Chitinase A | Q9Z493 |
Nucleotide sequence accession numbers.
Sequence data for CttA have been deposited in the Swiss-Prot/TrEMBL database under accession number Q4A3Y3. Data pertaining to the sca cluster are deposited in the EMBL database under accession number AJ278969.
RESULTS
CBPs in R. flavefaciens.
Cultures of R. flavefaciens strains 17, 007C, and 007S (a cotton adhesion-deficient mutant) were grown for 10 days in basal Hungate-Stack medium (see Materials and Methods) with microcrystalline cellulose (Avicel) as the sole added energy source. CBPs were eluted from the residual cellulose by using heat and a zwitterionic detergent (CHAPS) after carefully removing cells and nonspecific protein interactions with a succession of washes. The CBP fraction was then subjected to two-dimensional gel electrophoresis. Figure 1 shows the occurrence of three major protein spots (spots 1, 2, and 3), of 100 to 150 kDa, in R. flavefaciens strains 17 (Fig. 1A) and 007C (Fig. 1B). Notably, R. flavefaciens strain 007S showed a total absence of spot 2 (∼130 kDa) (Fig. 1C).
FIG. 1.
CBPs in R. flavefaciens. (A) Three-dimensional view of a two-dimensional gel separation showing the three major spots eluted from residual cellulose from a culture of R. flavefaciens 17. The image was generated using Bio-Rad PDQuest v 7.1. (B) Separation of the same protein fraction from R. flavefaciens 007C. (C) Separation of proteins from R. flavefaciens non-cotton-degrading mutant 007S. Numbers indicate the positions of the protein molecular markers. Spots 1, 2, and 3 are indicated.
Spot 2 from R. flavefaciens 17 was excised following two-dimensional gel electrophoresis (Fig. 1A) and subjected to proteolytic digestion with trypsin. The resulting peptides were resolved using MALDI-TOF analysis. Peptide fingerprinting (http://ca.expasy.org/tools/aldente/) showed no significant hits in public domain databases. However, when the peptide masses were compared with theoretical peptide masses for gene products predicted from available R. flavefaciens 17 DNA sequences, using the program PeptideMass (http://ca.expasy.org/tools/peptide-mass.html), one clear match was obtained with a gene product of unknown function encoded by the sca cluster. Thus, ORF3 from the sca cluster (25) matched the experimental mass fingerprint generated from MALDI-TOF analysis of spot 2, with a coverage of 51.05% (Fig. 1A). ORF3 from the sca cluster in R. flavefaciens 17 is hereafter called cttA, referring to the absence of the encoded protein from the cotton adhesion-deficient strain R. flavefaciens 007S (Fig. 1C).
To confirm the presence of CttA in R. flavefaciens 007C, peptide sequencing analysis was performed using ES-MS/MS on the corresponding spots from R. flavefaciens 007C and 17 (Fig. 1B). This revealed a striking similarity between CttA proteins from the two strains. The sequence of the cttA gene from 007C was also established after amplifying R. flavefaciens 007C chromosomal DNA, using primers specific for the R. flavefaciens 17 sca cluster. This confirmed the very close sequence similarity (>97% identical residues) between the two strains (data not shown).
Spots 1 and 3 (Fig. 1A) were also subjected to MALDI-TOF and ES-MS/MS peptide analysis. Some of the peptide sequences generated from spot 3 by using ES-MS/MS produced matches in BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) with the family 48 glycoside hydrolase from Ruminococcus albus (10) (Cel48A) and the family 3 CBM from Cel9J of Clostridium cellulolyticum (7), and two peptides, the Xyn11B and Xyn11D proteins, matched dockerin modules from R. flavefaciens 17 ScaC (2, 27). This strongly suggests that spot 2 represents an abundant family 48 cellulase that has its own CBM as well as a dockerin module.
Similar analyses carried out on spot 1 produced sequences matching that of the cellulosomal structural protein ScaA. The molecular mass detected by previous one-dimensional gel electrophoresis of ScaA (26), however, differed significantly from the estimated value in this experiment (∼120 versus ∼150 kDa, respectively). It is tempting to speculate that this protein could be a novel, previously unidentified scaffoldin protein.
Sequence of the cttA gene.
Sequencing downstream of ScaB previously revealed two open reading frames (ORFs) transcribed on the same strand (25). The second of these ORFs (Fig. 2A) was reported to encode a protein, ScaE, that is covalently linked to the R. flavefaciens cell surface peptidoglycan and interacts with the C-terminal dockerin module of the major cellulosomal structural protein ScaB, thus anchoring the cellulosome to the cell surface.
FIG. 2.
Diagrammatic representations of the sca operon (A) and the modular architecture of CttA (B) from R. flavefaciens 17. The location of the cttA gene in the sca operon is shown by the arrow with a diamond pattern. For details of sca gene sequences, see EMBL accession no. AJ278969. The diagram depicting CttA shows the deduced modular organization of the gene product. Two regions that contain the newly proposed CBMs are shown in gray. T-rich linker, threonine-rich linker sequence; SP, signal peptide; X, X module; Doc, dockerin module. The four recombinant proteins used in binding studies (see the text) are also indicated.
The unknown ORF3 that precedes scaE is now known to correspond to the CBP CttA (Fig. 2A). CttA has a conventional gram-positive signal peptide with a predicted site for proteolytic processing located at Ala29 and Val30, calculated using the Neural Network algorithm of the program SignalP (http://www.cbs.dtu.dk/services/SignalP/) (Fig. 2B). The molecular mass of the mature peptide is estimated to be 75.8 kDa. The protein migrates in a gel at a much higher mass (ca. 130 kDa), however, suggesting that it may be subjected to posttranslational modifications, such as glycosylation. Glycosylation of extracytoplasmic proteins in R. flavefaciens 17 appears to be common (12). The mature CttA protein contains an unknown N-terminal region followed by a 101-aa threonine-rich linker and a C-terminal region.
The first 381 residues of the mature CttA protein showed no close similarities to other protein sequences in the database. In view of the close association with cellulose, it was hypothesized that these regions might contain novel CBMs. These postulated functional interactions of CttA were explored by overexpressing portions of the protein in E. coli, as described below.
The C terminus of CttA shows extensive sequence homology with the C-terminal XDoc modular dyad of the R. flavefaciens 17 cellulosomal structural protein ScaB (37.4% identity, with 66.7% similarity over a 195-residue stretch). The XDoc module of ScaB carries an unconventional dockerin that includes three inserts preceded by an X module of unknown function, and the dyad has been shown to be responsible for interaction with the cognate cohesin module of the cell wall-associated protein ScaE (25).
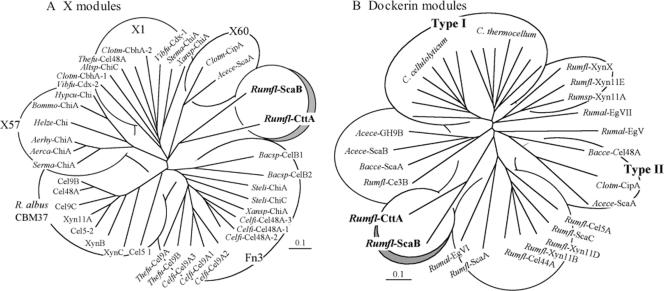
The X module portion of the dyad was subjected to ClustalX analysis against previously described modules of known (CBM37) and unknown (Fn3 and X modules) functions, and a phylogenetic tree was constructed (Fig. 3A). The results demonstrate that the CttA X module and the X module from ScaB belong to a separate branch of the tree and are distinct from other types of X module. It is interesting that the two loosely related X60 modules from the C. thermocellum CipA scaffoldin and from Acetivibrio cellulolyticus ScaA also appear immediately upstream of a dockerin, and the presence of the X60 module in the resultant XDoc dyad is essential for strong binding to the appropriate cohesin counterpart (1; R. Haimovitz and Y. Barak, unpublished results).
FIG. 3.
Phylogenetic relationships of the CttA-XDoc modular dyad in R. flavefaciens 17 in comparison with other related modules. (A) The X module of CttA (aa 512 to 621 of the deduced gene product) was mapped against a variety of previously described modules of known (CBM37) and unknown (Fn3 and X modules) functions. The CttA X module maps together with the conserved module from ScaB on a separate branch of the tree, in loose association with previously observed X1 and X60 modules. (B) The dockerin module of CttA (aa 622 to the C terminus of the deduced gene product) maps on a separate branch of the dockerin tree, together with the ScaB dockerin and distinct from other known R. flavefaciens dockerins as well as from the type I and type II dockerins. Bars, 0.1% amino acid substitutions. See Table 2 for the sources of the sequences and the abbreviations used in this figure. Multiple modules derived from a single protein are enumerated according to their positions relative to the N terminus of the polypeptide chain. Xansp-ChiA contains both an X1 module and an Fn3 domain.
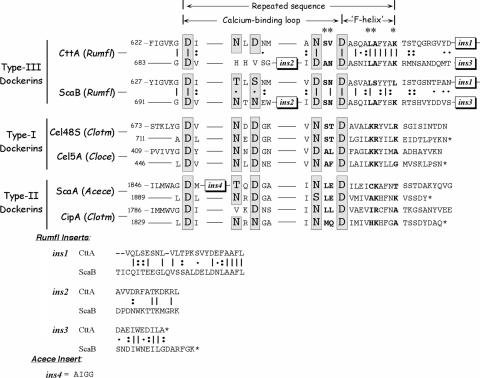
An additional phylogenetic tree was constructed (Fig. 3B), which served to compare the unconventional dockerin portion of the CttA dyad with other known dockerin modules from R. flavefaciens as well as with type I dockerins from clostridial cellulosomal enzymes and the known type II dockerins, derived from the C. thermocellum and A. cellulolyticus scaffoldins and a Bacteroides cellulosolvens enzyme. Like the upstream X modules, the dockerin portions of the CttA and ScaB XDocs both map together on a branch distinct from those of the other known dockerins. In fact, the dockerin sequences of the CttA and ScaB proteins are highly conserved, and both contain similar conserved insertions (Fig. 4). The first insertion is 23 residues long in CttA (26 residues in ScaB) and is positioned at the end of the linker segment that connects the two calcium-binding dockerin loops, thus extending the linker relative to those of other types of dockerin. The first calcium-binding loop is of the standard type, with calcium-coordinating Asp and Asn residues appearing at the expected positions. The second calcium-binding loop, however, is atypical—two of the characteristic Asp and/or Asn residues are missing, and like the case in ScaB, a 13-residue (second) insertion is embedded within the loop. Finally, a 10-residue extension occurs at the C terminus, which is somewhat shorter than the conserved insertion at the ScaB terminus. The two distinctive internal insertions are absent from any of the other known dockerins, including those derived from other R. flavefaciens cellulosomal proteins.
FIG. 4.
Comparison of amino acid sequences of the C-terminal dockerin modules of R. flavefaciens (Rumfl) CttA and ScaB and their relationship to selected type I and type II dockerins. Sequences ins1, ins2, and ins3 represent designated conserved inserts unique to the CttA and ScaB dockerins. A 110-residue “X module” precedes the sequence shown in the figure. The degree of conservation of each position within the repeated ScaB sequence is indicated as follows: vertical lines denote identity, colons indicate that the residues are conserved, and dots indicate that the residues are semiconserved, as defined by the EBI server (http://www2.ebi.ac.uk/clustalw/). The presumed calcium-coordinating residues are shown in a larger font and highlighted in gray, and suspected specificity residues are shown in bold and labeled at the top with an asterisk. A four-residue insert (ins4) is also present in the A. cellulolyticus (Acece) ScaA sequence. The GenBank accession codes for C. thermocellum (Clotm) Cel48A and CipA, C. cellulolyticum (Cloce) Cel5A, and A. cellulolyticus ScaA are L06942, L08665, M93096, and AF155197, respectively.
Interaction of the CttA XDoc with the ScaE cohesin.
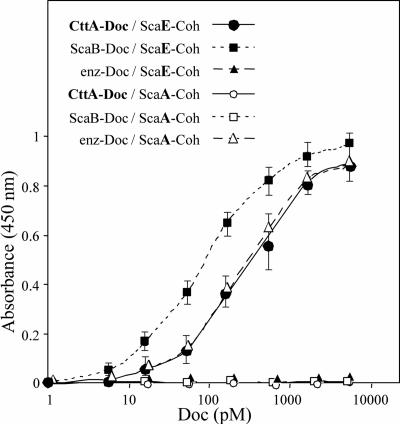
The interaction of the CttA XDoc with cohesin-borne scaffoldin proteins in the R. flavefaciens cellulosome was analyzed on microtiter plates, using a highly efficient matching fusion protein system in conjunction with an enzyme-linked affinity assay (3). The C-terminal XDoc region of CttA, comprising the ScaB-like dockerin and its upstream X module, was overexpressed in E. coli, fused to xylanase T-6 from G. stearothermophilus as a XynDoc chimeric protein. The XDoc of ScaB (25) was similarly produced as a XynDoc fusion protein to serve as a positive control for interaction with the R. flavefaciens ScaE cohesin. A previously constructed XynDoc fusion protein containing the dockerin of the enzyme Cel44A (25) was used as a negative control. The ScaE cohesin was expressed as a fusion protein downstream of the scaffoldin-borne C. thermocellum family 3a CBM. Likewise, a CBM-based fusion protein of cohesin 3 from R. flavefaciens ScaA was produced as a counterpart negative control (25); this cohesin is known to interact with the Cel44A dockerin but fails to interact with the XDoc module from ScaB (25). The CBM-Coh and XynDoc target proteins can be expressed in relatively large quantities, and both types of carrier protein impart stability to the fusion partner while providing a standardized functional setting (3).
As shown in Fig. 5, the CBM-based ScaE-Coh probe (ScaE-Coh) bound very strongly to the xylanase-borne CttA-XDoc protein. The observed affinity of binding was somewhat less than that observed for the positive control, i.e., the interaction between ScaE-Coh and ScaB-XDoc, but was very similar to that for the Cel44A-borne dockerin (enz-Doc) binding with the ScaA-based cohesin (ScaA-Coh). The interaction between ScaE-Coh and CttA-XDoc is selective, since CttA-XDoc (like its ScaB relative) essentially fails to recognize ScaA-Coh, and conversely, ScaE-Coh fails to interact with the Cel44A dockerin (enz-Doc) (Fig. 5).
FIG. 5.
Interaction of XDoc dyad from CttA with recombinant cohesin from ScaE. Binding of the CttA XDoc to the ScaE cohesin was achieved using a matching fusion protein system and an affinity-based enzyme-linked immunosorbent assay according to the method of Barak et al. (3). Wells of microtiter plates were coated with the desired CBM-Coh construct (the ScaE cohesin or cohesin 3 from ScaA) at a concentration of 0.3 μM, and the XynDoc fusion proteins, containing XDoc from CttA or ScaB or the dockerin from the R. flavefaciens cellulosomal enzyme Cel44A (enz-Doc), were examined at incremental concentrations. The amount of bound dockerin was determined using a rabbit anti-Xyn antibody followed by a peroxidase-conjugated secondary antibody, and color formation was determined colorimetrically. Error bars indicate the respective standard deviations from the means for the designated points.
Substrate binding by recombinant CttA fragments.
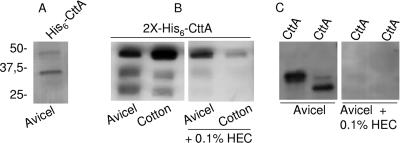
The N-terminal region of the mature CttA protein was further analyzed for its predicted role as a carbohydrate-binding protein. The gene coding for the 381-aa N-terminal portion of the mature CttA protein was cloned into a vector fused to the coding region for an N-terminal polyhistidine tag. The resulting recombinant protein product, termed His6-CttA, was then analyzed by SDS-PAGE and found to be partially cleaved by E. coli native proteases. Nevertheless, in an absorption assay using His6-CttA and insoluble carbohydrate, it was found that both the full-length recombinant protein product and its N-terminal proteolytic fragment, containing the N-terminal polyhistidine tag, bound avidly to cellulose (Fig. 6A). Taking advantage of this unexpected proteolytic event in E. coli during heterologous protein expression, we decided to repeat the expression of the N-terminal 381-aa region, but with the addition of a second C-terminal polyhistidine tag, to produce a protein termed 2X-His6-CttA. The second polyhistidine tag was designed to allow recovery of the C-terminal portion of the protein after proteolytic cleavage in E. coli and to facilitate studies on its ability to bind to insoluble carbohydrate. Figure 6B shows the results obtained when 2X-His6-CttA was adsorbed to two microcrystalline cellulose substrates, namely, Avicel and dewaxed cotton. This figure also shows that the two presumed proteolytic products from 2X-His6-CttA bound to the tested carbohydrate as well as to the uncleaved (∼46 kDa) 2X-His6-CttA protein product. The binding proved to be specific, since it was greatly inhibited by preincubating the insoluble carbohydrate with HEC.
FIG. 6.
Binding of recombinant proteins derived from CttA to crystalline cellulose. For the assay, the recombinant products were incubated with Avicel or cotton, as indicated, and then washed, and the bound material was recovered by heating in Laemmli buffer. CBPs were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membranes, and polyhistidine-tagged proteins were revealed by chemiluminescence using Ni-conjugated HRP. (A) Binding of the recombinant product His6-CttA (46 kDa) and its proteolytic derivative to Avicel. (B) Binding of the double-polyhistidine-tagged product 2X-His6-CttA and its proteolytic derivatives to Avicel and cotton and inhibition of binding by the addition of 0.2% HEC (hydroxyethyl cellulose). (C) Binding of recombinant 32-kDa and 25-kDa derivatives—CttA1 and CttA2, respectively—to Avicel and inhibition of their binding by the addition of 0.2% HEC.
The results of this experiment suggest that the mature 381-aa N-terminal region of CttA is composed of two novel CBMs. In order to determine the boundaries of each CBM within the N-terminal region of CttA, MALDI-TOF analysis of the two proteolytic fragments from 2X-His6-CttA was carried out. The results of MALDI-TOF analysis of the larger band provided peptide masses matching the amino acid sequence between F88 and K162, which is located within the first estimated 20 kDa of the overexpressed protein. On the other hand, MALDI-TOF analysis of the smaller band revealed peptide masses equivalent to the amino acid sequence located between G324 and R407, contained in the last estimated 10 kDa of 2X-His6-CttA.
The occurrence of two tandem CBMs in the modular architecture of CttA was investigated further by the cloning of two additional regions of the cttA gene, coding for a 32-kDa N-terminal region of the mature CttA protein and the adjacent 25-kDa region. The protein products were designated CttA1 and CttA2, for the 32- and 25-kDa protein products, respectively. The results of the binding assay with the new proteins are depicted in Fig. 6C. As anticipated from the previous analysis, both proteins were able to bind microcrystalline cellulose and were inhibited by the addition of HEC to the buffer medium prior to protein binding, further corroborating the occurrence of two novel CBMs in CttA.
DISCUSSION
It is widely accepted that the first step in the microbial degradation of highly recalcitrant and insoluble cellulosic substrates involves the adhesion to cellulose of either free enzymes or cell surface-attached cellulosome complexes. Not only have CBMs been implicated in increasing the effective concentration of enzymes on the surface of cellulosic material (4, 6, 33), but there is also evidence suggesting a more active role for certain CBMs in disrupting crystalline cellulose, thereby enhancing the catalytic activity of glycoside hydrolases (11). CBMs are very commonly found as part of the modular architecture of the major enzymes involved in the degradation of plant cell walls. CBMs are also found in nonhydrolytic scaffolding proteins within the cellulosomes of Clostridium spp. and are considered to be the major factor in the binding of the cellulosome complex, and of the bacterial cell, to the substrate (4). Curiously, no scaffolding protein from R. flavefaciens has been shown to carry a CBM. On the other hand, the cellulose-binding CttA protein from R. flavefaciens identified here is encoded by the sca gene cluster and represents the first case of a nonhydrolytic surface protein whose only function seems to be binding of the cell to cellulose.
The hydrophobic interaction of aromatic amino acid side chains, i.e., those of tryptophan, tyrosine, and less commonly, phenylalanine, with carbohydrate residues has been found to be ubiquitous among CBMs from different families (8). The 381-aa N-terminal region of the mature CttA protein contains many aromatic residues (0.78, 3.66, and 2.62 mol% of tryptophan, tyrosine, and phenylalanine, respectively). Although a three-dimensional predicted structural model could not be constructed due to the lack of an available protein template, the abundance of aromatic residues in the N-terminal portion of CttA could explain its ability to bind cellulose.
In earlier work, cultures of another R. flavefaciens strain, 007, were found to have lost the ability to degrade cotton cellulose following serial subcultivation with cellobiose as the energy source (30, 31). The cotton adhesion-deficient derivative, 007S, showed little change in the ability to degrade other forms of cellulose, giving, for example, 65.1% and 50.2% dry weight loss from Avicel and Sigmacel, respectively, after 5 days of incubation, compared with 70.1% and 60.1%, respectively, for the cotton-degrading strain 007C (31). These changes were tentatively attributed to regulatory mutations affecting attachment to cotton cellulose, but despite morphological differences in the cell surface, observable by EM, between the two strains, the underlying changes at the molecular level remained unsolved (30).
Figure 1B shows that the major CBPs of R. flavefaciens 007C are very similar to those of strain 17, shown in Fig. 1A. Furthermore, ES-MS/MS analysis of the 130-kDa CttA protein from strain 007C showed that it has high similarity with that from strain 17 (90% identity). Strain 007S, however, lacks the newly identified 130-kDa CttA protein among its major CBPs. The reason for the absence of this protein, however, remains unknown. The cttA and scaE genes from the 007C and 007S strains were sequenced following PCR amplification and were found to be identical to each other and very similar (97% amino acid identity) to their homologues from strain 17. A recent comparative analysis between the sca gene clusters of two strains of R. flavefaciens, 17 and FD-1, demonstrated a similar array of scaffoldin-encoding genes, including a homolog of cttA, which was also present in the same location in the sca cluster of genes in FD-1 (16). The conservation of cttA genes across different strains of R. flavefaciens tends to confirm that CttA plays an important role in this species.
The evidence from this work demonstrates that R. flavefaciens displays a unique mode of binding to cellulose via a novel nonhydrolytic protein which specializes in binding to cellulose. We infer that CttA contains two tandem novel CBMs and is anchored to the cell surface via interaction of its dockerin module with the covalently cell surface-attached ScaE protein. The importance of CttA for cell adhesion in R. flavefaciens needs to be addressed fully in further experiments. The apparent absence of the CttA protein in the cotton adhesion-deficient strain R. flavefaciens 007S could mean that CttA is required for the degradation of this particularly recalcitrant form of crystalline cellulose. However, since strain 007S is able to degrade other forms of cellulose (e.g., Avicel), additional CBMs with different target specificities must be sufficient to achieve the proximity necessary for the initial step of cellulose degradation for these substrates. The ScaA scaffoldin itself lacks a CBM within its modular architecture (26), so the presence of ScaA in the CBP fraction from R. flavefaciens must be attributed to its interactions with dockerin-containing proteins that carry CBMs, such as Cel44A. Another major R. flavefaciens protein eluted from the residual cellulose showed, after peptide sequencing, a multimodular architecture with a module sharing homology to a family 48 glycoside hydrolase, a CBM, and a dockerin module similar to those present in other cellulosomal catalytic subunits known to interact with the structural protein ScaA. Alternatively, there is also the possibility that another, as yet unidentified scaffoldin, similar to but larger than ScaA and carrying an integral CBM, may exist.
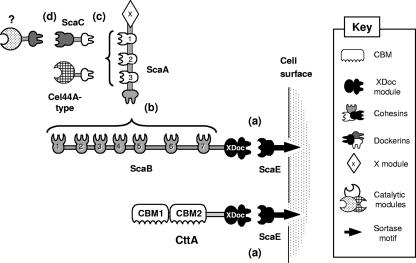
Our current view of the cellulosome system in R. flavefaciens strain 17 is presented schematically in Fig. 7. Both CttA and ScaB are attached to the cell envelope via the same component—ScaE—which is anchored covalently to the gram-positive peptidoglycan via a sortase signal motif. The other cellulosomal components detected thus far are also attached to the bacterial cell surface via the ScaB cohesins, using the same ScaE anchoring scaffoldin. This arrangement of the R. flavefaciens system alters our general view of bacterial cellulosomes, which are significantly more versatile and divergent than heretofore considered. CttA thus represents a noncatalytic cell surface protein which is separate from other cellulosome components, whose sole function appears to be involved in direct binding of the bacterial cell to cellulosic substrates.
FIG. 7.
Schematic overview of the proposed cellulosome system in R. flavefaciens strain 17. The system is characterized by at least four different cohesin-dockerin specificities, designated a to d. (a) The conserved XDoc dyad of CttA, like that of ScaB, interacts with the ScaE cohesin. (b) The seven ScaB cohesins interact with the ScaA dockerin, thereby increasing the number of components that are incorporated into the R. flavefaciens cellulosome. (c) The ScaA cohesins bind either directly to a group of dockerin-containing enzymes (Cel44A-like) or to the dockerin of ScaC, whose divergent cohesin recognizes and incorporates into the cellulosome a different set of dockerin-containing enzymes and other components (d).
Acknowledgments
We thank Pauline Young and Donna Henderson (Rowett Research Institute) for DNA sequencing and Garry Rucklidge, Martin Reid, and Gary Duncan (Rowett Research Institute) for protein analysis. We also thank Ilya Borovok (Tel Aviv University) for valuable discussions and for his suggestion of CttA as a name for the Orf3 protein.
The Rowett Research Institute is supported by the Scottish Executive Environment and Rural Affairs Department. Research grants 394/03 and 422/05 from the Israel Science Foundation (Jerusalem) and a grant from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel, are gratefully acknowledged. M.T.R. is supported by an EU Framework V GEMINI grant (QLRT-2001-02056). T.C. was supported by a Marie Curie fellowship program and a FEBS short-term fellowship program.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Adams, J. J., G. Pal, Z. Jia, and S. P. Smith. 2006. Mechanism of bacterial cell-surface attachment revealed by the structure of cellulosomal type II cohesin-dockerin complex. Proc. Natl. Acad. Sci. USA 103:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aurilia, V., J. C. Martin, S. I. McCrae, K. P. Scott, M. T. Rincon, and H. J. Flint. 2000. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology 146:1391-1397. [DOI] [PubMed] [Google Scholar]
- 3.Barak, Y., T. Handelsman, D. Nakar, A. Mechaly, R. Lamed, Y. Shoham, and E. A. Bayer. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491-501. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, E. A., J. P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., L. J. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes—structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 6.Beguin, P., and J. P. Aubert. 1994. The biological degradation of cellulose. FEMS Microbiol. Rev. 13:25-58. [DOI] [PubMed] [Google Scholar]
- 7.Belaich, A., G. Parsiegla, L. Gal, C. Villard, R. Haser, and J. P. Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. J. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 10.Devillard, E., D. B. Goodheart, S. K. Karnati, E. A. Bayer, R. Lamed, J. Miron, K. E. Nelson, and M. Morrison. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Din, N., H. G. Damude, N. R. Gilkes, R. C. Miller, Jr., R. A. Warren, and D. G. Kilburn. 1994. C1-Cx revisited: intramolecular synergism in a cellulase. Proc. Natl. Acad. Sci. USA 91:11383-11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, S. Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint, H. J., C. A. McPherson, and J. Bisset. 1989. Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and beta(1-3,1-4)glucanase activities. Appl. Environ. Microbiol. 55:1230-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hungate, R. E., and R. J. Stack. 1982. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl. Environ. Microbiol. 44:79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jindou, S., I. Borovok, M. T. Rincon, H. J. Flint, D. A. Antonopoulos, M. E. Berg, B. A. White, E. A. Bayer, and R. Lamed. 2006. Conservation and divergence in cellulosome architecture between two strains of Ruminococcus flavefaciens. J. Bacteriol. 188:7971-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julliand, V., A. de Vaux, L. Millet, and G. Fonty. 1999. Identification of Ruminococcus flavefaciens as the predominant cellulolytic bacterial species of the equine cecum. Appl. Environ. Microbiol. 65:3738-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirby, J., J. C. Martin, A. S. Daniel, and H. J. Flint. 1997. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol. Lett. 149:213-219. [DOI] [PubMed] [Google Scholar]
- 19.Krause, D. O., B. P. Dalrymple, W. J. Smith, R. I. Mackie, and C. S. McSweeney. 1999. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep. Microbiology 145:1797-1807. [DOI] [PubMed] [Google Scholar]
- 20.Kudo, H., K. J. Cheng, and J. W. Costerton. 1987. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can. J. Microbiol. 33:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Larue, R., Z. Yu, V. A. Parisi, A. R. Egan, and M. Morrison. 2005. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 7:530-543. [DOI] [PubMed] [Google Scholar]
- 23.Michalet-Doreau, B., I. Fernandez, and G. Fonty. 2002. A comparison of enzymatic and molecular approaches to characterize the cellulolytic microbial ecosystems of the rumen and the cecum. J. Anim. Sci. 80:790-796. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, K. E., S. H. Zinder, I. Hance, P. Burr, D. Odongo, D. Wasawo, A. Odenyo, and R. Bishop. 2003. Phylogenetic analysis of the microbial populations in the wild herbivore gastrointestinal tract: insights into an unexplored niche. Environ. Microbiol. 5:1212-1220. [DOI] [PubMed] [Google Scholar]
- 25.Rincon, M. T., T. Cepeljnik, J. C. Martin, R. Lamed, Y. Barak, E. A. Bayer, and H. J. Flint. 2005. Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the cell surface. J. Bacteriol. 187:7569-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rincon, M. T., S. Y. Ding, S. I. McCrae, J. C. Martin, V. Aurilia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rincon, M. T., J. C. Martin, V. Aurilia, S. I. McCrae, G. J. Rucklidge, M. D. Reid, E. A. Bayer, R. Lamed, and H. J. Flint. 2004. ScaC, an adaptor protein carrying a novel cohesin that expands the dockerin-binding repertoire of the Ruminococcus flavefaciens 17 cellulosome. J. Bacteriol. 186:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rincon, M. T., S. I. McCrae, J. Kirby, K. P. Scott, and H. J. Flint. 2001. EndB, a multidomain family 44 cellulase from Ruminococcus flavefaciens 17, binds to cellulose via a novel cellulose-binding module and to another R. flavefaciens protein via a dockerin domain. Appl. Environ. Microbiol. 67:4426-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoseyov, O., Z. Shani, and I. Levy. 2006. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 70:283-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart, C. S., S. H. Duncan, and H. J. Flint. 1990. The properties of Ruminococcus flavefaciens which differ in their ability to degrade cotton cellulose. FEMS Microbiol. Lett. 72:47-50. [DOI] [PubMed] [Google Scholar]
- 31.Stewart, C. S., S. H. Duncan, C. A. McPherson, A. J. Richardson, and H. J. Flint. 1990. The implications of the loss and regain of cotton-degrading activity for the degradation of straw by Ruminococcus flavefaciens strain-007. J. Appl. Bacteriol. 68:349-356. [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 34.Varel, V. H., and J. T. Yen. 1997. Microbial perspective on fiber utilization by swine. J. Anim. Sci. 75:2715-2722. [DOI] [PubMed] [Google Scholar]
- 35.Wedekind, K. J., H. R. Mansfield, and L. Montgomery. 1988. Enumeration and isolation of cellulolytic and hemicellulolytic bacteria from human feces. Appl. Environ. Microbiol. 54:1530-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]