Abstract
Adherent-invasive Escherichia coli (AIEC) isolated from Crohn's disease patients is able to adhere to and invade intestinal epithelial cells and to replicate in mature phagolysosomes within macrophages. Here, we show that the dsbA gene, encoding a periplasmic oxidoreductase, was required for AIEC strain LF82 to adhere to intestinal epithelial cells and to survive within macrophages. The LF82-ΔdsbA mutant did not express flagella and, probably as a consequence of this, did not express type 1 pili. The role of DsbA in adhesion is restricted to the loss of flagella and type 1 pili, as forced contact between bacteria and cells and induced expression of type 1 pili restored the wild-type phenotype. In contrast, the dsbA gene is essential for AIEC LF82 bacteria to survive within macrophages, irrespective of the loss of flagella and type 1 pilus expression, and the survival ability of LF82-ΔdsbA was as low as that of the nonpathogenic E. coli K-12, which was efficiently killed by macrophages. We also provide evidence that the dsbA gene is needed for LF82 bacteria to grow and survive in an acidic and nutrient-poor medium that partly mimics the harsh environment of the phagocytic vacuole. In addition, under such stress conditions dsbA transcription is highly up-regulated. Finally, the CpxRA signaling pathway does not play a role in regulation of dsbA expression in AIEC LF82 bacteria under conditions similar to those of mature phagolysosomes.
Crohn's disease (CD) is an inflammatory bowel disease that is likely the result of a genetic predisposition that leads to a mucosal immune regulatory cell defect, barrier defects, and susceptibility to environmental triggers (40). There is increasing evidence that the mucosa-associated flora are important in the pathogenesis of CD (25, 50, 57, 59). Some characteristic pathological elements of CD, including aphthous ulcers of the mucosa, mural abscesses, and macrophage and epithelioid cell granulomas, also occur in well-recognized infectious diseases such as shigellosis, salmonellosis, and Yersinia enterocolitis, in which invasiveness is an essential virulence factor of the bacteria involved (74). These pathogenic bacteria have not been found to be associated with CD. High levels of antibodies directed against Escherichia coli outer membrane protein C (OmpC) are observed in CD patients, and E. coli DNA has been detected in 80% of microdissected granulomas of CD patients, which suggests that E. coli may be involved in CD lesions (47, 56). Independent studies have reported the presence of increased numbers of bacteria belonging to the E. coli species cells and the presence of E. coli with virulence properties, termed adherent-invasive E. coli (AIEC), colonizing the ileal mucosa of CD patients (8, 18, 19, 43, 64).
AIEC strains are able to survive and replicate extensively in large vacuoles within macrophages without triggering host cell death. Infected macrophages secrete large amounts of tumor necrosis factor alpha (18, 27). We recently reported that, after phagocytosis, AIEC bacteria are internalized in phagosomes that transit along the classical endocytic pathway and mature into active phagolysosomes in which bacteria are exposed to acidic pH and the degradative activity of cathepsin D (10). The intravacuolar microenvironment of phagolysosomes is crucial for the full virulence expression of AIEC reference strain LF82, because the acidic pH of phagolysosomes is required for expression of the stress protein HtrA and for replication of AIEC LF82 bacteria within macrophages (9, 10).
Intracellular pathogens have evolved numerous strategies to survive within phagocytic cells, and many of these strategies involve proteins that are exported from the cytoplasm to either the periplasm or the outer membrane or are secreted out of the cell (11, 13, 26, 38). Some proteins are transported or assembled by means of specialized secretory systems, but many of these proteins pass through the periplasm, where they undergo some degree of folding into their native conformation. Many proteins, especially membrane and exported proteins, are stabilized by intramolecular disulfide bridges between cysteine residues, without which they are misfolded, unstable, and often inactive (1, 45). In E. coli, disulfide bond formation is catalyzed by the Dsb proteins (36). DsbA is a 21-kDa periplasmic protein with a CXXC motif in its active site. It interacts with reduced substrate proteins, catalyzing the oxidation of their cysteine residues to disulfide bonds (2). In a catalytic cascade pathway, the activity of DsbA is maintained by the function of the inner membrane protein DsbB, which oxidizes DsbA (1, 30) and which is reoxidized directly by membrane-bound ubiquinones (39). DsbC and DsbG are the periplasmic components of the isomerization pathway. These proteins reshuffle mispaired multiple disulfide bonds (6, 53). The active sites of DsbC and DsbG are maintained in the reduced form by the inner membrane protein DsbD, which transfers electrons from the cytoplasmic protein thioredoxin to DsbC and DsbG (37, 46, 53, 54).
The role of DsbA in virulence has been addressed for several pathogens. It is crucial in the biogenesis of toxins such as the Vibrio cholerae toxin (49, 62), E. coli heat-labile and heat-stable toxins produced by enterotoxigenic E. coli strains (48, 72), and the Bordetella pertussis toxin (61) and of multimeric structures on the bacterial surface involved in virulence, such as flagella in E. coli (16), fimbriae in V. cholerae (33), Neisseria meningitidis (65), enteropathogenic E. coli (23), or uropathogenic E. coli (35), and components of the type III secretory machinery in Yersinia pestis (34), Shigella flexneri (67, 69), Pseudomonas spp. (32) and Salmonella enterica serovar Typhimurium (44). DsbA is also required for the systemic stages of an E. coli K1 infection, although it is not known which factors are posttranslationally modified (29). In this study, we demonstrate that dsbA is required for adhesion of intestinal epithelial cells and for intramacrophagic survival of the CD-associated AIEC reference strain LF82. In addition, we provide evidence that the dsbA gene is essential for AIEC LF82 bacteria to grow and survive in an acidic and nutrient-poor medium that partly mimics the harsh environment encountered by bacteria within the phagocytic vacuole and that dsbA transcription is highly up-regulated under such stress conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown routinely in Luria-Bertani (LB) broth or on LB agar plates (Institut Pasteur Production) overnight at 37°C. When required, appropriate antibiotics were added to the media at the following final concentrations: ampicillin (Ap), 50 μg/ml; kanamycin (Km), 50 μg/ml; chloramphenicol (Cm), 25 μg/ml. Strain LF82 was isolated from a chronic ileal lesion of a patient with CD. E. coli strain JM109 was used as host strain for cloning experiments. E. coli strain K-12 C600 was used as a negative control for macrophage survival assays. E. coli BW25141 was the recipient of the Kmr cassette for the construction of the mutant.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| LF82 | E. coli isolated from an ileal biopsy sample of a patient with CD | 19 |
| LF82-ΔdsbA | LF82 isogenic mutant with dsbA gene deleted | This study |
| LF82-ΔfliC | LF82 isogenic mutant with fliC gene deleted | 3 |
| LF82-ΔcpxR | LF82 isogenic mutant with cpxR gene deleted | 9 |
| LF82-ΔhtrA | LF82 isogenic mutant with htrA gene deleted | 9 |
| BW25141 | E. coli strain harboring plasmid pSC140 (template for Kmr) | 20 |
| K-12 C600 | Nonpathogenic E. coli strain | Laboratory stock |
| Plasmids | ||
| pHSG575 | E. coli cloning vector; Cmr | Biolabs |
| pBAD33 | E. coli cloning vector; Cmr | 31 |
| pFPV25·1 | Vector for constitutive GFP expression; Apr | 66 |
| pKOBEG | pBAD cloning vector harboring λ phage redγβα operon; Cmr | 14 |
| pPBI01 | pHSG575 harboring the entire fim operon of E. coli K-12 strain J96 | 7 |
| pPBI11 | pBAD33 harboring the complete dsbA gene of LF82 | This study |
| JE7 | Cosmid pHC79 harboring the entire fim operon of AIEC strain LF82 | 7 |
Cell lines and cell cultures.
The murine macrophage-like cell line J774 (ATCC TIB67) was maintained in an atmosphere containing 5% CO2 at 37°C in RPMI 1640 medium (BioWhittaker; Cambrex Bio Sciences Verviers, Belgium) supplemented with 10% (vol/vol) fetal calf serum (FCS) (BioWhittaker) and 1% l-glutamine (Life Technologies, Cergy-Pontoise, France).
Intestine-407 cells (derived from human intestinal embryonic jejunum and ileum) were purchased from Flow Laboratories Inc., McLean, VA. Cultured cells were maintained in an atmosphere containing 5% CO2 at 37°C in modified Eagle medium (BioWhittaker) supplemented with 10% (vol/vol) FCS, 1% nonessential amino acids, 1% l-glutamine, 200 U of penicillin, 50 mg of streptomycin, 0.25 mg of amphotericin B per liter, and 1% minimal essential medium vitamin mix X-100 (Life Technologies).
Assays of adhesion and invasion of Intestine-407 cells and bacterial survival and replication within J774 macrophages.
The assays of bacterial invasion of Intestine-407 epithelial cells and bacterial survival and replication within macrophages were performed with a gentamicin protection assay, as described previously (8, 27). Briefly, monolayers were seeded in 24-well tissue culture plates (Polylabo, Strasbourg, France) with 2 × 105 cells per cm2 for Intestine-407 cells and 1 × 105 cells per cm2 for J774 macrophages and incubated for 20 h. Monolayers were infected in 1 ml of the cell culture medium without antibiotics and with heat-inactivated FCS at a multiplicity of infection of 10 bacteria per cell for adhesion and invasion assays and of 100 bacteria per cell for assays of bacterial uptake, survival, and replication within macrophages.
For assays of adhesion and invasion of Intestine-407 cells, after a 3-h incubation period at 37°C, monolayers were washed five times in phosphate-buffered saline (PBS) (pH 7.2). The epithelial cells were then lysed with 1% Triton X-100 (Sigma Chemical Co., St Louis, MO) in deionized water. Samples were diluted and plated on Mueller-Hinton agar plates to determine the numbers of cell-associated bacteria (adherent and intracellular bacteria). To determine the numbers of intracellular bacteria, fresh cell culture medium containing 100 μg of gentamicin per ml (Sigma) was added for 1 h to kill extracellular bacteria. The monolayers were then lysed with 1% Triton X-100, and the bacteria were quantified as described above. When needed, adhesion and invasion assays were performed after centrifugation for 8 min at 1,000 × g.
For assays of bacterial uptake, survival, and replication within macrophages, infected monolayers were centrifuged at 1,000 × g for 10 min and then incubated for 10 min at 37°C. The monolayers were washed twice in PBS, and the numbers of intracellular bacteria were determined after 40-min and 24-h incubation periods at 37°C in fresh culture medium containing 20 μg of gentamicin per ml.
Construction of the isogenic mutant and transcomplementation assay.
An isogenic mutant deleted of the dsbA gene was generated with a PCR product by the method described by Datsenko and Wanner (20). The basic strategy was to replace a chromosomal sequence with a selectable antibiotic resistance gene (Km) generated by PCR. This PCR product was generated with primers IMdsbA-1 and IMdsbA-2 (Table 2) with 50-nucleotide extensions that are homologous to regions adjacent to the dsbA gene and a template E. coli strain harboring the Km resistance gene. In addition, strain LF82 was transformed with plasmid pKOBEG, a plasmid encoding λ-Red proteins from phage λ, which protect linear DNA in bacteria, expressed under the control of an inducible promoter in the presence of 1 mM l-arabinose. This plasmid was maintained in bacteria at 30°C with 25 μg/ml of Cm.
TABLE 2.
Oligonucleotides and PCR product sizes
| Primer | Oligonucleotide sequence (5′→3′) | PCR product size (bp) | Use |
|---|---|---|---|
| IMdsbA-1 | AGAACCCCCTTTGCAATTAACACCTATGTATTAATCGGAGAGAGTAGATCGTAGGCTGGAGCTGCTTCG | 1,573 | LF82-ΔdsbA isogenic mutant construction |
| IMdsbA-2 | TAATAAAAAAGCCCGTGAATACTCACGGGCTTTATGTAATTTACATTGAACATATGAATATCCTCCTTAGTTC | ||
| DsbA-1 | CTGCCGGAAGGCGTGAAC | 237 | Allelic replacement of dsbA in LF82-ΔdsbA and RT-PCR |
| DsbA-2 | GCTGTTCCACGCCGCGTC | ||
| HindIII-dsbA | CCCAAGCTTGGGGTGAATACTCACGGGCTTTAAG | 706 | Transcomplementation of LF82-ΔdsbA |
| XbaI-dsbA | CCCGGGTCTAGATAATCGGAGAGAGTAGATCATG | ||
| k1 | CAGTCATAGCCGAATAGCCT | Kmr cassette amplification (20) | |
| k2 | CGGTGCCCTGAATGAACTGC | ||
| kt | CGGCCACAGTCGATGAATCC | ||
| FIME | GCAGGCGGTTTCTTACGGGG | 750 | OFF-oriented invertible element |
| INV | GAGGTGATGTGAAATTAATTTAC | ||
| FIMA | GATGCGGTACGAACCTGTCC | 450 | ON-oriented invertible element |
| INV | GAGGTGATGTGAAATTAATTTAC | ||
| htrA-1 | GCGAACTGATCGGTATCAAC | 168 | qRT-PCR |
| htrA-2 | GAGTTCAGCTCAGTCCCCA | ||
| yihE3 | CGCCCTGAACGTTGGACAGC | 208 | qRT-PCR |
| yihE4 | CGCCCAACCGCTTCCATCTG | ||
| yihE3 | CGCCCTGAACGTTGGACAGC | 822 | RT-PCR |
| yihE5 | CTACTCTCTCCGATTAATACATAG | ||
| 16S-1 | ATGACCAGCCACACTGGAAC | 157 | qRT-PCR |
| 16S-2 | CTTCCTCCCCGCTGAAAGTA |
Strain LF82/pKOBEG was grown at 30°C with 1 mM l-arabinose to induce Red expression. When the optical density at 620 nm (OD620) reached 0.5, the bacterial culture was incubated for 10 min at 42°C to stop the replication of the thermosensitive replication plasmid pKOBEG. Bacteria were washed three times with 10% glycerol, and PCR products were electroporated. The isogenic mutant was selected on LB agar containing 50 μg/ml of Km. The replacement of the dsbA gene by the Km resistance cassette in the LF82-ΔdsbA isogenic mutant was confirmed by PCR (Table 2).
A 706-bp PCR product obtained with primers HindIII-dsbA and XbaI-dsbA and containing the dsbA open reading frame was cloned into the pBAD33 vector under the control of an l-arabinose-inducible promoter and designated pPBI11 (Tables 1 and 2). This construct was used for transcomplementation experiments with LF82-ΔdsbA, following culture performed in medium containing 0.2% l-arabinose.
Yeast cell aggregation assay.
Commercial baker's yeast (Saccharomyces cerevisiae) was suspended in PBS (4 mg [dry weight] per ml). Bacterial strains were grown overnight at 37°C without agitation in LB broth, washed, and resuspended in PBS at an OD620 of 0.4. Equal volumes of yeast cell suspension and bacterial suspension were mixed on a glass slide. Aggregation was monitored visually.
Motility assay.
Bacterial strains were grown overnight at 37°C without agitation in LB broth, and 2 μl of the culture was inoculated into the center of 0.3% LB agar plates. The plates were incubated at 37°C for 18 h, and motility was assessed quantitatively by examining the circular swimming motion of the growing, motile bacterial cells.
TEM. (i) Negative staining.
For transmission electron microscopy (TEM), bacteria were grown overnight in LB broth at 37°C without shaking, placed for 1 min on carbon-Formvar copper grids (Electron Microscopy Sciences, Hatfield, England,) and negatively stained for 1 min with phosphotungstic acid pH 6.0.
(ii) Immunolabeling.
Gold immunolabeling was performed by the method of Levine (41). A drop of bacteria grown overnight in LB broth at 37°C without shaking was placed on a carbon-Formvar copper grid. Excess liquid was removed, and the grid was placed face down on a suitable dilution of antiserum raised against type 1 pili for 15 min. After 30 washings in wash solution (PBS plus 1% bovine serum albumin and 1% Tween 20), the grid was placed on a drop of gold-labeled goat anti-rabbit serum (BB International, Cardiff, United Kingdom) for 15 min. After a further thorough washing, the grid was negatively stained with phosphotungstic acid, pH 6.0. Grids were examined with a Hitachi H-7650 TEM.
Confocal microscopy.
J774 macrophages were seeded on glass cover slides in 24-well plates at a density of 0.5 × 105 cells per cm2 and were grown for 18 h in an atmosphere containing 5% CO2 at 37°C. After infection with green fluorescent protein (GFP)-expressing bacteria, as described above, the cells were washed with PBS to eliminate extracellular bacteria and fixed with 3% paraformaldehyde for 10 min. Subsequently, they were washed with PBS, incubated 5 min with 0.1 M glycine, washed with PBS, and permeabilized with 0.1% Triton X-100 for 20 min. After washes with PBS, the slides were incubated twice for 10 min with PBS-0.2% gelatin. The actin cytoskeleton was stained for 15 min with tetramethyl rhodamine isocyanate (TRITC)-labeled phalloidin (Sigma). Monolayers were then washed with PBS and distilled water and mounted on glass slides with a Mowiol solution (Calbiochem, Darmstadt, Germany). Slides were examined with a Zeiss LSM 510 Meta confocal microscope.
Growth ability in acidic and nutrient-poor medium.
Overnight bacterial cultures were harvested, washed, and resuspended in the same volume of acidic and nutrient-poor medium (9) containing 100 mM bis-Tris, 0.1% Casamino Acids, 0.16% glycerol, and 10 μM MgCl2. The pH was adjusted to 5.8 with HCl. Cultures were then diluted 1:50 in the medium, and bacteria were grown at 37°C with shaking. At different times of culture, 50 μl of the culture was diluted in physiologic water and plated onto Mueller-Hinton agar plates to determine the number of CFU per milliliter.
Colony immunoblotting.
The rabbit antiserum raised against purified type 1 pilus preparations was a generous gift from Maryvonne Moulin-Schouleur (INRA-Centre de Tours, UR 86, Pathologie Bactérienne, Nouzilly, France). Bacteria were grown overnight at 37°C in LB broth without shaking. A aliquot of 1 ml of culture at an OD of 0.1 was centrifuged, and the pellet of bacteria was suspended in PBS. A 3-μl sample was spotted onto a nitrocellulose membrane (Amersham International, Buckinghamshire, England), and the membrane was treated as previously described (3).
RNA manipulation, RT, and real-time RT-PCR.
Total RNAs were extracted from bacteria and treated with DNase (Roche Diagnostics, Mannheim, Germany) to remove any contaminating genomic DNA. For yihE transcript analysis, reverse transcription (RT)-PCR was performed with the standard Two Step RT-PCR-GO kit (MP Biomedicals, Illkirch, France). Briefly, cDNA was synthesized with 1 μg of total RNA and random octamers, and PCR was done with 5 μl of cDNA and specific primers to 16S rRNAs and yihE mRNAs (yihE3 and yihE5 [Table 2]). PCR products were visualized after electrophoresis on an agarose gel. For quantification, real-time quantitative RT-PCR (qRT-PCR) was performed with a Light Cycler instrument (Roche Diagnostics), and quantification of the htrA mRNAs, dsbA mRNAs, yihE mRNAs, or 16S rRNA (as a control) was performed with specific primers (Table 2) and RNA master SYBR green I (Roche Diagnostics) with 0.5 μg of total RNA.
Statistical analysis.
Student's t test was used for comparison of the two groups of data. All experiments were repeated at least three times. A P value of less than or equal to 0.05 was considered statistically significant.
RESULTS
The AIEC LF82-ΔdsbA mutant is not able to survive within J774 macrophages.
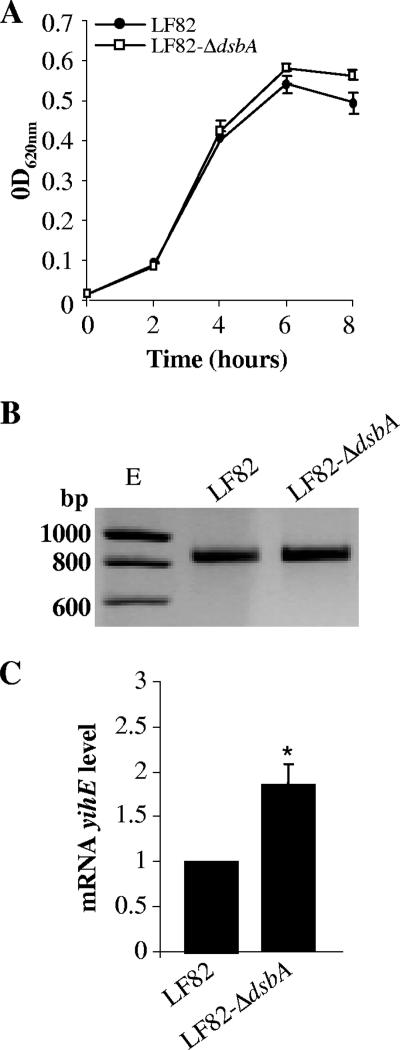
Screening of the Tn5phoA mutant library of the CD-associated AIEC reference strain LF82 indicated that three mutants having an insertion of the transposon into the dsbA gene were significantly attenuated in the ability to resist macrophage killing (9). In order to study the role of the dsbA gene in the virulence of the AIEC strain LF82, an isogenic deletion mutant of the dsbA gene was constructed as described in Materials and Methods. We checked whether deletion of the dsbA gene in strain LF82 could interfere with bacterial growth. The growth curves for the wild-type strain LF82 and the LF82-ΔdsbA mutant in the bacterium-cell incubation medium used for J774 macrophage infection (RPMI supplemented with 10% heat-inactivated FCS) were similar at all time points tested (Fig. 1A), indicating that DsbA was not essential for bacterial growth in rich culture medium.
FIG. 1.
Deletion of the dsbA gene affects neither the growth of bacteria nor yihE expression in the LF82-ΔdsbA mutant. (A) Growth of the wild-type strain LF82 and of LF82-ΔdsbA in RPMI medium supplemented with 10% heat-inactivated FCS. (B) Analysis of yihE mRNAs of LF82 and LF82-ΔdsbA by two-step RT-PCR using primers annealing the 5′and 3′ ends of the yihE cDNA. (C) yihE mRNA level of LF82-ΔdsbA relative to that of the wild-type strain LF82 determined by real-time RT-PCR experiments after growth of the bacteria in LB medium. Only experiments showing the same levels of 16S rRNA for each sample were taken into account.
The dsbA gene can be transcribed alone or together with the upstream gene yihE (5), and the yihE gene can influence global gene expression (42) and is involved in Salmonella enterica serovar Typhimurium and Shigella flexneri virulence (5, 24, 63). In AIEC strain LF82, yihE lies upstream of dsbA, and the upstream sequences of both yihE and dsbA are similar to those of E. coli K-12 strains (data not shown).
We checked that the dsbA knockout in AIEC strain LF82 did not affect yihE expression. The presence of full-length yihE transcripts was observed in LF82-ΔdsbA (Fig. 1B). In contrast to a potential decrease in yihE mRNA transcripts, which could have resulted from dsbA gene deletion, we observed an increase in the yihE mRNA level in LF82-ΔdsbA compared to that of the wild-type strain LF82 (Fig. 1C).
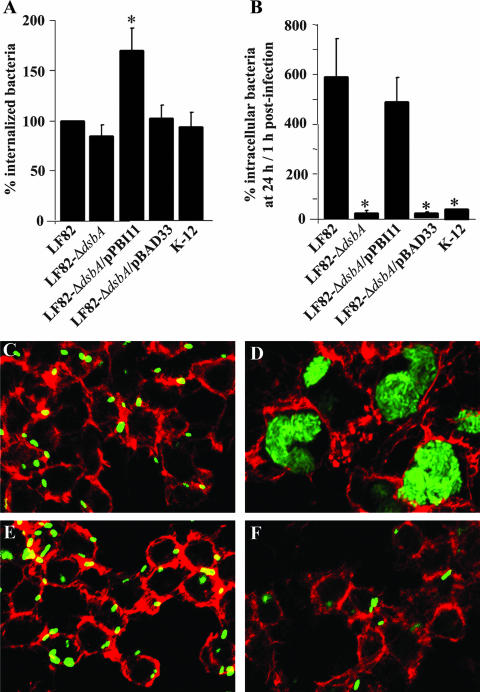
The phenotype of LF82-ΔdsbA was analyzed within J774 macrophages at 1 h and 24 h postinfection. No significant differences in the internalization levels of the wild-type strain and the dsbA null mutant were observed, as the percentage of intracellular bacteria at 1 h postinfection was 81.7% ± 7.8% for LF82-ΔdsbA compared to the wild-type strain LF82, taken as 100% (Fig. 2A). Transcomplementation experiments were performed with the dsbA gene cloned into plasmid vector pBAD33, forming pPBI11. Interestingly, the percentage of intracellular bacteria observed at 1 h postinfection for LF82-ΔdsbA transcomplemented with pPBI11 was significantly increased compared to the wild-type strain LF82, which is suggestive of a dose effect of DsbA due to differences in the number of dsbA gene copies.
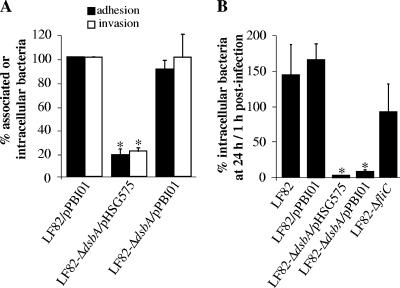
FIG. 2.
The dsbA gene is required for survival of strain LF82 within J774 macrophages. (A) Uptake of the LF82, LF82-ΔdsbA, LF82-ΔdsbA transformed with the cloned dsbA gene (pPBI11), and LF82-ΔdsbA harboring the vector alone (pBAD33). Results are expressed as the number of intracellular bacteria after 1 h of gentamicin treatment relative to that obtained for strain LF82, taken as 100%. (B) Bacterial survival and replication after 24 h of gentamicin treatment. Results are expressed as the number of intracellular bacteria at 24 h relative to that obtained at 1 h after gentamicin exposure, taken as 100%. The nonpathogenic E. coli K-12 strain C600, which is efficiently killed by J774, was used as a negative control. Data are means ± standard errors of the means for four separate experiments. *, P < 0.05. (C to F) Confocal microscopic examinations of J774 macrophages infected with wild-type strain LF82 and LF82-ΔdsbA. Bacteria were transformed with plasmid pFPV25.1 for constitutive expression of GFP. The actin cytoskeleton of J774 cells was stained with TRITC-labeled phalloidin. Shown are J774-A1 macrophages infected with the LF82 wild-type strain (C and D) and with LF82-ΔdsbA (E and F) at 1 h postinfection (C and E) and at 24 h postinfection (D and F).
As a consequence of deletion of the dsbA gene, LF82-ΔdsbA was drastically impaired in the ability to survive within macrophages (Fig. 2B). Percentages of intracellular bacteria at 24 h postinfection compared to those at 1 h postinfection, taken as 100%, were 19.1% ± 5.9% for LF82-ΔdsbA and 593.1% ± 149.6% for the wild-type strain LF82. This indicates that LF82-ΔdsbA was unable to resist the bactericidal activity of J774 macrophages. Its survival ability was as low as that of the nonpathogenic E. coli K-12 strain C600, which is efficiently killed by J774 macrophages. Transcomplementation of LF82-ΔdsbA with the cloned wild-type dsbA gene (plasmid pPBI11) restored intramacrophagic bacterial survival to a level similar to that displayed by the wild-type strain. Transformation of LF82-ΔdsbA with vector pBAD33 alone did not restore the ability of the mutant to survive within phagocytic cells (Fig. 2B). The absence of bacterial survival of intramacrophagic LF82-ΔdsbA at 24 h postinfection was confirmed by confocal microscopic examination of macrophages infected with wild-type strain LF82 and LF82-ΔdsbA harboring plasmid pFPV25.1 for constitutive expression of GFP. At 1 h postinfection, the wild-type strain LF82 and LF82-ΔdsbA showed similar entry into J774 macrophages (Fig. 2C and 2E). At 24 h postinfection, AIEC LF82-infected macrophages exhibited large vacuoles containing numerous bacteria (Fig. 2D) whereas dsbA-negative mutant-infected macrophages showed vacuoles containing only a few bacteria (Fig. 2F). These experiments demonstrate that the dsbA gene is crucial for the survival of AIEC LF82 bacteria within J774 macrophages.
The AIEC LF82-ΔdsbA mutant shows a decreased ability to adhere to Intestine-407 intestinal epithelial cells.
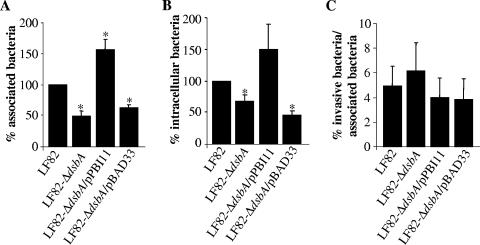
The phenotype of LF82-ΔdsbA was analyzed with intestinal epithelial cells. Quantitative adhesion assays showed that LF82-ΔdsbA was significantly reduced in the ability to adhere to Intestine-407 epithelial cells, having a 49.4% ± 8.2% residual adhesion level compared to wild-type strain LF82, taken as 100% (Fig. 3A). In addition, the LF82-ΔdsbA mutant was also significantly attenuated in the ability to invade Intestine-407 epithelial cells, with a 67.2% ± 9.6% residual invasion level compared to the wild-type strain LF82 (Fig. 3B). However, the percentage of invasive bacteria relative to adherent bacteria for LF82-ΔdsbA was not significantly different compared to that obtained for the wild-type strain LF82 (Fig. 3C), indicating that the decreased ability of LF82-ΔdsbA to invade intestinal epithelial cells is mostly due to a decreased ability to adhere to intestinal epithelial cells. Interestingly, the residual adhesion level of LF82-ΔdsbA transcomplemented with the cloned dsbA gene (pPBI11) was 156.1% ± 16.5% and was significantly increased compared to that of strain LF82, as a possible consequence of an increased number of dsbA gene copies (Fig. 3A). Transcomplementation of LF82-ΔdsbA with the vector pBAD33 alone did not restore the ability of the mutant to adhere to Intestine-407 epithelial cells. Thus, these experiments demonstrate that the dsbA gene plays a role in the ability of AIEC strain LF82 to adhere to intestinal epithelial cells.
FIG. 3.
Phenotype of the LF82-ΔdsbA mutant in Intestine-407 epithelial cells. (A) Adhesion of LF82, LF82-ΔdsbA, LF82-ΔdsbA transformed with the cloned dsbA gene (pPBI11), and LF82-ΔdsbA harboring the vector alone (pBAD33) to Intestine-407 cells. Cell-associated bacteria were quantified after a 3-h infection period. The results are expressed as levels of cell-associated bacteria (adherent plus intracellular) relative to those obtained for wild-type strain LF82, taken as 100%. (B) Bacterial invasion of Intestine-407 cells was determined after gentamicin treatment for an additional 1 h. The results are expressed as levels of intracellular bacteria relative to those obtained for wild-type strain LF82, taken as 100%. (C) Percentage of intracellular bacteria relative to adherent bacteria. Data are means ± standard errors of the means for four separate experiments. *, P < 0.05.
The LF82-ΔdsbA mutant does not express flagella and type 1 pili.
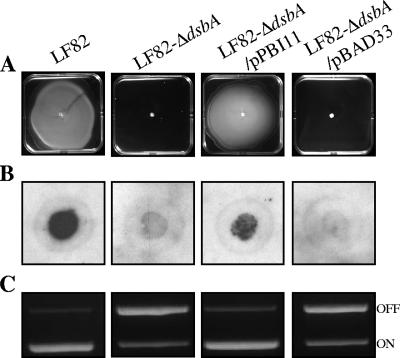
It has been previously reported that, in E. coli, DsbA participates in the posttranslational modification of a protein involved in flagellar synthesis by catalyzing disulfide bond formation in the P-ring protein FlgI (16). In order to investigate the involvement of DsbA in AIEC strain LF82 flagellar synthesis, motility assays were performed on 0.3% agar plates (Fig. 4A). The LF82-ΔdsbA mutant was nonmotile on a soft agar plate, and transcomplementation with the cloned dsbA restored the motility of LF82-ΔdsbA to a level similar to that of wild-type strain LF82. In addition, TEM examination of bacteria indicated that, unlike the wild-type strain LF82 or LF82-ΔdsbA transcomplemented with dsbA, LF82-ΔdsbA did not synthesize flagella (Fig. 5).
FIG. 4.
Lack of expression of type 1 pili and flagella in LF82-ΔdsbA. Experiments were performed with strain LF82, LF82-ΔdsbA, LF82-ΔdsbA transformed with the cloned dsbA gene (pPBI11), and LF82-ΔdsbA harboring the vector alone (pBAD33) (A) Motility was visualized on a 0.3% agar plate as a halo of radial diffusion of bacteria around the primary inoculum after 16 h at 37°C. (B) Colony immunoblotting using polyclonal antibodies raised against purified type 1 pili. (C) Determination by PCR analysis of the invertible element orientation of the fim operon. A 450-bp product revealed the phase-ON orientation, and a 750-bp product revealed the phase-OFF orientation of the invertible element (58).
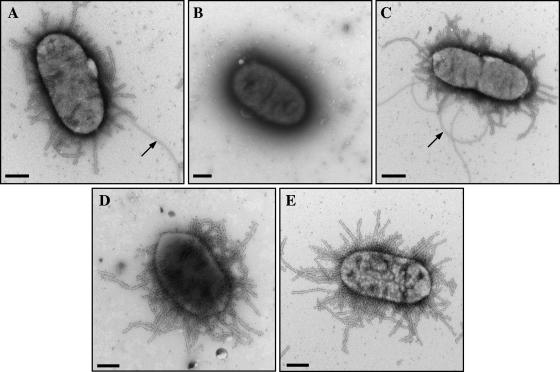
FIG. 5.
TEM of gold immunolabeling of LF82 bateria (A), LF82-ΔdsbA (B), and LF82-ΔdsbA transformed with plasmid pPBI11 harboring cloned dsbA (C), with plasmid pPBI01 harboring the cloned E. coli K-12 fim operon (D), or with cosmid JE7 harboring the cloned LF82 fim operon (E) using polyclonal antibodies raised against purified type 1 pili. Arrows indicate a flagellar structure. Bars, 0.5 μm.
It has also been previously reported that DsbA participates in the posttranslational modification of fimbrial structures, such as bundle-forming pili in enteropathogenic E. coli, Pap pili in uropathogenic E. coli, and toxin-coregulated pili in Vibrio cholerae (70). Moreover, a decreased ability of AIEC strain LF82 to adhere to and invade intestinal epithelial cells has already been observed and is characteristic of type 1 pilus-negative mutants (7). Type 1 pilus expression of LF82-ΔdsbA was assessed by colony immunoblotting using polyclonal antibodies raised against type 1 pili and by determining the ability of the bacteria to aggregate yeast cells via binding to d-mannose residues. As shown in Fig. 4B and Table 3, LF82-ΔdsbA was unable to synthesize type 1 pili and was also unable to aggregate yeast cells. In addition, gold immunolabeling assays using anti-type 1 pilus antibodies indicated an absence of type 1 pili on the bacterial surface of LF82-ΔdsbA in contrast to the wild-type strain LF82 (Fig. 5). Transcomplementation of LF82-ΔdsbA with the clone dsbA gene (pPBI11) fully restored type 1 pilus expression, while transformation with the vector alone (pBAD33) did not restore the wild-type phenotype (Fig. 4B and 5 and Table 3). Altogether, these results demonstrate that LF82-ΔdsbA is impaired in both flagellar and type 1 pilus expression.
TABLE 3.
Expression of type 1 pili as determined by yeast aggregation
| Strain | Yeast aggregation resulta |
|---|---|
| LF82 | +++ |
| LF82-ΔdsbA | − |
| LF82-ΔdsbA/pPBI11 | +++ |
| LF82-ΔdsbA/pBAD33 | − |
| LF82-ΔdsbA/pPBI01 | +++ |
| LF82-ΔdsbA/JE7 | ++ |
Aggregation was monitored visually.
The absence of AIEC LF82 type 1 pilus expression is not due to lack of oxidizing disulfide bonds of Fim proteins in the DsbA null mutant.
To determine whether DsbA could be involved in oxidizing disulfide bonds in one of the Fim proteins required for secretion or assembly of type 1 pili, LF82-ΔdsbA was transformed either with plasmid pPBI01, harboring the K-12 fim operon, or with cosmid JE7, harboring the entire LF82 fim operon, as we previously reported that AIEC strain LF82 produces a variant of type 1 pili compared to those of E. coli K-12 (7). As shown by gold immunolabeling assays using polyclonal antibodies raised against purified type 1 pili and by determination of the ability of the bacteria to aggregate yeast cells, LF82-ΔdsbA/JE7 and LF82-ΔdsbA/pPBI01 expressed functional type 1 pili able to aggregate yeast cells (Fig. 5 and Table 3). These results indicate that functional E. coli K-12 type 1 pili or AIEC LF82 type 1 pilus variants can be synthesized in AIEC strain LF82 in the absence of DsbA.
Type 1 pilus expression is mediated by a process called phase variation, in which the bacteria switch between piliated and nonpiliated states under the control of a switch-invertible element, located upstream of the type 1 pilus-encoding operon. PCR experiments using two sets of primers specific to the phase-ON and phase-OFF orientations of the invertible element clearly demonstrated that the decrease in type 1 pilus expression in LF82-ΔdsbA correlated with a preferential switch of the invertible element to the phase-OFF orientation (Fig. 4C). Transcomplementation of LF82-ΔdsbA with the cloned dsbA gene (pPBI11) restored a switch of the invertible element mostly to the phase-ON orientation. Thus, the absence of type 1 pilus expression in the LF82-ΔdsbA mutant corresponds to phase variation, as was previously reported for the LF82-ΔfliC mutant (3).
The loss of the ability of the LF82-ΔdsbA mutant to adhere to Intestine-407 intestinal epithelial cells is linked only to the absence of type 1 pilus and flagellar expression.
To assess the involvement of type 1 pili and flagella in the decreased ability of the LF82-ΔdsbA mutant to adhere to and invade intestinal epithelial cells, quantitative adhesion and invasion assays were performed with induced type 1 pilus expression and forced contact between bacteria and Intestine-407 epithelial cells performed by a centrifugation step. The adhesion and invasion levels of the LF82-ΔdsbA mutant transformed with pPBI01 were 89.5% ± 7.3% and 98.9% ± 19.1%, respectively, not significantly different (P > 0.05) from those of strain LF82, also transformed with pPBI01 (Fig. 6A). Thus, the decreases in the adhesion and invasion levels of LF82-ΔdsbA were related only to the absence of motility and type 1 pilus expression.
FIG. 6.
Effects of induced type 1 pilus expression and forced contact between bacteria and host cells on LF82-ΔdsbA interactions with intestinal epithelial cells and macrophages. (A) Adhesion and invasion abilities of LF82-ΔdsbA transformed with pPBI01, harboring the entire fim operon, with Intestine-407 epithelial cells. Experiments were performed with an initial centrifugation step to establish close contact between bacteria and epithelial cells to bypass the absence of bacterial motility. (B) Intramacrophagic survival ability of LF82 and LF82-ΔdsbA with or without induced type 1 pilus expression obtained by transformation with pPBI01, harboring the entire fim operon, and LF82-ΔfliC. See the legends to Fig. 2 and 3. Data are means ± standard errors of the means for four separate experiments. *, P < 0.05.
The involvement of DsbA in intramacrophagic survival of LF82 bacteria is independent of type 1 pilus and flagellar expression.
As the decreased ability of the DsbA-negative mutant to survive within macrophages could be related to the lack of type 1 pilus expression, we performed experiments with the dsbA null mutant transformed with the cloned fim operon (pPBI01). The survival ability of the transformed mutant was compared with that of strain LF82, also transformed with pPBI01 (Fig. 6B). The percentages of intracellular bacteria at 24 h postinfection compared to those at 1 h postinfection, taken as 100%, were 8.9% ± 1.8% for LF82-ΔdsbA/pPBI01 and 160.2% ± 22.3% for LF82/pPBI01, indicating that the encoding fim operon plasmid pPBI01 did not rescue the ability of LF82-ΔdsbA to survive within macrophages. This was not due to a growth defect dependent on the presence of the plasmid pPBI01, since transformation of strain LF82 with pPBI01 did not modify its ability to grow in LB broth or cell culture medium (data not shown) or to survive and replicate within macrophages (Fig. 6B). Altogether, these results indicate that in the DsbA null mutant, in addition to the lack of type 1 pilus expression essential for adhesion to and invasion of intestinal epithelial cells, another virulence factor(s) that needs to be posttranslationally modified by DsbA and that is essential for the survival of LF82 bacteria within macrophages must be identified.
We reported previously that flagella play a role in the virulence of AIEC strain LF82 that is not only restricted to motility but related to coordinate expression of invasive determinant (3). In order to test whether the decreased survival ability of LF82-ΔdsbA could result from the down-regulation of virulence gene expression due to a lack of flagella, the intracellular behavior of the LF82-ΔfliC mutant was analyzed in macrophages (Fig. 6B). No significant differences in the percentages of intracellular bacteria at 24 h postinfection compared to those at 1 h postinfection were observed between the wild-type strain LF82 and the LF82-ΔfliC mutant. Thus, this result demonstrates that the lack of flagella cannot account for the inability of the DsbA-negative mutant to survive within macrophages.
DsbA is required for the growth and survival of AIEC LF82 bacteria in acidic and nutrient-poor medium.
A previous study has shown that intramacrophagic AIEC LF82 bacteria reside in an acidic phagolysosomal compartment (10). A low-pH (pH 5.8) and nutrient-poor medium containing glycerol and Casamino Acids was used to partly mimic the phagocytic vacuole content (9). The growth of LF82-ΔdsbA was compared with that of the wild-type strain LF82 under these in vitro stress conditions. Growth rates were measured by determining the number of CFU per ml over a 24-h period (Fig. 7A). After 3 h of growth, the number of CFU per milliliter measured for LF82-ΔdsbA was significantly lower (P < 0.05) than that of the wild-type strain, with 12.1 × 107 ± 0.7 × 107 CFU per ml for LF82-ΔdsbA and 18.8 × 107 ± 1.0 × 107 CFU per ml for the wild-type strain LF82. After 6 h of incubation, the number of CFU per milliliter determined for LF82-ΔdsbA was 3.3-fold greater than that obtained after 3 h of incubation, indicating that this mutant was able to replicate. However, its growth was still significantly lower (P < 0.05) than that of the wild-type strain. From 6 h to 24 h of growth, the wild-type strain continued to replicate efficiently in acidic and nutrient-poor medium, but, interestingly, lysis of the LF82-ΔdsbA bacteria was observed. There was a significant 2.3-fold decrease (P < 0.05) in the number of CFU per milliliter of LF82-ΔdsbA after 24 h of growth compared to that observed for this mutant after 6 h of growth. Transcomplementation of LF82-ΔdsbA with plasmid pPBI11 harboring a cloned dsbA gene restored bacterial growth similar to that of the wild-type strain. These results show that the dsbA gene is required for the growth and survival ability of strain LF82 in an acidic and nutrient-poor medium.
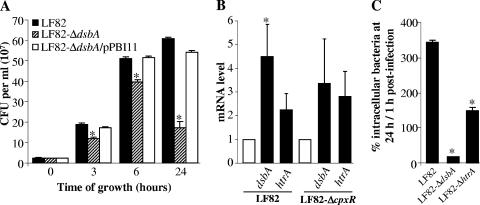
FIG. 7.
Involvement of DsbA in bacterial growth and dsbA gene expression in AIEC strain LF82 under in vitro phagocytic vacuole stress conditions. (A) Growth of the wild-type strain LF82, LF82-ΔdsbA, and LF82-ΔdsbA transcomplemented with plasmid pPBI11, harboring the dsbA gene, in acidic and nutrient-poor medium. (B) dsbA and htrA mRNA levels of the wild-type strain LF82 and the LF82-ΔcpxR mutant determined by RT-PCR experiments after growth of the bacteria in acidic and nutrient-poor medium relative to that of bacteria grown in LB broth. Only experiments showing the same levels of 16S rRNA for each sample were taken into account. Data are means ± standard errors of the means for three separate experiments. *, P < 0.05. (C) Intracellular survival ability of LF82, LF82-ΔdsbA, and LF82-ΔhtrA within macrophages. See the legend to Fig. 2 for experiment setup details. Data are means ± standard errors of the means for three separate experiments. *, P < 0.05.
dsbA gene expression is up-regulated in AIEC LF82 bacteria grown under in vitro acidic and nutrient-poor stress conditions.
To study the transcriptional activation of the dsbA gene under in vitro conditions that partly mimic the phagocytic vacuole content, dsbA mRNA levels were measured by real-time PCR in strain LF82 after growth of bacteria in acidic and nutrient-poor medium. The level of dsbA mRNA increased 4.5 ± 1.4-fold when LF82 bacteria were grown in acidic and nutrient-poor medium relative to growth in LB medium (Fig. 7B). Hence, transcription of the dsbA gene was up-regulated in strain LF82 when bacteria grew in vitro under stress conditions that partly mimicked the stress encountered within the phagocytic vacuole. We reported previously that LF82 htrA transcription is activated under such stress conditions (9). Compared to the level of htrA mRNAs, that of dsbA mRNAs was even higher in LF82 bacteria grown under phagocyte stress conditions. It is noteworthy that the behaviors of LF82-ΔdsbA and LF82-ΔhtrA mutants are different within macrophages. The htrA mutant, unlike the dsbA mutant, is able to survive macrophage killing (Fig. 7C).
In Escherichia coli K-12, the two-component regulatory system CpxRA controls transcription of the dsbA gene through a CpxR binding site located in the yihE promoter region (17, 51, 52). To analyze the involvement of this regulatory system, dsbA mRNA levels were measured in the LF82-ΔcpxR mutant by real-time PCR after growth of the bacteria in acidic and nutrient-poor medium. The levels of dsbA mRNAs were 3.4 ± 1.9-fold greater in the LF82-ΔcpxR mutant grown in acidic and nutrient-poor medium than in bacteria grown in LB broth, and they were not significantly different from those measured in the wild-type strain LF82 (Fig. 7B). We also observed that the increased level of dsbA mRNAs in a CpxR null mutant was of a magnitude similar to that of htrA mRNAs. Thus, these results indicate that the two-component regulatory system CpxRA does not play a role in htrA and dsbA expression in AIEC strain LF82 under conditions that partly mimic the content of the phagocytic vacuole; this finding is not surprising, since it has been reported that acidic pH turns off the CpxRA system (22).
DISCUSSION
The ileal mucosa of patients with CD is abnormally colonized by pathogenic AIEC strains, which have the ability to survive and extensively replicate into active phagolysosomes within macrophages (10, 27). Screening of the Tn5phoA mutant library of the CD-associated AIEC reference strain LF82 indicated that three mutants having an insertion of the transposon into the dsbA gene were significantly attenuated in the ability to resist macrophage killing (9). The activity of DsbA has already been reported to be necessary for many virulence phenotypes, including colonization, adhesion, invasion, and/or intracellular survival of various pathogens, such as Salmonella enterica (44), E. coli K1 (29), enteropathogenic E. coli (73), Proteus mirabilis (12), and Shigella flexneri (67, 69, 71). To confirm the role of DsbA in the virulence of AIEC strain LF82, a mutant with the dsbA gene deleted was constructed. The LF82-ΔdsbA mutant was unable to survive within macrophages. In addition to its attenuated phenotype within macrophages, LF82-ΔdsbA was affected in the ability to adhere to and subsequently to invade intestinal epithelial cells, demonstrating that DsbA is essential for multiple virulence functions in AIEC strain LF82. In addition, we observed that the level of yihE transcripts in the LF82-ΔdsbA mutant was significantly higher than in the wild-type strain LF82. In E. coli, yihE lies upstream of dsbA and is transcribed together with the latter (5). YihE is highly expressed upon activation of the Cpx stress response pathway as a result of the binding of phosphorylated CpxR to a conserved DNA binding motif (21) in the yihE promoter region (51). In the absence of the DsbA oxidoreductase, unfolded proteins can accumulate in the periplasm and induce the CpxR envelope stress response (52). Thus, we speculate that the increase in yihE transcripts is a result of activation of the Cpx stress response.
The LF82-ΔdsbA mutant does not express flagella. A defect in flagellar assembly was expected, as DsbA is involved in disulfide bond formation in the P-ring protein FlgI in E. coli K-12 (16). In addition, the dsbA-negative AIEC LF82 mutant does not synthesize type 1 pili, which suggests that in AIEC strains the posttranslational oxidation of disulfide bonds of these surface structures could be carried out by DsbA. Indeed, DsbA has been reported to contribute to the posttranslational oxidation of disulfide bonds of various fimbrial structures, such as Pap pili in uropathogenic E. coli (35), bundle-forming pili in enteropathogenic E. coli (23), and toxin-coregulated pili in Vibrio cholerae (62). Interestingly, transformation with a cloned K-12 fim operon or LF82 fim operon allows LF82-ΔdsbA to express functional type 1 pili, suggesting that DsbA does not exert a direct action on type 1 pilus subunit posttranslational modification. The decrease in type 1 pilus expression in LF82-ΔdsbA is correlated with a preferential switch of the invertible element to the phase-OFF orientation. As it has been previously reported that the absence of flagella induces a down-regulation of type 1 pilus expression in strain LF82, resulting from a preferential switch toward the phase-OFF position of the invertible DNA element located upstream of the fim operon (3), the absence of piliation of the DsbA-negative LF82 mutant could result from the absence of flagella observed in this mutant.
Since an LF82 dsbA null mutant does not express type 1 pili and flagella, we analyzed their respective roles in the decreased adhesion and invasion of LF82-ΔdsbA. Transformation of LF82-ΔdsbA with a cloned fim operon to induce expression of type 1 pili and forced contact between bacteria and cells restore its abilities to adhere to and invade intestinal epithelial cells. These results indicate that the role of DsbA in AIEC LF82 adhesion and subsequent invasion is restricted to its action on flagellar expression and, owing to the lack of flagella, on type 1 pilus expression. DsbA is therefore not involved in posttranslational oxidation of disulfide bonds of any virulence factors, including those so far documented as playing a role in the ability of AIEC strain LF82 to adhere to and invade intestinal epithelial cells (3, 4, 55).
LF82 bacteria require DsbA to resist macrophage killing. The survival level of LF82-ΔdsbA is not different from that of a nonpathogenic E. coli K-12 strain, which is efficiently killed by macrophages, and transcomplementation of the LF82-ΔdsbA mutant with the cloned dsbA gene fully restored the survival ability of this mutant within macrophages. The inability of intracellular LF82-ΔdsbA bacteria to survive within macrophages could be linked to the absence of functional HtrA protease, since it has been recently reported that the lack of a disulfide bond in the HtrA periplasmic protease results in an unfolded and unstable HtrA protein (60). However, the phenotypes of LF82-ΔdsbA and LF82-ΔhtrA are not similar, since, in the absence of HtrA, intramacrophagic bacteria are not able to replicate but are able to survive (9) and, in the absence of DsbA, the intramacrophagic bacteria cannot survive. Moreover, the absence of type 1 pilus or flagellar expression cannot explain the loss of resistance to macrophage killing. Indeed, we show in the present study that the LF82-ΔfliC mutant was not impaired in the ability to survive and replicate within macrophages. We also observed that a type 1 pilus-negative LF82 derivative mutant showed a replication rate of intramacrophagic bacteria similar to that of the wild-type strain (M. A. Bringer, A. L. Glasser, and A. Darfeuille-Michaud, submitted for publication).
We previously reported that LF82 bacteria survive and replicate into active phagolysosomes (10). Within phagolysosomes, LF82 bacteria are exposed to drastic conditions, including low pH. Our findings showed that LF82-ΔdsbA was unable to grow as efficiently as the wild-type strain LF82 in an acidic and nutrient-poor medium partly mimicking the stress encountered within the phagocytic vacuole. This result is in accordance with previous data showing that an E. coli K-12 dsbA mutant exhibits growth defects on minimal medium (2). In addition, the number of viable bacteria for the LF82-ΔdsbA mutant decreased after 24 h of culture under such stress conditions. Thus, the DsbA oxidoreductase is essential for AIEC bacteria to resist killing in the hostile environment of phagocytic vacuoles. Interestingly, transcription of the dsbA gene was highly up-regulated in strain LF82 when bacteria grew in vitro in acidic and nutrient-poor medium, conditions that partly mimicked stresses encountered within the phagocytic vacuole, compared to bacteria grown in rich medium. This suggests that DsbA is specifically activated under phagolysosomal conditions to allow AIEC bacteria to survive under these harsh environmental conditions. This up-regulation of dsbA gene expression could be due to nutrient-poor growth conditions and/or to acidic growth conditions. It has been previously reported that dsbA transcription is induced under minimal medium growth conditions and under basic growth conditions (28) in Salmonella enterica serovar Typhimurium, an intracellular pathogen that is able to survive and multiply within macrophages, and that in E. coli K-12 the level of DsbA is increased at basic pH (68). Thus, we hypothesize that up-regulation of dsbA in strain LF82 is mainly due to nutrient starvation rather than to low pH. In addition, our study shows that there was no difference in bacterial growth between the wild-type LF82 strain and LF82-ΔdsbA when cultures were performed in rich medium, such as RPMI supplemented with FCS. A possible explanation is that in the absence of the DsbA oxidoreductase, unfolded proteins can accumulate in the periplasm, inducing cell death. Indeed, in DsbA-negative bacteria, the HtrA protease is not folded and therefore cannot fulfill its function in the removal of irreversibly damaged or abnormal proteins from cellular envelope (15). A second hypothesis, which does not exclude the latter, is that DsbA contributes in AIEC strain LF82 toward posttranslationally modifying specific virulence determinants involved in the survival of intramacrophagic bacteria.
In summary, DsbA plays a central role in CD-associated E. coli pathogenesis, as deletion of dsbA in AIEC strain LF82 results in pleiotropic effects on adhesion to and subsequent invasion of intestinal epithelial cells, via the loss of flagellar and type 1 pilus expression, and as well as on the ability of AIEC bacteria to survive within macrophages. In addition, we observed that the expression of htrA (9) and dsbA is up-regulated under the acidic and nutrient-poor stress conditions encountered by intramacrophagic bacteria. This coordinated regulation of dsbA and htrA genes is of great benefit for AIEC bacteria, as HtrA, which is essential for the intramacrophagic replication of AIEC bacteria (9), needs to be posttranslationally modified by DsbA. We are currently investigating the presence of other specific virulence factors posttranslationally modified by DsbA and/or a specific regulator(s) playing a central role in the orchestration of the intramacrophagic survival/replication of AIEC bacteria, a pathogenic trait essential for the release of large amounts of tumor necrosis factor alpha.
Acknowledgments
This study was supported by grants from the Ministère de l'Education Nationale, de la Recherche et de la Technologie (EA 3844) and from INRA (USC 2018) and by grants from the Association F. Aupetit (AFA) and Institut de Recherche des Maladies de l'Appareil Digestif (IRMAD, Laboratoire Astra France).
We thank Maryvonne Moulin-Schouleur (INRA, Nouzilly, France) for antibodies raised against type 1 pili. The confocal microscope was the property of IFR Santé Auvergne. We thank Christelle Drégnaux and Claire Szczepaniak (CICS, Université d'Auvergne, France) for technical assistance with TEM. The technical assistance of Benoit Chassaing is gratefully acknowledged.
Footnotes
Published ahead of print on 20 April 2007.
REFERENCES
- 1.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, J. C., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 3.Barnich, N., J. Boudeau, L. Claret, and A. Darfeuille-Michaud. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781-794. [DOI] [PubMed] [Google Scholar]
- 4.Barnich, N., M. A. Bringer, L. Claret, and A. Darfeuille-Michaud. 2004. Involvement of lipoprotein NlpI in the virulence of adherent invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect. Immun. 72:2484-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belin, P., and P. L. Boquet. 1994. The Escherichia coli dsbA gene is partly transcribed from the promoter of a weakly expressed upstream gene. Microbiology 140:3337-3348. [DOI] [PubMed] [Google Scholar]
- 6.Bessette, P. H., J. J. Cotto, H. F. Gilbert, and G. Georgiou. 1999. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem. 274:7784-7792. [DOI] [PubMed] [Google Scholar]
- 7.Boudeau, J., N. Barnich, and A. Darfeuille-Michaud. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272-1284. [DOI] [PubMed] [Google Scholar]
- 8.Boudeau, J., A. L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bringer, M. A., N. Barnich, A. L. Glasser, O. Bardot, and A. Darfeuille-Michaud. 2005. HtrA stress protein is involved in intramacrophagic replication of adherent and invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect. Immun. 73:712-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bringer, M. A., A. L. Glasser, C. H. Tung, S. Meresse, and A. Darfeuille-Michaud. 2006. The Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell. Microbiol. 8:471-484. [DOI] [PubMed] [Google Scholar]
- 11.Bruggemann, H., C. Cazalet, and C. Buchrieser. 2006. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr. Opin. Microbiol. 9:86-94. [DOI] [PubMed] [Google Scholar]
- 12.Burall, L. S., J. M. Harro, X. Li, C. V. Lockatell, S. D. Himpsl, J. R. Hebel, D. E. Johnson, and H. L. Mobley. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72:2922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Celli, J. 2006. Surviving inside a macrophage: the many ways of Brucella. Res. Microbiol. 157:93-98. [DOI] [PubMed] [Google Scholar]
- 14.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 16.Dailey, F. E., and H. C. Berg. 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 18.Darfeuille-Michaud, A., J. Boudeau, P. Bulois, C. Neut, A. L. Glasser, N. Barnich, M. A. Bringer, A. Swidsinski, L. Beaugerie, and J. F. Colombel. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127:412-421. [DOI] [PubMed] [Google Scholar]
- 19.Darfeuille-Michaud, A., C. Neut, N. Barnich, E. Lederman, P. Di Martino, P. Desreumaux, L. Gambiez, B. Joly, A. Cortot, and J. F. Colombel. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405-1413. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 22.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnenberg, M. S., H. Z. Zhang, and K. D. Stone. 1997. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability—a review. Gene 192:33-38. [DOI] [PubMed] [Google Scholar]
- 24.Edwards-Jones, B., P. R. Langford, J. S. Kroll, and J. Yu. 2004. The role of the Shigella flexneri yihE gene in LPS synthesis and virulence. Microbiology 150:1079-1084. [DOI] [PubMed] [Google Scholar]
- 25.Elson, C. O. 2000. Commensal bacteria as targets in Crohn's disease. Gastroenterology 119:254-257. [DOI] [PubMed] [Google Scholar]
- 26.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 27.Glasser, A. L., J. Boudeau, N. Barnich, M. H. Perruchot, J. F. Colombel, and A. Darfeuille-Michaud. 2001. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 69:5529-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goecke, M., C. Gallant, P. Suntharalingam, and N. L. Martin. 2002. Salmonella typhimurium DsbA is growth-phase regulated. FEMS Microbiol. Lett. 206:229-234. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez, M. D., C. A. Lichtensteiger, and E. R. Vimr. 2001. Adaptation of signature-tagged mutagenesis to Escherichia coli K1 and the infant-rat model of invasive disease. FEMS Microbiol. Lett. 198:125-128. [DOI] [PubMed] [Google Scholar]
- 30.Guilhot, C., G. Jander, N. L. Martin, and J. Beckwith. 1995. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc. Natl. Acad. Sci. USA 92:9895-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha, U. H., Y. Wang, and S. Jin. 2003. DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 71:1590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu, S. H., J. A. Peek, E. Rattigan, R. K. Taylor, and J. L. Martin. 1997. Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae. J. Mol. Biol. 268:137-146. [DOI] [PubMed] [Google Scholar]
- 34.Jackson, M. W., and G. V. Plano. 1999. DsbA is required for stable expression of outer membrane protein YscC and for efficient Yop secretion in Yersinia pestis. J. Bacteriol. 181:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob-Dubuisson, F., J. Pinkner, Z. Xu, R. Striker, A. Padmanhaban, and S. J. Hultgren. 1994. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc. Natl. Acad. Sci. USA 91:11552-11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111-135. [DOI] [PubMed] [Google Scholar]
- 37.Katzen, F., and J. Beckwith. 2000. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769-779. [DOI] [PubMed] [Google Scholar]
- 38.Knodler, L. A., and O. Steele-Mortimer. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587-599. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi, T., S. Kishigami, M. Sone, H. Inokuchi, T. Mogi, and K. Ito. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 94:11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucharzik, T., C. Maaser, A. Lugering, M. Kagnoff, L. Mayer, S. Targan, and W. Domschke. 2006. Recent understanding of IBD pathogenesis: implications for future therapies. Inflamm. Bowel Dis. 12:1068-1083. [DOI] [PubMed] [Google Scholar]
- 41.Levine, M. M., P. Ristaino, G. Marley, C. Smyth, S. Knutton, E. Boedeker, R. Black, C. Young, M. L. Clements, C. Cheney, et al. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, M. S., J. S. Kroll, and J. Yu. 2001. Influence of the yihE gene of Shigella flexneri on global gene expression: on analysis using DNA arrays. Biochem. Biophys. Res. Commun. 288:91-100. [DOI] [PubMed] [Google Scholar]
- 43.Martin, H. M., B. J. Campbell, C. A. Hart, C. Mpofu, M. Nayar, R. Singh, H. Englyst, H. F. Williams, and J. M. Rhodes. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127:80-93. [DOI] [PubMed] [Google Scholar]
- 44.Miki, T., N. Okada, and H. Danbara. 2004. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J. Biol. Chem. 279:34631-34642. [DOI] [PubMed] [Google Scholar]
- 45.Missiakas, D., and S. Raina. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Missiakas, D., F. Schwager, and S. Raina. 1995. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 14:3415-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mow, W. S., E. A. Vasiliauskas, Y. C. Lin, P. R. Fleshner, K. A. Papadakis, K. D. Taylor, C. J. Landers, M. T. Abreu-Martin, J. I. Rotter, H. Yang, and S. R. Targan. 2004. Association of antibody responses to microbial antigens and complications of small bowel Crohn's disease. Gastroenterology 126:414-424. [DOI] [PubMed] [Google Scholar]
- 48.Okamoto, K., H. Yamanaka, M. Takeji, and Y. Fuji. 2001. Region of heat-stable enterotoxin II of Escherichia coli involved in translocation across the outer membrane. Microbiol. Immunol. 45:349-355. [DOI] [PubMed] [Google Scholar]
- 49.Peek, J. A., and R. K. Taylor. 1992. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 89:6210-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podolsky, D. K. 2002. Inflammatory bowel disease. N. Engl. J. Med. 347:417-429. [DOI] [PubMed] [Google Scholar]
- 51.Pogliano, J., A. S. Lynch, D. Belin, E. C. Lin, and J. Beckwith. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11:1169-1182. [DOI] [PubMed] [Google Scholar]
- 52.Raivio, T. L. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 53.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rietsch, A., P. Bessette, G. Georgiou, and J. Beckwith. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rolhion, N., N. Barnich, L. Claret, and A. Darfeuille-Michaud. 2005. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 187:2286-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryan, P., R. G. Kelly, G. Lee, J. K. Collins, G. C. O'Sullivan, J. O'Connell, and F. Shanahan. 2004. Bacterial DNA within granulomas of patients with Crohn's disease—detection by laser capture microdissection and PCR. Am. J. Gastroenterol. 99:1539-1543. [DOI] [PubMed] [Google Scholar]
- 57.Sartor, R. B., H. C. Rath, S. N. Lichtman, and E. A. van Tol. 1996. Animal models of intestinal and joint inflammation. Baillière's Clin. Rheumatol. 10:55-76. [DOI] [PubMed] [Google Scholar]
- 58.Schwan, W. R., H. S. Seifert, and J. L. Duncan. 1992. Growth conditions mediate differential transcription of fim genes involved in phase variation of type 1 pili. J. Bacteriol. 174:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shanahan, F. 2002. Crohn's disease. Lancet 359:62-69. [DOI] [PubMed] [Google Scholar]
- 60.Skorko-Glonek, J., A. Sobiecka-Szkatula, and B. Lipinska. 2006. Characterization of disulfide exchange between DsbA and HtrA proteins from Escherichia coli. Acta Biochim. Pol. 53:585-589. [PubMed] [Google Scholar]
- 61.Stenson, T. H., and A. A. Weiss. 2002. DsbA and DsbC are required for secretion of pertussis toxin by Bordetella pertussis. Infect. Immun. 70:2297-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun, D., M. Lafferty, J. Peek, and R. Taylor. 1997. Domains within the Vibrio cholerae toxin coregulated pilin subunit that mediate bacterial colonization. Gene 192:79-85. [DOI] [PubMed] [Google Scholar]
- 63.Suntharalingam, P., H. Spencer, C. V. Gallant, and N. L. Martin. 2003. Salmonella enterica serovar Typhimurium rdoA is growth phase regulated and involved in relaying Cpx-induced signals. J. Bacteriol. 185:432-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swidsinski, A., A. Ladhoff, A. Pernthaler, S. Swidsinski, V. Loening-Baucke, M. Ortner, J. Weber, U. Hoffmann, S. Schreiber, M. Dietel, and H. Lochs. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44-54. [DOI] [PubMed] [Google Scholar]
- 65.Tinsley, C. R., R. Voulhoux, J. L. Beretti, J. Tommassen, and X. Nassif. 2004. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J. Biol. Chem. 279:27078-27087. [DOI] [PubMed] [Google Scholar]
- 66.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 67.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc. Natl. Acad. Sci. USA 92:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yohannes, E., D. M. Barnhart, and J. L. Slonczewski. 2004. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. J. Bacteriol. 186:192-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, J., B. Edwards-Jones, O. Neyrolles, and J. S. Kroll. 2000. Key role for DsbA in cell-to-cell spread of Shigella flexneri, permitting secretion of Ipa proteins into interepithelial protrusions. Infect. Immun. 68:6449-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu, J., and J. S. Kroll. 1999. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 1:1221-1228. [DOI] [PubMed] [Google Scholar]
- 71.Yu, J., E. E. Oragui, A. Stephens, J. S. Kroll, and M. M. Venkatesan. 2001. Inactivation of DsbA alters the behaviour of Shigella flexneri towards murine and human-derived macrophage-like cells. FEMS Microbiol. Lett. 204:81-88. [DOI] [PubMed] [Google Scholar]
- 72.Yu, J., H. Webb, and T. R. Hirst. 1992. A homologue of the Escherichia coli DsbA protein involved in disulphide bond formation is required for enterotoxin biogenesis in Vibrio cholerae. Mol. Microbiol. 6:1949-1958. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, H. Z., and M. S. Donnenberg. 1996. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 21:787-797. [DOI] [PubMed] [Google Scholar]
- 74.Zumla, A., and D. G. James. 1996. Granulomatous infections: etiology and classification. Clin. Infect. Dis. 23:146-158. [DOI] [PubMed] [Google Scholar]