Abstract
Shewanella oneidensis MR-1 is purported to express outer membrane cytochromes (e.g., MtrC and OmcA) that transfer electrons directly to Fe(III) in a mineral during anaerobic respiration. A prerequisite for this type of reaction would be the formation of a stable bond between a cytochrome and an iron oxide surface. Atomic force microscopy (AFM) was used to detect whether a specific bond forms between a hematite (Fe2O3) thin film, created with oxygen plasma-assisted molecular beam epitaxy, and recombinant MtrC or OmcA molecules coupled to gold substrates. Force spectra displayed a unique force signature indicative of a specific bond between each cytochrome and the hematite surface. The strength of the OmcA-hematite bond was approximately twice that of the MtrC-hematite bond, but direct binding to hematite was twice as favorable for MtrC. Reversible folding/unfolding reactions were observed for mechanically denatured MtrC molecules bound to hematite. The force measurements for the hematite-cytochrome pairs were compared to spectra collected for an iron oxide and S. oneidensis under anaerobic conditions. There is a strong correlation between the whole-cell and pure-protein force spectra, suggesting that the unique binding attributes of each cytochrome complement one another and allow both MtrC and OmcA to play a prominent role in the transfer of electrons to Fe(III) in minerals. Finally, by comparing the magnitudes of binding force for the whole-cell versus pure-protein data, we were able to estimate that a single bacterium of S. oneidensis (2 by 0.5 μm) expresses ∼104 cytochromes on its outer surface.
Dissimilatory-iron-reducing microorganisms, like Shewanella oneidensis MR-1, are able to generate energy by electron transport-coupled reduction of Fe(III)-bearing minerals. In contrast to soluble electron acceptors (e.g., oxygen), which can diffuse into the microbial cell and be reduced at the inner cytoplasmic membrane, Fe(III) exists predominantly as an insoluble solid phase (e.g., ferrihydrite, goethite, and hematite) in the environment. Therefore, metal-reducing bacteria must shuttle electrons from the internal cytoplasmic membrane to iron located outside the bacterium's cell wall. One hypothesis is that S. oneidensis MR-1 uses a sophisticated protein network to shuttle electrons from the cytoplasmic membrane across the periplasmic space and through the outer membrane to the extracellular surface of the bacterium, where a putative iron reductase catalyzes terminal electron transfer directly to Fe(III) in the structure of a mineral (1, 27, 28, 32, 35, 42).
The terminal iron reductase is believed to be composed of outer membrane, decaheme c-type cytochromes. In particular, two outer membrane c-type cytochromes, MtrC (locus tag SO1778; also known as OmcB) and OmcA (locus tag SO1779), have been shown to play an important role in the reduction of metals like Fe(III) (3, 35-37). Previous studies have shown that the disruption of mtrC or omcA has no effect on the ability of S. oneidensis MR-1 to reduce soluble electron acceptors, including oxygen, nitrate, nitrite, and anthraquinone-2,6-disulfonate (3, 36, 38). However, mutants defective in either mtrC or omcA exhibit reduced rates of solid MnO2 reduction and mutants defective in mtrC also show a diminished capacity to reduce Fe(III) (3, 36, 38). Moreover, it was recently shown that both MtrC and OmcA can independently reduce Fe(III) chelated to nitrilotriacetic acid in vitro (45).
While these studies demonstrate MtrC's and OmcA's involvement in Fe(III) reduction, it is still unclear whether either cytochrome can transfer an electron to a Fe(III) oxide through direct physical contact. A prerequisite for this type of reaction would be the formation of a stable bond between the cytochrome and an iron oxide surface that would presumably facilitate electronic coupling between the cytochrome and the iron oxide. Evidence supporting this idea has been provided by studies examining the binding forces between living cells of S. oneidensis MR-1 and solid iron oxides (29, 32). These studies suggest that S. oneidensis MR-1 actively synthesizes and/or localizes proteins (including MtrC and OmcA) at the mineral interface to function in iron oxide binding and/or reduction.
The present work was undertaken to examine whether it was plausible that S. oneidensis MR-1 uses the outer membrane cytochromes MtrC and OmcA for direct cytochrome-mediated Fe(III) binding. Atomic force microscopy (AFM) was used to probe forces between the iron oxide mineral hematite (Fe2O3) and purified recombinant MtrC and OmcA from S. oneidensis MR-1. The approach force measurements were interpreted with a model describing the steric force between hematite and a cytochrome-coated substrate. The retraction curves, or force “spectra,” were compared to a model that predicts the theoretical force-extension relationship for a cytochrome molecule that forms a specific bridging bond with the surface of hematite.
We found that both MtrC and OmcA are able to form a specific bond with hematite. The force of this attractive interaction is nearly twice as large for the OmcA-hematite bond as for the MtrC-hematite bond. Conversely, the frequency with which OmcA forms a bond with hematite is only one-half that for the MtrC-hematite bond. Furthermore, the bond between MtrC and hematite appears to be more resilient, as we were able to observe numerous folding/unfolding reactions for MtrC molecules that formed a bridging bond with the hematite-coated tip. These observations reveal that each cytochrome has unique binding attributes which would presumably allow each to serve a unique role in a putative, terminal Fe(III) reductase.
Finally, the force spectra for the cytochrome-hematite bonds were compared to previously published spectra collected for a living S. oneidensis MR-1 bacterium and an iron oxide surface (29, 32). The force-distance profiles for the pure proteins were remarkably similar to those for the living bacterium, supporting the hypothesis that S. oneidensis MR-1 targets specific c-type cytochromes to the cell surface, where these proteins make direct contact with the Fe(III) oxide. By comparing the magnitudes of forces between the whole-cell and pure-protein spectra, we were able to obtain a rough estimate of the number of MtrC and/or OmcA molecules on a single bacterium of S. oneidensis MR-1.
MATERIALS AND METHODS
Purification and preparation of recombinant MtrC and OmcA.
The recombinant cytochrome proteins were expressed in S. oneidensis MR-1 and purified as described previously (44, 45, 51). Each contained a Cys4/V5/His6 tag at its carboxy terminus to facilitate protein purification and covalent attachment to gold-coated coverslips via cysteine residues (45, 51). Purified proteins were stored at −20°C in a buffer solution containing 20 mM HEPES, pH 7.6, 5 mM β-mercaptoethanol (reducing agent), protease inhibitor, 150 mM NaCl, 1% octyl β-d-glucopyranoside (OGP), and 10% glycerol. Due to the presence of a lipoprotein consensus sequence for both OmcA and MtrC, OGP was required to solubilize the cytochromes (44, 45, 51). Protein concentrations were determined using the 2DQuant kit from Amersham Biosciences (Uppsala, Sweden).
Preparation of proteins for AFM analysis.
Recombinant MtrC and OmcA were covalently attached to gold substrates via C terminus Cys residues as described by Wigginton et al. (51). Briefly, the recombinant proteins were dialyzed and equilibrated in 1× phosphate-buffered saline (PBS) buffer (10 mM sodium phosphate buffer, 150 mM NaCl), pH 7.4, containing 15 mM OGP using Microcon (Millipore Corporation) centrifugal filters (molecular weight cutoff, 10,000) to a concentration of 10 to 100 μg/ml. This solution was spotted onto a gold-covered coverslip (Molecular Imaging Corp.), allowing the Cys residues located at the protein's carboxy terminus to form a covalent bond with the gold substrate. After this, the buffer solution was carefully removed and rinsed with fresh PBS buffer (pH 7.4), and then the protein-functionalized gold substrate was left to equilibrate in this buffer at room temperature. The protein-coated substrates were then used in an AFM as described below.
Synthesis of hematite-functionalized AFM tips.
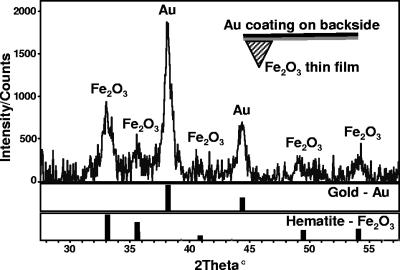
Si3N4 cantilevers (Digital Instruments) were ultrasonically cleaned in acetone and isopropyl alcohol, mounted on sample holders, and placed within a UV-ozone cleaner for 3 min prior to insertion into ultrahigh vacuum. After introduction into the deposition chamber, the cantilevers were cleaned with atomic oxygen from an electron cyclotron resonance oxygen plasma source at a chamber pressure of ∼2 × 10−5 torr for 15 min to rid the cantilevers of any residual adventitious carbon. The Fe2O3 films were grown in excess atomic oxygen using a Fe metal evaporation rate of ∼0.1 A/s, which has been shown previously to produce single crystalline epitaxial α-Fe2O3 grown on α-Al2O3(0001) substrates (23). The cantilevers were continuously rotated during oxygen plasma-assisted molecular beam epitaxial growth to prevent any unintended line-of-sight shadowing caused by the profile of the AFM tip. Following growth, the Fe flux from the electron beam evaporator was eliminated utilizing a shutter and reduced to zero while the coated cantilevers were bathed in atomic oxygen. Finally, the oxygen flux was stopped, and the cantilevers were removed from the deposition chamber. The surface coating on the cantilever was confirmed to be solely composed of hematite by X-ray diffraction analysis (Fig. 1). A total of eight replicate hematite tips were used in the AFM experiments described below.
FIG. 1.
X-ray diffraction pattern of an AFM cantilever that was coated with a hematite thin film using oxygen plasma-assisted molecular beam epitaxy. Gold (Au) and hematite (Fe2O3) references are shown for comparison. The Au signal originates from a coating applied to the “backside” (see schematic inset) of the cantilever by the manufacturer. The Au coating increases laser reflectivity for the AFM's optical lever detection, but it does not interfere with measurements of the force between the protein and hematite.
AFM force spectroscopy.
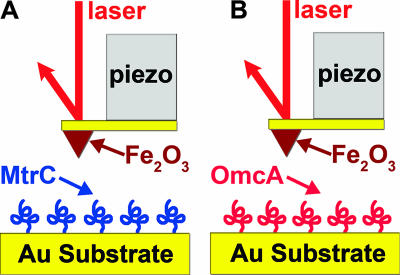
Force measurements were made with either a Veeco Bioscope AFM with a NanoScope IV controller or an Asylum Research MFP3D AFM at room temperature. Figure 2 is a schematic of the experimental setup. Raw data from the AFM were collected as the output of the photodiode detector, which is directly proportional to the deflection of the cantilever, as a function of the position of the tip, which is translated by a piezoelectric scanner. These raw data were plotted as so-called voltage displacement curves and then converted into force-distance curves according to a well-established protocol (10, 11). The force-distance curves were analyzed with SPIP (Image Metrology) and Igor Pro (WaveMetrics) software (30).
FIG. 2.
Schematic describing the experimental setup used to examine the interactions between hematite (Fe2O3) and purified S. oneidensis MR-1 outer membrane cytochromes MtrC (A) and OmcA (B). MtrC and OmcA are drawn arbitrarily here since their three-dimensional structures are not known. Purified recombinant proteins were covalently attached to a gold substrate via terminal Cys residues and were probed with a hematite-functionalized AFM tip in PBS buffer at pH 7.4.
Force measurements began within 15 to 30 min of preparing the cytochrome-functionalized gold substrates. Two different sample substrates were used for each cytochrome. The tip of a hematite-coated cantilever was brought into contact with a cytochrome-functionalized substrate (approach force curves) and pushed against the sample until the cantilever registered a deflection of 50 nm. The tip was then pulled from the substrate surface, thereby generating a retraction force curve or “force spectra.” Each approach-retraction cycle took 1 second (i.e., scan rate of 1 Hz).
Up to 100 force curves were collected on each of five different spots for each of the four cytochrome samples (two samples of MtrC and two samples of OmcA). The cantilevers of the hematite-functionalized tips had spring constant values between 0.06 and 0.07 N m−1, as determined by the thermal method of Hutter and Bechhoefer (20). The tip radius, reported by the manufacturer (Veeco), is 20 to 60 nm. The z-piezoelectric scanner of the AFM was calibrated as described previously (30). The vertical travel distance of the z-piezoelectric scanner was 2,000 nm.
Theoretical analysis of force-distance curves.
A theory developed for grafted polymers was used to model steric interactions for the “sharp” hematite-coated tip approaching a “flat” cytochrome-coated surface. This model predicts a repulsive force when a polymer-coated substrate approaches another surface. The origin of the repulsion is due to the unfavorable entropy associated with the confinement (or compression) of the polymer chains at the interface (21). The theoretical force-distance (F-x) relationship is given as (6, 7, 31)
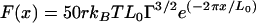 |
(1) |
where r is the radius of the tip (in m), kB is Boltzmann's constant (1.381 × 10−23 J K−1), T is temperature (in K), L0 is the equilibrium thickness of the polymer (in m), and Γ is the density of the polymer on the surface (in m−2).
The worm-like chain (WLC) model was used to create theoretical retraction curves for cytochromes (i.e., MtrC and OmcA) that form a bridging bond with hematite. This model describes the entropic restoring force generated when a linear polymer, such as a polypeptide, is mechanically extended or unfolded (17). The force-extension (F-x) relationship of the WLC model is given as (2, 33, 34, 43)
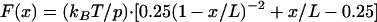 |
(2) |
where p is the polypeptide's persistence length (in meters), L is the polypeptide's contour length (in meters), and the other parameters are as in equation 1.
The persistence length embodies the stiffness or elasticity of a polymer and for purified protein molecules has been determined to be between 0.3 and 1.0 nm (8, 22, 33, 34, 40, 41, 43, 49). It is significant that this length is approximately equivalent to the average distance between consecutive α carbon atoms in a polypeptide chain, which is approximately 0.4 nm (33, 41). The contour length is the fully extended length of an entire polypeptide or segment of a polypeptide and can be calculated as the product of the length of an amino acid and the total number of amino acids in the polypeptide's primary sequence (8, 30, 32, 41). Equation 2 can therefore be used to predict the unfolding trajectory of a protein (e.g., MtrC or OmcA) for which the primary amino acid sequence is known.
RESULTS
Measurements of force between MtrC and hematite.
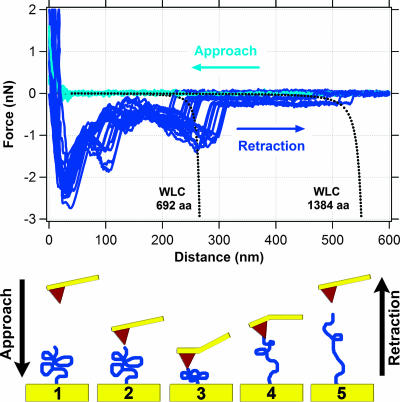
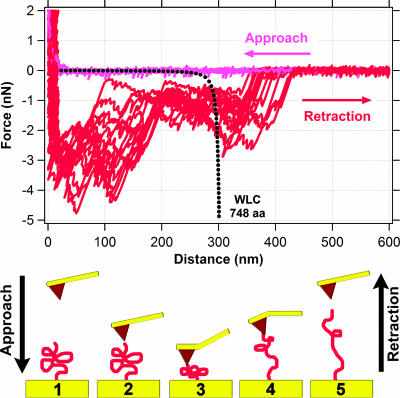
Figure 3 shows typical approach and retraction curves for measurements of force between hematite and MtrC. Although data were collected out to 2,000 nm, Fig. 3 shows force data only for distances of 0 to 600 nm because no detectable interaction was observed at distances >600 nm. Force approach curves show that the cantilever experiences no force until it comes to within ∼30 nm of the cytochrome-functionalized gold substrate. At this range, a repulsive force (positive sign) between the hematite and MtrC is detected. This repulsive force continues to increase to a maximum of ∼2 nN.
FIG. 3.
Measurements of force between hematite and MtrC. The top graph shows 14 approach (light blue) and 14 retraction (dark blue) force curves. The dotted black lines correspond to the theoretical force-extension relationship for polypeptides composed of 692 (single MtrC molecule) and 1,384 (putative MtrC dimer) amino acids as predicted by the WLC model (equation 2). The bottom schematic illustrates one approach/retraction cycle, starting at frame 1 and ending at frame 5.
All retraction curves exhibited an initial “jump from contact” event at ∼25 nm (Fig. 3). The magnitude of the adhesion force was, in general, around 2 nN. When the tip snapped back towards its resting position, it created a “blind window” in the force curve, within which no structure could be observed. Hence, details within the first 50 to 75 nm are hidden by the “jump from contact” event. This is an unavoidable consequence of using AFM because the tip will “jump from contact” when the spring constant of the cantilever exceeds the actual force gradient between the tip and the sample.
A total of 668 of 881 (76%) retraction curves exhibited a feature that resembled a “sawtooth” (e.g., see the region from 150 to 300 nm in Fig. 3). These features represent attractive interactions between the cytochrome and hematite. Most retraction curves exhibited multiple sawteeth in a single force profile. However, some retraction curves contained only a single sawtooth. The length scale of these types of attractive interactions extended out to 225 to 300 nm (Fig. 3). In addition to these prominent sawtooth force signatures, the retraction profiles for MtrC occasionally exhibited a relatively weak, long-range attractive force that extended out to ∼500 nm (Fig. 3).
Sawteeth such as those shown in Fig. 3 are often attributed to bridging bonds that form when a polymer links two surfaces (see, e.g., references 6 and 43). Therefore, we attempted to capture the sawtooth force signature by allowing MtrC to be tethered to the hematite tip during subsequent approach-retraction cycles without physically breaking the bond between the protein and mineral. This was accomplished by decreasing the travel distance of the z-piezoelectric scanner to 200 or 275 nm.
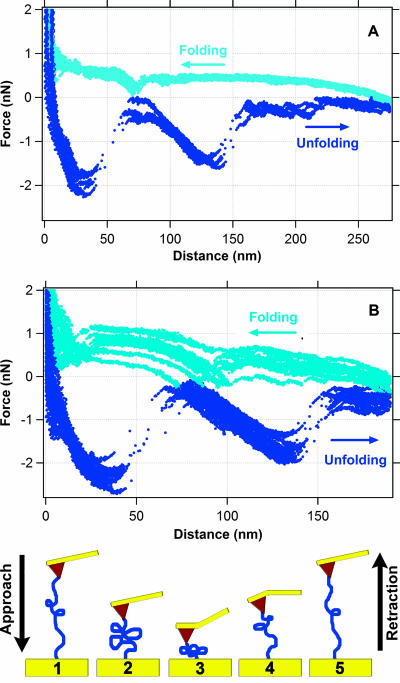
As shown in Fig. 4, pulling the MtrC polypeptide chain triggers a series of sawtooth-like features. After the polypeptide was stretched to 200 or 275 nm, the force was relaxed as the hematite tip was allowed to approach the MtrC substrate. The approach curves reveal sawteeth that mirror the unfolding trajectory (Fig. 4). We were able to repeat this process over a 4-min period and detect 240 continuous unfold-refold events before the bond between MtrC and hematite was broken. When this happened, a new spot was selected on the MtrC substrate and the experiment was continued. Each new spot yielded nearly identical force trajectories, indicating that the data for the interaction between MtrC and hematite were reproducible.
FIG. 4.
Folding and unfolding trajectories observed for MtrC during approach (light blue) and retraction (dark blue) cycles. Here, the hematite-coated tip was not fully retracted as described for Fig. 3, but rather only to a distance of 275 nm (A) or 200 nm (B). Under these conditions, the MtrC molecule does not detach from the tip but rather maintains a physical bond with the hematite-functionalized tip (depicted in the bottom schematic), allowing the observation of unfolding and refolding pathways of the polypeptide.
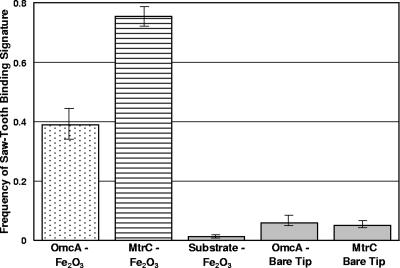
A number of control experiments were conducted to confirm the specificity of the bond between MtrC and hematite. Measurements of force between (i) an uncoated gold substrate and a hematite-functionalized tip and (ii) an MtrC-functionalized gold substrate and a bare silicon nitride AFM tip (i.e., a tip that was not coated with hematite) were conducted. In each case, most retraction curves were featureless profiles fluctuating around zero force. For control experiment i, only 6 out of 487 retraction curves (1%) exhibited a sawtooth-like feature, and for control experiment ii, only 32 of 633 retraction curves (5%) exhibited a sawtooth-like feature (Fig. 5). Moreover, the trajectories of the 6 and 32 sawteeth observed for control experiments i and ii, respectively, did not match the trajectories of the sawteeth observed in spectra of the force between MtrC and hematite.
FIG. 5.
Plot showing the frequency of observing a binding event (i.e., sawtooth force signature) between S. oneidensis MR-1 cytochromes and hematite (Fe2O3). Shown are the average values (±standard errors) for OmcA-functionalized substrates and hematite-coated tips (n = 816), MtrC-functionalized substrates and hematite-coated tips (n = 881), bare substrates that were not coated with cytochromes and hematite-coated tips (n = 487), OmcA-functionalized substrates and bare Si3N4 tips (n = 468), and MtrC-functionalized substrates and bare Si3N4 tips (n = 633). These values include only those force curves for which the tip was completely separated from the polypeptide during an approach-retraction cycle, and they do not include folding-refolding cycles like those shown in Fig. 4.
In addition to these control experiments, we conducted experiments using buffer containing free MtrC to determine if the observed binding events indeed arise from specific MtrC-hematite interactions. This type of control experiment (i.e., injection of “free” protein) has been described previously (19, 46). When free MtrC (0.2 μM) was added to the AFM experiment, sawteeth disappeared from the retraction curves. Apparently, free MtrC binds to the hematite-functionalized tip, thus preventing recognition of the MtrC molecules on the gold substrate.
Measurements of force between OmcA and hematite.
Experiments measuring the force between OmcA and hematite were conducted in the same manner described above with the exception that OmcA was substituted for MtrC. Figure 6 shows typical approach and retraction curves for hematite and OmcA. Similar to the MtrC data, no interactions were observed at distances >600 nm.
FIG. 6.
Measurements of the force between hematite and OmcA. The top graph shows 14 approach (light red) and 14 retraction (dark red) force curves. The dotted black line corresponds to the theoretical force-extension relationship for a polypeptide composed of 748 amino acids (single OmcA molecule), as predicted by the WLC model (equation 2). The bottom schematic illustrates one approach/retraction cycle, starting at frame 1 and ending at frame 5.
Approach curves for OmcA are similar to those for MtrC. There is a repulsive force that begins when the hematite is brought to within ∼20 to 30 nm of the OmcA-functionalized substrate (Fig. 6). The repulsive force continues to increase to a maximum of ∼2 nN at contact.
Similar to the MtrC data, retraction curves for OmcA exhibit an initial “jump from contact” event, but the adhesion force is stronger (3 to 4 nN; Fig. 6). The retraction curves for OmcA also show sawtooth-like force signatures (Fig. 6); however, the frequency of these events is only 39% (318 of 816). The length scale of the attractive interactions between OmcA and hematite extended to 325 to 425 nm (Fig. 6).
As described above, unfolding/folding events were observed for MtrC that was tethered to the hematite tip. Therefore, we conducted a similar experiment for OmcA and hematite. The travel distance of the z-piezoelectric scanner was decreased to less than 400 nm, thereby allowing OmcA the opportunity to form a stable physical bond with the hematite-functionalized tip. However, after numerous attempts we were unable to observe unfolding-refolding events like those that were detected for MtrC bound to hematite.
OmcA control experiments included AFM measurements of (i) an uncoated gold substrate and a hematite-functionalized tip and (ii) an OmcA-functionalized gold substrate and a bare silicon nitride AFM tip. Most retraction curves were essentially featureless horizontal lines fluctuating around zero force. For control experiment i, 1% of retraction curves exhibited sawteeth, and for control experiment ii, only 27 of 468 retraction curves (6%) exhibited sawteeth (Fig. 5). Moreover, the trajectories of the sawteeth observed for control experiments i and ii did not match the trajectories of the sawteeth observed in spectra for the force between OmcA and hematite.
As described above for MtrC, we used a buffer containing free OmcA to determine the specificity of the cytochrome-hematite bond. Again, we did not observe sawteeth when 0.2 μM free OmcA was added to the AFM experiment. The results of these control experiments are consistent with those obtained previously by Xiong et al. (52) in which the authors used dynamic light scattering and fluorescence correlation spectroscopy to show that purified OmcA has a specific preference for binding to hematite particles (52).
DISCUSSION
Steric forces at the cytochrome-hematite interface.
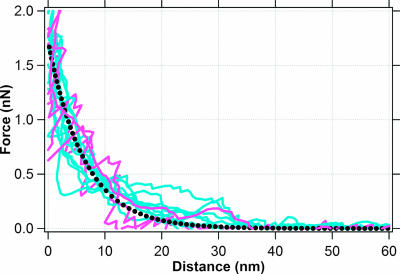
The approach force curves for both MtrC and OmcA reveal a repulsive force as these cytochromes are brought into contact with hematite (Fig. 3 and 6). Because these measurements involve a polymer-coated surface, equation 1 was used to determine whether steric forces may explain this repulsion. Figure 7 compares the approach curves for the MtrC-hematite and OmcA-hematite pairs with a theoretical force-distance relationship for an AFM tip approaching a cytochrome-coated surface having a thickness (L0) of 40 nm and density (Γ) of 2.3 × 1016 molecules m−2. The approach forces are indeed consistent with steric interactions between the hematite tip and cytochrome substrate.
FIG. 7.
Force curves for hematite approaching a MtrC-functionalized substrate (light blue) or an OmcA-functionalized substrate (light red). The dotted black curve is the theoretical steric force (equation 1) between a hematite-coated tip (60-nm radius) and a surface coated with a protein (thickness = 40 nm; density = 2.3 × 1016 molecules m−2).
Further, the values for the thickness and density of the cytochrome coating are consistent with values found by Wigginton et al. (51). Specifically, Wigginton et al. provide an AFM image that shows a 20-nm-thick coating of MtrC on a gold substrate. This value is less than the thickness that was used to model the steric force shown in Fig. 7. However, Wigginton et al. collected their AFM image in air whereas our force curves were collected on hydrated proteins within an aqueous solution. Wigginton et al. (51) also provide scanning tunneling microscopy (STM) images, which reveal the packing density of MtrC and OmcA on gold substrates. According to these STM images, the density values are 1.2 × 1016 and 2.6 × 1016 molecules m−2 for OmcA and MtrC, respectively. The cytochrome density used in Fig. 7 is consistent with these STM data. It is important to note that there is a typographical error in the caption of the STM images published by Wigginton et al. (51). The image corresponding to OmcA is actually an STM image of MtrC and vice versa (N. Wigginton, personal communication).
Force spectra of the cytochrome-hematite bond.
While the approach curves show a repulsive steric force, many of the retraction curves (Fig. 5) reveal that a stable, attractive bond forms between hematite and each of the cytochromes. This may, at first glance, seem rather surprising. However, this type of hysteresis in approach and retraction curves is often observed in force measurements with polymers (see, e.g., reference 6).
As discussed above, the attractive interactions between the cytochromes and hematite manifest themselves as sawtooth-shaped force extension profiles. Others have shown that this type of “force signature” is the result of the mechanical unfolding or stretching of a polypeptide that forms a direct, bridging bond between two surfaces (8, 15, 16, 25, 26, 30, 33, 41, 47, 48). To determine whether MtrC and OmcA could produce the observed sawtooth force profiles, the retraction curves were compared to the theoretical unfolding trajectories of these cytochromes as predicted by the WLC model (equation 2).
Figure 3 shows the predicted WLC force trajectory for a polypeptide composed of 692 amino acids, which is the number of amino acids in recombinant MtrC. At low force, the model predicts a linear relationship between force and extension of the polypeptide. This is shown as a more-or-less horizontal line extending out to a distance of ∼200 nm. This trajectory dramatically changes to a nonlinear relationship between force and extension as the polypeptide approaches its fully extended length (Fig. 3, between 225 and 275 nm). Figure 3 also shows the predicted WLC force trajectory for a polypeptide composed of 1,384 amino acids, which would be the number of amino acids in a recombinant MtrC dimer.
For comparison, Fig. 3 plots a number of retraction curves selected to span the range of observations for hematite on a MtrC-functionalized substrate. While these curves are extremely similar to one another, slight variations in trajectory can be observed for the individual traces. These variations are due in part to the fact that we used several different hematite tips and MtrC substrates in our AFM experiments (see Materials and Methods). Variations in the retraction curves may also be due to the measurement itself. For example, Farshchi-Tabrizi et al. (14) demonstrated that even when two relatively simple, hard surfaces are brought into contact under precisely the same conditions there will often be variation in the resulting retraction curves. They speculate that such variations are due to the measurement itself, which induces structural changes in the sample's previous contact area. It is possible that such events also occur with our soft biological samples. Nonetheless, the level of reproducibility in the MtrC-hematite force spectra is clearly significant.
The observed retraction profiles are more complex than the WLC-modeled extension of MtrC. However, the rupture events for the sawteeth (i.e., the “snapback” to zero force) occur at a distance of ∼250 nm, which is very similar to the predicted contour length of MtrC (277 nm, for p = 0.4 nm). This remarkable likeness indicates that our AFM measurements are indeed probing a specific bond that forms between MtrC and hematite. It should also be noted that we observed another notable sawtooth binding event at a distance of ∼500 nm. The significance of this will be discussed in more detail below.
Although the WLC model and observed force spectroscopy have similar rupture lengths, there is at least one notable difference between them with respect to MtrC on hematite. The WLC model predicts only one single unfolding event (i.e., one sawtooth), while the observed force profiles exhibit up to three unfolding events within a single retraction profile. Other publications have shown that a single polypeptide chain can yield multiple sawteeth in its unfolding trajectory if that protein has more than one mechanically stable domain within its structure (8, 22, 30, 41).
To determine whether MtrC might have multiple, mechanically stable domains, we attempted to unfold and refold MtrC without completely rupturing the bond between MtrC and hematite. As shown in Fig. 4, we were able to successfully unfold (retraction curves) and sequentially refold (approach curves) MtrC as it was tethered to the hematite-coated tip through a bridging bond. The fact that we were able to observe this fold-unfold cycle in up to 240 consecutive attempts indicates a very resilient bond between MtrC and hematite. During reversible unfolding/refolding cycles (Fig. 4) qualitatively similar force-distance trajectories could be observed for both retraction and approach measurements. Interestingly, the trajectories of folding and unfolding curves appear as mirror images of one another, consisting of successive convex or concave paths reflected across the line defining zero force (Fig. 4). While a detailed analysis of these force curves is beyond the scope of this work, the observation of nearly identical sets of unfolding/folding force-distance trajectories indicates the existence of specific substructures within the MtrC polypeptide itself that are apparently essential for mineral recognition and binding.
A similar type of analysis was conducted for the OmcA-hematite retraction curves. Figure 6 shows the predicted WLC force trajectory for a polypeptide composed of 748 amino acids, which is the number of amino acids in recombinant OmcA. For comparison, Fig. 6 also shows a number of retraction curves for the OmcA-hematite pair. The retraction curves exhibit qualitatively similar sawtooth-like features extending outward to distances of 325 to 425 nm. As was the case for MtrC and hematite, the rupture event between OmcA and hematite occurs at a distance that is approximately equal to the predicted contour length for OmcA (299 nm, for p = 0.4 nm).
Similar to those for MtrC, the retraction profiles for OmcA exhibit multiple sawtooth events rather than the single sawtooth predicted by the WLC model. This suggests that OmcA also has a number of mechanically stable domains within its overall structure. However, unlike what was found for MtrC, we were unable to sequentially unfold and refold OmcA without completely breaking its bond to hematite. Perhaps, this indicates that the OmcA-hematite bond is not as resilient as the MtrC-hematite bond.
The above discussion focuses solely on the sawtooth features in the retraction curves. As noted above, these “force signatures” demonstrate that a specific bond forms after hematite comes into physical contact with each cytochrome. However, we have neglected to discuss the “jump from contact” feature observed in the retraction curves.
The “jump from contact” adhesion event detected in the retraction curves for MtrC and OmcA (Fig. 3 and 6) has a very different force profile than the sawtooth events characteristic of specific protein-mineral bonds. The magnitudes of this attractive force between MtrC and OmcA and hematite are approximately 2 nN and 4 nN, respectively. Such adhesion is commonly noted in force measurements and likely originates from nonspecific forces such as the van der Waals force (30, 33).
Relevance to electron transfer reactions between S. oneidensis MR-1 and Fe(III) oxides.
In the past, we hypothesized that sawtooth-like signatures for the force between S. oneidensis MR-1 and an Fe(III) oxide indicate that surface-exposed outer membrane proteins form a specific bond with the mineral surface (29, 32). Recently, we showed that specific sawtooth force trajectories could be attributed to the formation of a bond between an engineered protein expressed on the surface of a gram-negative bacterium cell and a solid substrate in situ (30). The work presented herein demonstrates that bonds between MtrC and hematite (Fig. 3) or OmcA and hematite (Fig. 6) yield sawtooth force signatures.
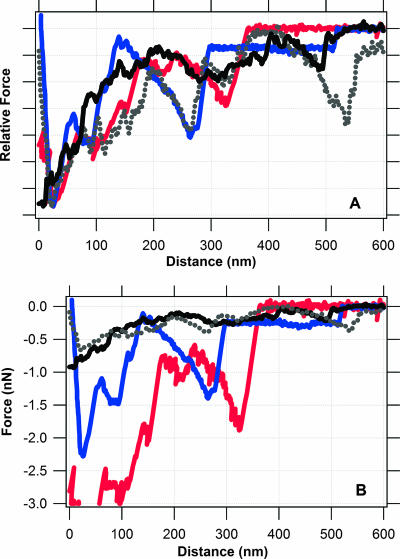
Figure 8 was constructed to determine whether force spectra collected with whole cells of S. oneidensis MR-1 correspond to the data collected with the purified cytochromes. This figure shows retraction curves collected for the following pairs: MtrC-hematite profiles from Fig. 3, OmcA-hematite profiles from Fig. 6, and retraction curves for a living S. oneidensis MR-1 cell and goethite (FeOOH) taken from references 29 and 32. It is worth noting that the whole-cell and pure-protein force spectra were collected independently using two different AFMs. Furthermore, the whole-cell data were collected by probing an iron oxide surface with a biologically active force probe (i.e., live S. oneidensis MR-1 on the AFM probe), whereas the data presented herein were collected by essentially placing the iron oxide on the AFM tip and coating a substrate with purified cytochrome protein.
FIG. 8.
Spectra for force-distance relationship between an Fe(III) oxide and either a purified cytochrome or S. oneidensis MR-1. Traces correspond to retraction curves for an Fe(III) oxide and each of the following: S. oneidensis MR-1 grown with Fe(III) as the terminal electron acceptor (black and gray curves), recombinant MtrC (blue curve), and recombinant OmcA (red curve). Curves for the purified cytochromes come from this study. The solid black curve was taken from reference 32, and the dotted gray curve was taken from reference 29. (A) Relative force-distance spectra where the y axis has been normalized such that the maximum force in each retraction curve is set to a value of 1. This procedure does not alter the distance data; it simply plots force on a relative scale. (B) Unaltered versions of the force spectra presented in panel A for comparison.
There is a very strong correlation between the force signatures observed for the purified proteins and the whole cells (Fig. 8A). The sawtooth force signature observed around 300 nm is present in both the whole-cell and cytochrome data. This correlation supports our original hypothesis that MtrC or OmcA of both on the surface of S. oneidensis form a directed, specific bond with the surface of iron oxides. It is very difficult to say with certainty that the whole-cell sawtooth observed near 300 nm is due solely to one or the other cytochrome. As noted above, both cytochromes are able to form a specific bond with hematite, but each protein has unique binding attributes. On the one hand, OmcA binds to hematite with a stronger force than MtrC (compare retraction curves in Fig. 3 and 6). On the other hand, MtrC forms a bond with hematite more frequently (Fig. 5) and the nature of this bond is much more resilient (Fig. 4) than that with OmcA. It is therefore more likely that either cytochrome could be the origin of the “whole-cell” sawtooth near 300 nm, as each cytochrome provides a unique function in binding and/or reduction of Fe(III) in minerals.
One notable difference in the protein spectra versus the whole-cell spectra is the magnitude of force for the cytochrome signature at ∼300 nm. The cytochrome binding forces are significantly “stronger” than those revealed by the whole-cell data (Fig. 8B). This may be related to some type of shielding event that occurs when a cytochrome is present within the outer membrane of a whole cell, for example, due to the presence of lipopolysaccharides within the outer membrane. Another possible explanation is that there is a greater density of cytochromes on the gold substrate than on the outer surface of an S. oneidensis bacterium. If this is true, we can obtain a rough estimate of the number of MtrC and/or OmcA molecules exposed on the surface of S. oneidensis.
According to the steric model (Fig. 7), the cytochrome density on the gold substrate is approximately 2 × 1016 molecules m−2. For the 300-nm sawtooth, this packing density corresponds to a binding force of ∼1 nN for MtrC and ∼2 nN for OmcA (Fig. 3 and 6). This force is roughly three to five times greater than the binding force for the same sawtooth observed with the whole cell (Fig. 8B). For the hypothetical case when there are three (or five) protein chains in parallel, each with identical extension ratios between the tip and sample, the observed force would be three (or five) times as great as if one chain formed a bridging bond between the tip and sample (5). If we make this assumption, then the density of MtrC and OmcA on the whole cell should be approximately one-third to one-fifth that of the gold-coated cytochrome substrate. This corresponds to a density of 4 × 1015 to 7 × 1015 MtrC (or OmcA) molecules per m2 of cell surface. For a rod-shaped bacterium that is 0.5 by 2.0 μm, the size of an S. oneidensis cell (50), we obtain an estimate of 13,000 to 22,000 MtrC (or OmcA) molecules exposed on the outer surface of the cell.
We are aware of the fact that a bond's loading rate can have a significant impact on the observed binding force (4, 12, 13, 18). However, the loading rates are similar for both sets of data: 130 nN sec−1 and 100 to 140 nN sec−1 for the pure-protein and whole-cell spectra, respectively. Furthermore, our estimate of ∼104 cytochromes per bacterium is consistent with other work that shows that OmpA and Lpp, two of the major outer membrane proteins in gram-negative bacteria, occur at about 105 to 106 molecules per cell (9, 24, 39). Therefore, our estimate provides a reasonable benchmark of the number of MtrC and/or OmcA cytochromes on the surface of a single bacterium of S. oneidensis grown under anaerobic conditions with Fe(III) as the terminal electron acceptor.
There is another striking similarity between the whole-cell and protein data at a length scale of ∼500 nm (Fig. 8). At this scale, the data for S. oneidensis MR-1 exhibit a notable sawtooth binding event. While this is not a classic sawtooth, the data for MtrC reveal a longer-range attractive force with a bond rupture event at ∼500 nm. As noted above, the predicted contour length of MtrC is ∼277 nm. Therefore, the attractive force detected at length scales greater than 300 nm cannot be attributed to a single molecule of MtrC. Perhaps, this longer-range binding feature originates from the unraveling of a dimer of MtrC. In a previous publication, mass peptide analysis identified both MtrC and OmcA in polypeptides having apparent Mrs of ≈150,000 and ≥210,000, as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (29). These results suggest that MtrC or OmcA can form oligomeric complexes in S. oneidensis MR-1. Others have also proposed a similar arrangement in which several outer membrane proteins, including MtrC and OmcA, form a functional “reductase complex” that catalyzes the terminal reduction of Fe(III) (3, 45). Additional AFM studies with mutagenically altered forms of MtrC or OmcA in combination with modeling or computer simulations will be needed to confirm/refine this model.
Acknowledgments
A portion of this research was performed as part of an EMSL Scientific Grand Challenge project at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy's (DOE) Office of Biological and Environmental Research (OBER) Program and located at Pacific Northwest National Laboratory (PNNL). PNNL is operated for the DOE by Battelle Memorial Institute under contract DE-AC05-76RLO1830. B.H.L. acknowledges the support of the U.S. DOE Office of Basic Energy Sciences (OBES) Geosciences Research Program. L.S. acknowledges the support of the U.S. DOE OBER Genomics-Genomes to Life Program. S.K.L. acknowledges financial support provided by the National Science Foundation and the DOE OBES Geosciences Research Program. Partial financial support for R.Y. was provided by the American Chemical Society.
We acknowledge the helpful comments provided by four anonymous reviewers and N. Wigginton.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Arnold, R. G., T. J. DiChristina, and M. R. Hoffmann. 1988. Reductive dissolution of Fe(III) oxides by Pseudomonas species 200. Biotechnol. Bioeng. 32:1081-1096. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, C. G., S. B. Smith, V. A. Bloomfield, and C. Bustamante. 1997. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA 94:6185-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beliaev, A. S., D. A. Saffarini, J. L. McLaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 4.Bell, G. 1978. Models for the specific adhesion of cells to cells. Science 200:618-627. [DOI] [PubMed] [Google Scholar]
- 5.Bemis, J. E., B. B. Akhremitchev, and G. C. Walker. 1999. Single polymer chain elongation by atomic force microscopy. Langmuir 15:2799-2805. [Google Scholar]
- 6.Butt, H. J., M. Kappl, H. Mueller, R. Raiteri, W. Meyer, and J. Ruhe. 1999. Steric forces measured with the atomic force microscope at various temperatures. Langmuir 15:2559-2565. [Google Scholar]
- 7.Camesano, T. A., and B. E. Logan. 2000. Probing bacterial electrosteric interactions using atomic force microscopy. Environ. Sci. Technol. 34:3354-3362. [Google Scholar]
- 8.Carrion-Vazquez, M., A. F. Oberhauser, S. B. Fowler, P. E. Marszalek, S. E. Broedel, J. Clarke, and J. M. Fernandez. 1999. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA 96:3694-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducker, W. A., T. J. Senden, and R. M. Pashley. 1991. Direct measurement of colloidal forces using an atomic force microscope. Nature 353:239-241. [Google Scholar]
- 11.Ducker, W. A., T. J. Senden, and R. M. Pashley. 1992. Measurements of forces in liquids using a force microscope. Langmuir 8:1831-1836. [Google Scholar]
- 12.Evans, E. 2001. Probing the relation between force lifetime and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105-128. [DOI] [PubMed] [Google Scholar]
- 13.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farshchi-Tabrizi, M., M. Kappl, Y. J. Cheng, J. Gutmann, and H. Butt. 2006. On the adhesion between fine particles and nanocontacts: an atomic force microscope study. Langmuir 22:2171-2184. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, J. M., and H. Li. 2004. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science 303:1674-1678. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, T. E., P. E. Marszalek, and J. M. Fernandez. 2000. Stretching single molecules into novel conformations using the atomic force microscope. Nat. Struct. Biol. 7:719-724. [DOI] [PubMed] [Google Scholar]
- 17.Flory, P. J. 1989. Statistical mechanics of chain molecules. Hanser Publisher, Munich, Germany.
- 18.Gergely, C., J. Voegel, P. Schaaf, B. Senger, M. Maaloum, J. K. Horber, and J. Hemmerle. 2000. Unbinding process of adsorbed proteins under external stress studied by atomic force microscopy spectroscopy. Proc. Natl. Acad. Sci. USA 97:10802-10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinterdorfer, P., W. Baumgartner, H. J. Gruber, K. Schilcher, and H. Schindler. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA 93:3477-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutter, J., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868-1873. [Google Scholar]
- 21.Israelachvili, J. N. 1992. Intermolecular and surface forces, 2nd ed. Academic Press, London, United Kingdom.
- 22.Kellermayer, M. S., S. B. Smith, H. L. Granzier, and C. Bustamante. 1997. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 276:1112-1116. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y. J., Y. Gao, and S. A. Chambers. 1997. Selective growth and characterization of pure, epitaxial alpha-Fe2O3(0001) and Fe3O4(001) films by plasma-assisted molecular beam epitaxy. Surf. Sci. 371:358-370. [Google Scholar]
- 24.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 25.Li, H., and J. M. Fernandez. 2003. Mechanical design of the first proximal Ig domain of human cardiac titin revealed by single molecule force spectroscopy. J. Mol. Biol. 334:75-86. [DOI] [PubMed] [Google Scholar]
- 26.Li, H., A. F. Oberhauser, S. D. Redick, M. Carrion-Vazquez, H. P. Erickson, and J. M. Fernandez. 2001. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA 98:10682-10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd, J. R. 2003. Microbial reduction of metals and radionuclides. FEMS Microbiol. Rev. 27:411-425. [DOI] [PubMed] [Google Scholar]
- 28.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 29.Lower, B. H., M. F. Hochella, and S. K. Lower. 2005. Putative mineral-specific proteins synthesized by a metal reducing bacterium. Am. J. Sci. 305:687-710. [Google Scholar]
- 30.Lower, B. H., R. Yongsunthon, F. P. Vellano III, and S. K. Lower. 2005. Simultaneous force and fluorescence measurements of a protein that forms a bond between a living bacterium and a solid surface. J. Bacteriol. 187:2127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lower, S. K. 2005. Directed natural forces of affinity between a bacterium and mineral. Am. J. Sci. 305:752-765. [Google Scholar]
- 32.Lower, S. K., M. F. Hochella, and T. Beveridge. 2001. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and alpha-FeOOH. Science 292:1360-1363. [DOI] [PubMed] [Google Scholar]
- 33.Mueller, H., H. J. Butt, and E. Bamberg. 1999. Force measurements on myelin basic protein adsorbed to mica and lipid bilayer surfaces done with the atomic force microscope. Biophys. J. 76:1072-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller, D. J., W. Baumeister, and A. Engel. 1999. Controlled unzipping of a bacterial surface layer with atomic force microscopy. Proc. Natl. Acad. Sci. USA 96:13170-13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, C. R., and J. M. Myers. 1997. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim. Biophys. Acta 1326:307-318. [DOI] [PubMed] [Google Scholar]
- 36.Myers, J. M., and C. R. Myers. 2002. Genetic complementation of an outer membrane cytochrome omcB mutant of Shewanella putrefaciens MR-1 requires omcB plus downstream DNA. Appl. Environ. Microbiol. 68:2781-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers, J. M., and C. R. Myers. 1998. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim. Biophys. Acta 1373:237-251. [DOI] [PubMed] [Google Scholar]
- 38.Myers, J. M., and C. R. Myers. 2001. Role for outer membrane cytochromes OmcA and OmcB of Shewanella putrefaciens MR-1 in reduction of manganese dioxide. Appl. Environ. Microbiol. 67:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neidhardt, F. C., R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, DC.
- 40.Oberdorfer, Y., H. Fuchs, and A. Janshoff. 2000. Conformational analysis of native fibronectin by means of force spectroscopy. Langmuir 16:9955-9958. [Google Scholar]
- 41.Oberhauser, A. F., P. E. Marszalek, M. Carrion-Vazquez, and J. M. Fernandez. 1999. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 6:1025-1028. [DOI] [PubMed] [Google Scholar]
- 42.Pitts, K. E., P. S. Dobbin, F. Reyes-Ramirez, A. J. Thomson, D. J. Richardson, and H. E. Seward. 2003. Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome MtrA. J. Biol. Chem. 278:27758-27765. [DOI] [PubMed] [Google Scholar]
- 43.Rief, M., J. M. Fernandez, and H. E. Gaub. 1998. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 81:4764-4767. [Google Scholar]
- 44.Shi, L., J. T. Lin, L. M. Markillie, T. C. Squier, and B. S. Hooker. 2005. Overexpression of multi-heme c-type cytochromes. BioTechniques 38:297-299. [DOI] [PubMed] [Google Scholar]
- 45.Shi, L., Z. Wang, B. Chen, D. A. Elias, M. U. Mayer, Y. A. Gorby, S. Ni, B. H. Lower, D. W. Kennedy, D. S. Wunschel, H. M. Mottaz, M. J. Marshall, E. A. Hill, A. S. Beliaev, J. M. Zachara, J. K. Fredrickson, and T. C. Squier. 2006. Isolation of high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes from Shewanella oneidensis MR-1. J. Bacteriol. 188:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stroh, C., H. Wang, R. Bash, B. Ashcroft, J. Nelson, H. Gruber, D. Lohr, S. M. Lindsay, and P. Hinterdorfer. 2004. Single-molecule recognition imaging microscopy. Proc. Natl. Acad. Sci. USA 101:12503-12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sulchek, T. A., R. W. Friddle, K. Langry, E. Y. Lau, H. Albrecht, T. V. Ratto, S. J. DeNardo, M. E. Colvin, and A. Noy. 2005. Dynamic force spectroscopy of parallel individual Mucin1-antibody bonds. Proc. Natl. Acad. Sci. USA 102:16638-16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Touhami, A., M. H. Jericho, J. M. Boyd, and T. J. Beveridge. 2006. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 188:370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tskhovrebova, L., J. Trinick, J. A. Sleep, and R. M. Simmons. 1997. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature 387:308-312. [DOI] [PubMed] [Google Scholar]
- 50.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 51.Wigginton, N. S., K. M. Rosso, B. H. Lower, L. Shi, and M. F. Hochella, Jr. 2007. Electron tunneling properties of outer-membrane decaheme cytochromes from Shewanella oneidensis. Geochem. Cosmochim. Acta 71:543-555. [Google Scholar]
- 52.Xiong, Y., L. Shi, B. Chen, M. U. Mayer, B. H. Lower, Y. Londer, S. Bose, M. F. Hochella, J. K. Fredrickson, and T. C. Squier. 2006. High-affinity binding and direct electron transfer to solid metals by the Shewanella oneidensis MR-1 outer membrane c-type cytochrome OmcA. J. Am. Chem. Soc. 128:13978-13979. [DOI] [PubMed] [Google Scholar]