Abstract
The competence transcription factor ComK plays a central role in competence development in Bacillus subtilis by activating the transcription of the K regulon. ComK-activated genes are characterized by the presence of a specific sequence to which ComK binds, a K-box, in their upstream DNA region. Each K-box consists of two AT-boxes with the consensus sequence AAAA-(N)5-TTTT, which are separated by a flexible spacer resulting in either two, three, or four helical turns between the starting nucleotides of the repeating AT-box units. In this study, the effects of potential determinants of ComK regulation in K-boxes were investigated by testing ComK's transcription activation and DNA-binding affinity on altered K-boxes with mutations either in the spacer between the AT-boxes or in the consensus sequence of the AT-boxes. The most striking result demonstrates the importance of the second thymine base in the AT-boxes. Mutation of this T into a guanine resulted in a threefold reduction in transcription activation and DNA binding by ComK. Transcription activation, as well as DNA binding, was almost completely abolished when both AT-boxes contained a T2-to-G mutation. This result was corroborated by in silico analyses demonstrating that a combination of mutations at the T2 positions of both AT-boxes is not found among any ComK-activated K-boxes, indicating that at least one consensus T2 position is required to maintain a functional K-box. The results suggest an important structural role for T2 in ComK binding, probably by its specific position in the minor groove of the DNA.
The development of genetic or natural competence is an adaptation process that enables bacterial cells to take up and integrate exogenous DNA into their genomes. This phenomenon has been studied extensively in the gram-positive soil bacterium Bacillus subtilis. Competence development depends on the presence of a key regulatory protein, i.e., the competence transcription factor ComK. During exponential growth, the presence of free ComK in the cell is prevented by both transcriptional and posttranslational control. Transcription of comK is repressed by the binding of AbrB, CodY, and Rok to the comK promoter (12, 14, 27), while any ComK that is produced is trapped by MecA and targeted for proteolytic degradation by ClpCP (32). At the end of the exponential growth phase, increased cell densities are sensed and interpreted by a quorum-sensing pathway. This results in the production of ComS, a small protein that can liberate ComK from the proteolytic complex (5, 8, 28). At the same time in growth, the repressing effect of CodY and AbrB is relieved in response to nutrient limitation. Free ComK can bind to its own promoter and activate gene transcription (33, 34). Furthermore, ComK can repress rok transcription by binding to the rok promoter, thereby relieving the repression by Rok of comK transcription (14). The ensuing autostimulation of comK transcription results in a rapid increase in the ComK concentration in the cell. In addition to stimulating the transcription of its own gene, ComK stimulates the transcription of the K regulon, which contains among others the late competence genes, encoding the DNA binding, uptake, and integration machinery (for a review, see references 6 and 13).
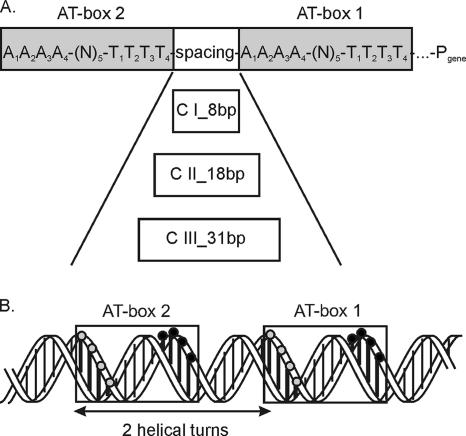
ComK activates transcription by binding to specific sequences, so-called K-boxes, in the upstream region of ComK-activated genes (Fig. 1). Each K-box consists of two AT-boxes with the consensus sequence AAAA-(N)5-TTTT. These boxes are separated by a spacer of a discrete number of helical turns, which positions both AT-boxes on the same side of the DNA helix. Based on the length of the spacer (two, three or four helical turns calculated between the first A's of both AT-boxes), K-boxes are divided into three different classes, i.e., those belonging to class I, class II, or class III promoters, respectively. Functional ComK is thought to act as a tetramer composed of two dimers, each binding to an AT-box. Tetramerization of the two dimers is accompanied by bending of the DNA (9). Previous research has demonstrated that the major role of ComK in transcription activation at the comG promoter is in stabilizing RNA polymerase binding, probably by facilitating contacts between the polymerase and the upstream DNA through bending of the DNA in the promoter region (29).
FIG. 1.
Overview of the ComK-binding site (K-box) in B. subtilis. (A) Schematic representation of a K-box. A K-box consists of two AT-boxes (shaded gray) with the consensus sequence AAAA-(N)5-TTTT. In this study, the AT-box closest to the −35 position is indicated as AT-box 1, while the box further upstream is called AT-box 2. In each AT-box, the positions of the four A's and T's are indicated with subscript numbers. The three distinguished classes of K-boxes (C I, C II, and C III) are indicated with the lengths of their respective spacers. The average AT content of the spacer region is over 60%. (B) Helical representation of a K-box. In this figure, a class I K-box with a spacer of two helical turns is depicted in a helical projection. The dark vertical bars represent the AT base pairs in the AT boxes. The gray circles represent the (A)4 stretches, and the black circles represent the (T)4 stretches in both AT-boxes.
In 2002, three transcriptome studies reported on defining the ComK regulon in B. subtilis 168 (3, 11, 22). In silico analyses demonstrated the presence of over 1,000 putative K-boxes in the genome (one-third in intergenic regions) which could in principle be functional, i.e., up to three base pair deviations from the consensus K-box sequence with a maximum of two deviations per AT-box (9). Based on the three transcriptome studies, it was shown that, under laboratory conditions, only 8% of the genes preceded by a K-box were indeed regulated by ComK, indicating that the sole presence of a K-box is not enough to predict regulation by ComK (11). Probably, additional elements in or near K-boxes are important for the activities of the boxes being regulated by ComK.
As is depicted in Fig. 1, different elements can be distinguished in a K-box, like the spacer between the AT-boxes and the consensus sequence of these boxes. In principle, these characteristics could serve as additional critical elements to determine whether a K-box is regulated and, when regulated, to what level ComK stimulates transcription at this particular box. In the present study, a search for critical determinants in a K-box was performed by investigating the effects of alterations in the lengths and GC contents of the spacers between the AT-boxes. Furthermore, the effects of point mutations in the consensus sequence of the AT-boxes were determined using the K-box of the comG operon as a model, as well as an idealized K-box. This K-box was chosen because the level of transcription of the comG operon is among the highest ComK-activated transcription levels and requires only ComK, minimizing the chance of interference of other regulatory proteins (9, 29). The effects on regulation by ComK were investigated by monitoring transcription activation in vivo using β-galactosidase assays and by determining the DNA-binding affinity of ComK for wild-type and mutant K-boxes in vitro by electrophoretic mobility shift assay (EMSAs). The results show the particular importance of the second thymine-adenine base pair in an AT-box for the regulation of the K-box by ComK, suggesting that this position is crucial for ComK-DNA interactions.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. B. subtilis strains were grown in minimal medium (35) supplemented with erythromycin (5 μg/ml) or spectinomycin (100 μg/ml). Escherichia coli strains were grown in TY medium (26) plus 0.2% glucose, ampicillin (100 μg/ml), or spectinomycin (100 μg/ml). Maltose-binding protein-ComK overexpression was induced with the IPTG (isopropyl-β-d-thiogalactopyranoside; 0.3 mM)-pMal protein fusion and purification system (NEB). Lactococcus lactis strains were grown in a twofold-diluted M17-based medium (Difco) supplemented with 0.5% glucose (GM17) and, if required, erythromycin (4 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant feature(s) | Reference or source |
|---|---|---|
| Strains | ||
| B. subtilis 168 | trpC2 | 1 |
| L. lactis NZ9000 | MG1363 pepN::nisRK | 18 |
| E. coli XL1-Blue | endA1 gyrA96 thi hsdR17 (rK− mK−) supE44 relA1 lac/F′ proAB lacLqlacZΔM15 Tn10 | Stratagene |
| Plasmids | ||
| pNZ8048 | Cmr, inducible expression vector containing the nisA promoter | 18 |
| pMal-ComK | Ampr, contains malE-comK fusion | 30 |
| pWK-Sp | Spr, contains amyE-flanking region for integrations into the amyE locus of B. subtilis 168 | W. K. Smits et al., unpublished |
| pILORI4 | Eryr, pIL252 carrying the multiple cloning site and promoterless lacZ of pORI13 | 20 |
| pILORI-G | Eryr, comG promoter-lacZ fusion | This work |
| pILORI-C1 | Eryr, contains idealized class I K-box | This work |
| pILORI-C2 | Eryr, contains idealized class II K-box | This work |
| pILORI-C3 | Eryr, contains idealized class III K-box | This work |
| pG-b1/2P-SpGC-middle | Eryr, contains perfect comG promoter K-box, with 60% overall GC content in spacer, GCs in middle of spacing | This work |
| pG-b1/2P-SpGC-end | Eryr, contains perfect comG promoter K-box, with 60% overall GC content in spacer and GCs at ends of spacer | This work |
| pG-wt | Eryr, contains wild-type common K-box of comG promoter | This work |
| pG-perfecta | Eryr, wild-type K-box of comG promoter mutated to perfect K-box sequence | This work |
| pG-b2P-b1Ax-Ga | Eryr, K-box with perfect AT-box 1 + AxG mutation in AT-box 2 (x = position 1, 2, 3, or 4) | This work |
| pG-b2P-b1Tx-Ga | Eryr, K-box with perfect AT-box 1 + TxG mutation in AT-box 2 (x = position 1, 2, 3, or 4) | This work |
| pG-b2A2G-b1AxG | Eryr, K-box with A2-G in AT-box 1 + AxG mutation in AT-box 2 (x = position 1 or 3) | This work |
| pG-b2A2G-b1TxG | Eryr, K-box with A2-G in AT-box 1 + TxG mutation in AT-box 2 (x = position 2, 3, or 4) | This work |
| pG-b2T2G-b1Pa | Eryr, K-box with T2G mutation in AT-box 2 as single bp change in a perfect K-box background | This work |
| pG-b2T2G-b1T2Ga | Eryr, K-box with T2--G mutations in both AT-boxes (perfect K-box background) | This work |
Plasmid used as a template to create a plasmid used for integration of the same reporter construct in the chromosome of B. subtilis 168.
DNA techniques, materials, and transformations.
Standard molecular biology methods were used as described previously (2). Enzymes were purchased from Roche, New England Biolabs, or Pharmacia. Radiolabeled nucleotides were obtained from Amersham. For plasmid isolations and PCR product purifications, the High Pure plasmid isolation kit and the High Pure PCR product purification kit (Roche), respectively, were used. B. subtilis was transformed as described by Anagnostopoulos and Spizizen (1). The transformation of L. lactis NZ9000 was achieved by electroporation using a Gene Pulser (Bio-Rad Laboratories, Richmond, CA) as described by Leenhouts and Venema (21).
PCR amplifications and plasmid constructions.
PCRs were performed as described previously (15). For amplification of PCR products, Pwo or Expand polymerase (Roche) was used. Unless specified otherwise, chromosomal DNA of B. subtilis 168 was used as a template. Plasmids and primers used in this study are listed in Table 1 and Table 2, respectively.
TABLE 2.
Primers used for the construction of different K-box mutants
| Plasmid | Primera | Sequenceb |
|---|---|---|
| Basic plasmids | ||
| pILORI-G | comG-start-BamHI | GATCGGATCCTTGATTACCTTTTCTTCTTTTTCTACAATATGCG |
| comG-end-XbaI | GATCTCTAGATTATGCCTCTTCAATCAAGTTTTTGC | |
| pILORI-Cx | oligo-start-EcoRI | GATCGAATTCCATCCGGCTCCGGCAGAATC |
| oligo-end-BamHI | GATCGGATCCAAAACGGCCTTTTGC | |
| Plasmids with mutations in boxes | ||
| pG-wt | comG-AT2-EcoRI | GATCGAATTCAGAATTGGTTTTTCAGCATATAACATCTCAC |
| pG-perfect | comG-perfect | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTC |
| pG-b2P-b1A1G | comG-b1A1G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACGAAATC |
| pG-b2P-b1A2G | comG-b1A2G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAGAATC |
| pG-b2P-b1A3G | comG-b1A3G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAAGATCACG |
| pG-b2P-b1A4G | comG-b1A3G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAAAGTCACG |
| pG-b2P-b1T1G | comG-b1T1G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAAAATCACGGTTTCCCTG |
| pG-b2P-b1T2G | comG-b1T2G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAAAATCACGTGTTCCCTG |
| pG-b2P-b1T3G | comG-b1T3G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAAAATCACGTTGTCCCTG |
| pG-b2P-b1T4G | comG-b1T4G | GATCGAATTCAAAATTGGTTTTTCAGCATATAACATCTCACAAAATCACGTTTGCCCTG |
| pG-b2T2G-b1P | comG-b2T2G | GATCGAATTCAAAATTGGTTGTTCAGCATATAACATCTC |
| Plasmids with mutations in spacer | ||
| pG-b1+2P-spGC-middle | comG-b2P-spGC-middle | GATCGAATTCAAAATTGGTTTTTCAGCACCTGGCATCTCACAAAATCACG |
| pG-b1+2P-spGC-end | comG-b2P-spGC-end | GATCGAATTCAAAATTGGTTTTTCCGCGTATAACGTCTCGCAAAATCACG |
Primers for plasmids with mutations in the boxes and spacer are forward primers which are used in combination with primer comG-end-XbaI.
Underlining indicates the part of the primer that anneals to the chromosome, and italics indicates restriction sites. Boldface nucleotides are mutated.
To investigate the effect of spacer length on regulation by ComK, synthetic DNA sequences containing an idealized K-box were designed. The K-box contained the perfect AT-boxes found in the promoters of addAB (AT-box 1) and comG (AT-box 2). The spacer sequence consisted of random DNA with an AT content similar to that of B. subtilis intergenic regions; 8 (CCCAAGGC), 18 (CTCTACCCAAGGCAGTGC), or 31 (CTCTACCAGCATACCAAGGTATGGTCAGTGC) bp of this random sequence were inserted between the AT-boxes to form a class I, II, or III K-box, respectively. The DNA upstream of the boxes contained 20 bp of DNA located at this position (directly upstream) relative to the AT-boxes in the comC promoter. The downstream DNA arises from the equivalent (downstream) position in the comG promoter. These oligonucleotide DNA fragments were used as templates for PCR amplifications with the primers oligo-start-EcoRI and oligo-end-BamHI, and after being digested with EcoRI and BamHI, they were cloned into the EcoRI/BamHI-digested plasmid pILORI-G, resulting in pILORI-C1, pILORI-C2, and pILORI-C3 as representatives of class I, II, and III K-boxes, respectively. pILORI-G contains a comG promoter-lacZ fusion, made as follows. The promoter region was amplified with the primers comG-start-BamHI and comG-end-XbaI. After digestion with BamHI and XbaI, the PCR fragment was cloned into the BamHI/XbaI-digested plasmid pILORI4, upstream of the promoterless lacZ gene.
Plasmids pG-b1+2P-SpGC-middle and pG-b1+2P-SpGC-end were constructed by amplification of the comG promoter region and K-box with primers comG-b2P-SpGC-middle and comG-b2P-SpGC-end, respectively, and combined with primer comG-end-XbaI. The PCR products were digested with EcoRI and XbaI and ligated into EcoRI/XbaI-digested pILORI4. In both plasmids, the GC content of the spacer is increased to 60%.
To investigate the effect of point mutations in the AT-box sequences, comG-lacZ fusion constructs were generated. The wild-type comG promoter fragment containing the common K-box was amplified with primers comG-AT2-EcoRI and comG-end-XbaI. To obtain an idealized K-box in the comG promoter, the guanine at position 2 in the A stretch of AT-box 2 was mutated into an adenine by amplifying the comG promoter fragment with primers comG-perfect and comG-end-XbaI. In both cases, the PCR products were digested with EcoRI and XbaI and ligated into EcoRI/XbaI-digested pILORI4, resulting in plasmids pG-wt and pG-perfect, respectively. Using the same strategy, point mutations were introduced in the AT-boxes. Plasmids were constructed using the primer combinations listed in Table 2. Single mutations were made in AT-box 1, while AT-box 2 remained perfect. The resulting plasmids were named pG-b2P-b1AxG and pG-b2P-b1TxG, with x referring to the position in the A or T stretch of AT-box 1 which is mutated into a G. Some single mutations were combined with the A2-to-G (A2-G) mutation in AT-box 2 that is present in the wild-type K-box of the comG promoter. In these cases, plasmids containing the single mutations were used as templates for PCR amplification with primers comG-AT2 and comG-end-XbaI, yielding a comG promoter fragment containing two mutations in the K-box. The created plasmids are named pG-b2A2G-b1AxG and pG-b2A2G-b1TxG, again with the x referring to the mutated position. Plasmid pG-b1T2G-b2P with only a T2-G mutation in AT-box 1 was constructed by amplification of the K-box region with primers comG-b1T2G and comG-end-XbaI. The same primers were used to construct plasmid pG-b1T2G-b2T2G, combining the T2-G mutation in AT-box 1 with a T2-G mutation in AT-box 2, using plasmid pG-b1T2G as the template. For all plasmids, cloning was performed in L. lactis NZ9000. Plasmids were checked by sequencing and transformed into B. subtilis 168.
To exclude the possibility that differences in observed transcription levels are due to variations in the numbers of copies of the reporter plasmids, an alternative strategy was developed, using chromosomal integrations for a representative series of mutants. For this purpose, all K-boxes with single-base-pair mutations, the perfect K-box control, and the T2 mutation series were cloned into plasmid pWK-Sp (W. K. Smits et al., unpublished), a cloning vector that allows integrations into the amyE locus of B. subtilis 168. To construct these pWKS plasmid series, the (mutant or perfect) K-box-comG promoter lacZ fragments were obtained by amplification with primers pG_F (5′-CGATGCATGCCATGGTA) and pG_R (5′-GCCACCTTCGTTTTCAGACTTTGC), using the original low-copy-number plasmids with the same mutations as the template (Table 1). The PCR products were digested with EcoRI and HindIII and ligated into EcoRI/HindIII-digested pWK-Sp. The resulting plasmids were transformed into E. coli MC1061 and, after isolation and sequencing, into B. subtilis 168. The obtained clones were verified for the absence of amylase activity on starch plates, indicating integration of the plasmids in the chromosomal amyE locus. All constructs were checked by DNA sequencing.
Transcription activation assays.
Transcription activation by ComK on wild-type and mutant K-boxes upstream of the comG promoter was tested in B. subtilis using fusions with lacZ as a reporter. Cultures were grown in minimal medium to stimulate competence development, and samples were taken from the transition point until 3 to 4 h into the stationary growth phase, at 1-hour intervals. Samples were analyzed for β-galactosidase activity as described by Israelsen et al. (16) and for protein expression levels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (19) and Western blotting (31). ComK was detected with a ComK-specific first antibody (34) and an anti-rabbit horseradish peroxidase-conjugated secondary antibody (Amersham) and visualized by chemiluminescence detection using the ECL Western blotting analysis system (Amersham).
EMSAs.
EMSAs were performed essentially as described previously (33). For this purpose, ComK was purified according to the method of Hamoen et al. (9). Double-stranded DNA probes were amplified with PCR, using the same primer combination as was used for the construction of the plasmids (Table 2). Probes for determining the effect of spacer length were amplified with primers oligo-start-EcoRI and comG-end-XbaI. Probes were end labeled with T4 polynucleotide kinase using [γ-32P]ATP. Proteins and DNA probes were premixed on ice in 20 μl binding buffer [20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 100 mM KCl, 0.5 mM dithiothreitol, 0.05 mg/ml poly(dI-dC), 0.05 mg/ml bovine serum albumin, and 8.7% glycerol]. Samples were incubated for 15 min at 37°C to allow protein-DNA complexes to form. Fifteen microliters of each sample was loaded on a nondenaturing 6% polyacrylamide gel. Gels were run in TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM EDTA) at 100 V, dried, and autoradiographed using phosphor screens. Protein shifts were quantified using the Quantity One software package.
RESULTS
Class III K-boxes are less active than class I and II boxes.
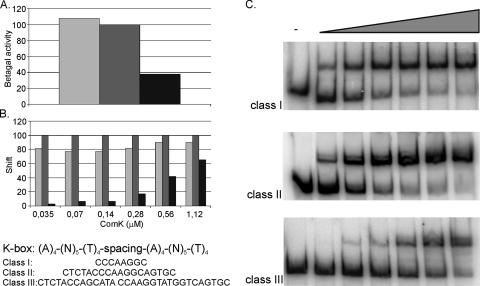
In the B. subtilis genome, three classes of K-boxes can be distinguished based on the length of the spacer between the AT-boxes, which amounts to either 8, 18, or 31 base pairs, corresponding to two, three, or four helical turns separating the start positions of the AT-boxes for class I, II, or III promoters, respectively. These spacers position both AT-boxes on the same side of the DNA helix, thereby enabling interactions between the ComK dimers that each bind to an AT-box (9). To investigate the effect of the spacer length on ComK binding and transcription activation, synthetic DNA fragments were developed and cloned upstream of a comG promoter-lacZ fusion, thereby replacing the original K-box of comG. These DNA fragments contained “perfect” AT-boxes, separated by 8, 18, or 31 random base pairs, resulting in representatives of a class I, II, or III promoter, respectively. β-Galactosidase assays demonstrated that ComK activates the transcription of all three classes but that the level of transcription is lower with a class III K-box (Fig. 2A), while there is little or no difference in transcription activation between a class I and a class II promoter. The same pattern is seen in DNA binding by ComK. Using EMSAs, ComK binding to the DNA of a K-box was investigated for a range of ComK concentrations (Fig. 2B). The percentage of shifted probe was determined for each ComK-DNA combination, demonstrating that the affinity of ComK for a K-box is lowest when the spacer spans four helical turns, while there is only a slight difference between a class I and a class II K-box (Fig. 2C). Binding of ComK at a class III K-box can ultimately result in shift levels comparable to shifts at a class I or II K-box, but only at higher ComK concentrations.
FIG. 2.
Effect of the length of the spacer on the regulation of a K-box by ComK. (A) β-Galactosidase (Betagal) assays were performed to investigate transcription activation by ComK on perfect K-boxes with different spacer lengths, representing a class I, II, or III promoter. The sequences of the spacers used to create a class I, II, or III K-box are depicted underneath the two diagrams. The sample for the depicted β-galactosidase assay was taken after 4 h in the stationary growth phase. β-Galactosidase activities are represented as percentages of the activity for the class II K-box. (B and C) EMSAs were performed to determine DNA binding by ComK to the three classes of K-boxes. Binding was tested for different ComK concentrations. (B) ComK shifts were quantified and are represented as percentages of the shift seen for the class II K-box for every ComK concentration. Light-gray bars, class I K-box; medium-gray bars, class II K-box; black bars, class III K-box. (C) Concentrations are schematically indicated by the wedge. ComK concentrations increased in twofold increments from 0.035 to 1.12 μM. Upper autoradiogram, class I K-box; middle autoradiogram, class II K-box; lower autoradiogram, class III K-box.
As control experiments, Western blotting and plasmid isolations were performed to check the levels of ComK and the plasmid contents for the different strains in all experiments. These assays demonstrated that levels of expression of ComK and plasmid contents are comparable for all strains tested (results not shown), indicating that differences in transcription activation are due to the mutations in the K-boxes and not to variation in ComK expression and/or plasmid copy numbers between the strains.
The spacer GC content does not affect regulation by ComK.
B. subtilis is an AT-rich bacterium, with an average AT base pair content in the genome sequence of about 60%; also, the DNA of the spacer region in a K-box is relatively AT rich. Binding of ComK to a K-box induces a bend in the DNA, which is required for transcription activation, as was shown at the comG promoter (29). In general, AT base pairs are regarded as more flexible in bending than the somewhat more rigid GCs (17, 24), suggesting that the AT content of the spacer region might be an important determinant for regulation by ComK. To investigate this possibility, the AT content was reduced from 60% to 40%, using the comG promoter K-box as a model. In order to focus only on the influence of the spacer sequence, the AT-boxes were changed into perfect consensus sequences. Furthermore, it could be imagined that the position of the GC base pairs along the spacer affects bending abilities and thereby regulation by ComK. To take this possibility into account, two constructs were tested: (i) a construct in which the ends of the spacer were enriched in GC content (the sequence of the spacer was CCGCGTATAACGTCTCGC, with mutations in bold) and (ii) a construct in which the AT-GC substitutions were positioned in the middle of the spacer (the sequence of the spacer was CAGCACCTGGCATCTCAC). Surprisingly, these changes in the spacer sequence did not significantly affect transcription activation by ComK compared with that of a K-box with a wild-type spacer (CAGCATATAACATCTCAC). β-Galactosidase assays showed only a slight increase (110% of the control level) in transcription when the spacer middle was enriched in GC base pairs and a very slight decrease (95% of the control level) when the GCs were introduced at the spacer ends. In addition, EMSAs demonstrated comparable levels of binding of ComK to control K-boxes and spacers in mutant K-boxes (results not shown).
Position T2 in AT-box 1 is critical for activation by ComK.
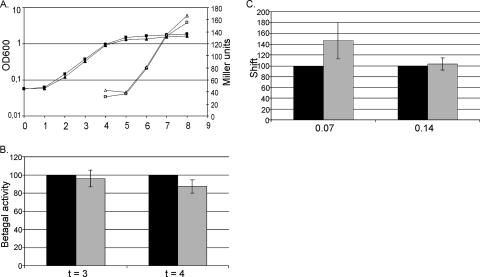
As described above, the characteristics of the spacer region do not determine whether a K-box is used for regulating the transcription of the downstream gene but rather to what extent the ComK-dependent promoter is activated. In a search for possibly other features of a K-box that do determine whether and to what level transcription is regulated from a K-box, the influence of mutations in the consensus sequence of the AT-boxes was investigated. To determine the effect of a single-base-pair mutation on ComK regulation, a clean test system was developed using the class II promoter of the comG operon as a model. In previous research, this promoter was used to investigate the role of ComK in transcription activation, since it is among the promoters most highly activated by ComK and transcription activation is dependent only on ComK (29). The wild-type comG promoter contains an additional AT-box at two helical turns upstream of the start site of the common K-box. Since the goal of this research is to focus on determinants of general importance for ComK regulation, the extra upstream AT-box was omitted and a comG promoter containing only the common K-box was fused with the lacZ gene in plasmid pG-wt. The K-box of comG has one base pair deviation from the consensus sequence, i.e., an A-G mutation at position 2 in AT-box 2. This mutation was repaired in plasmid pG-perfect, creating a perfect, idealized K-box. As shown in Fig. 3A and B, ComK activates transcription at both promoters to comparable levels. Furthermore, ComK showed similar levels of DNA binding on both K-boxes (Fig. 3C).
FIG. 3.
Comparison of the wild-type comG promoter K-box and a perfect K-box. (A) β-Galactosidase assays were performed to determine the levels of transcription activation by ComK at the wild-type K-box of the comG promoter and at an idealized, perfect K-box. Samples were taken from transition point until 4 h into the stationary growth phase. β-Galactosidase activities (open symbols) are shown in Miller units. Levels of the growth of the cultures were measured by determining the optical density at 600 nm (OD600) (filled symbols). Triangles, wild-type K-box; squares, perfect K-box. (B) Average β-galactosidase activities over four experiments were determined and are represented as percentages, with the value for the wild-type K-box set to 100%. The error bars show the standard deviations. The values for the last two time points are depicted, indicating little difference between both K-boxes. (C) EMSAs were performed to investigate DNA binding by ComK at the wild-type and perfect K-boxes. Binding was determined for different ComK concentrations. ComK shifts were quantified and are represented as percentages of the shift seen for the wild-type comG promoter K-box for 0.07 and 0.14 μM ComK. Black bars, wild-type comG promoter K-box; gray bars, perfect K-box.
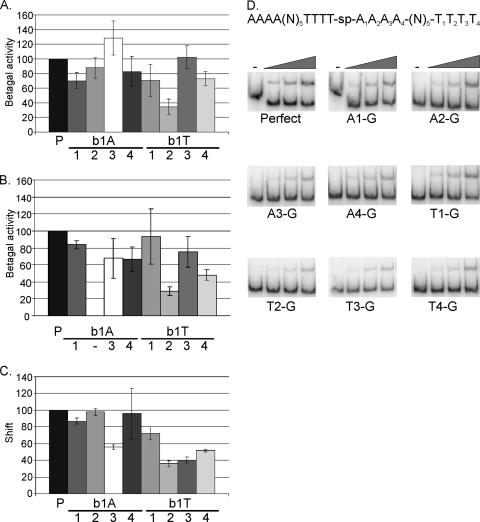
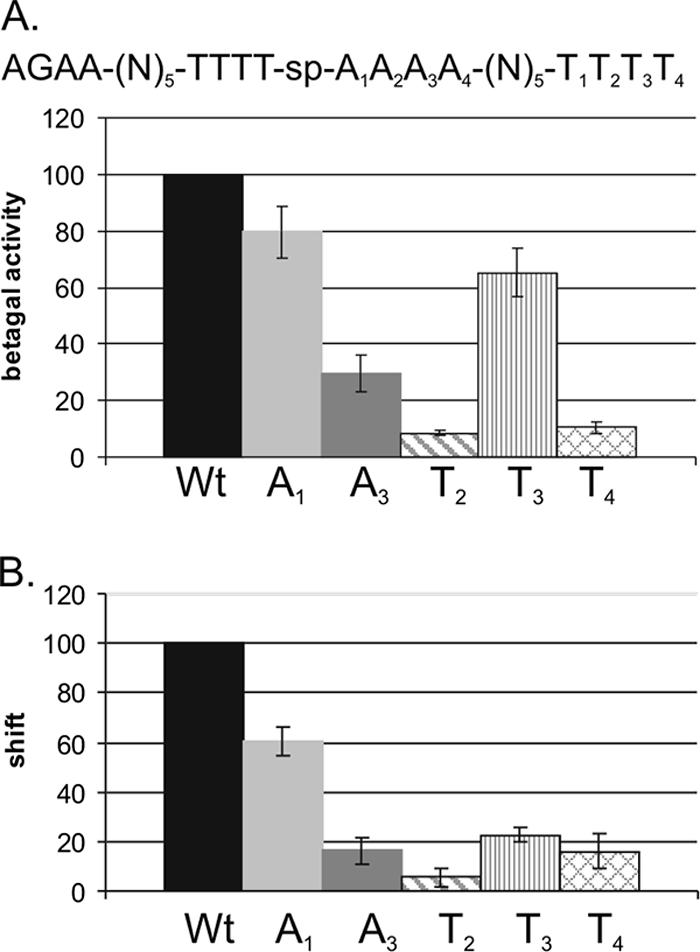
To investigate the effect of single-base-pair mutations in an AT-box on regulation by ComK, the idealized K-box of the comG promoter was subjected to a systematic, box-scanning mutagenesis introducing single G residues at each position in AT-box 1. Mutants were tested for transcription activation by β-galactosidase assays. As shown in Fig. 4A, most single mutations lower the level of transcription; the exceptions are A3-G and T3-G. Whereas mutations at most positions result in a relatively mild decrease of transcription levels to about 60 to 80% of the control level, introduction of a single G at the T2 position has a stronger effect, resulting in a threefold reduction compared to the level of transcription in pG-perfect.
FIG. 4.
Effect of single mutations on transcription activation and binding by ComK. (A) β-Galactosidase assays were performed to determine the effects of single-base-pair mutations on transcription activation by ComK in vivo, using different K-box-comG promoter-lacZ fusion constructs located on low-copy-number plasmids in B. subtilis. For all mutants, β-galactosidase (Betagal) activities are represented as percentages of the level of activity determined for the perfect K-box in the same experiment. The figure depicts the activities measured after 4 h of stationary growth. Error bars representing the standard deviation are included for all single-base-pair mutations. (A and B) P indicates the perfect K-box. Numbers 1 to 4 under “b1A” or “b1T” indicate the position that is replaced by a G in the A or T stretch of AT-box 1 (b1; the right AT-box, in which the A's and T's are named Ax or Tx) of the perfect K-box, used as the control in this experiment. (B) Details are the same as described for panel A, but in this case, the K-box-comG promoter-lacZ fusion constructs used were integrated into the amyE locus on the chromosome of B. subtilis. The A2-G mutant is missing, because the required mutation was not obtained. (C and D) To determine the effect of single-base-pair mutations on DNA binding by ComK, EMSAs were performed. ComK shifts for the mutant K-boxes were quantified and are represented as percentages of the shifts for the perfect K-box at different ComK concentrations. The standard deviation determined for the average over multiple experiments are depicted for each mutant. Quantifications are shown for the binding reaction with 0.07 μM ComK. For each gel in panel D, results for a blank (−) and three samples with ComK concentrations increasing in twofold increments from 0.018 to 0.07 μM are shown (indicated by the wedges). Perfect, the perfect K-box; Ax-G, A-G substitution at position x in the A-stretch of AT-box 1 (right AT-box); Tx-G, T-G substitution at position x in the T stretch of AT-box 1.
To exclude the possibility that the observed effects are caused by differences in the numbers of copies of the plasmids rather than that they reflect actual differences in ComK-dependent gene regulation, the same experiment was performed using K-box promoter-lacZ constructs integrated into the amyE locus of B. subtilis 168 in such a way that the reporters are present only in a single copy in each cell. For the A2-G mutation, a good construct was not obtained, so this K-box is not included in chromosomal integrations. For the other K-box mutants, the effects observed with the integrated reporters were similar to the effects obtained using the K-box promoter-lacZ constructs located on the low-copy-number plasmid (Fig. 4B). Relatively mild effects can be seen for most mutations, again with the exception of T2-G, which clearly reduces transcription activation from the K-box. This test confirms that the effects on transcription activation are caused by the single-base-pair mutation and not by potential copy number differences of the plasmids.
In addition to affecting transcription activation, single mutations in a K-box also affect DNA binding by ComK (Fig. 4C and D). The binding effects for each position usually correlate rather well with the effect seen on transcription, although binding at the A3-G mutation K-box is more comparable to transcription determined with the integrated reporter than that determined with the plasmid-located reporter. Except for T1, the T-stretch mutations show a clear reduction in binding by ComK. Like the effect of the mutation on transcription, a T-G substitution on position T2 shows the largest effect on ComK binding, indicating that this base pair position is most important for ComK-DNA interactions.
Transcription of and binding to constructs with double mutations in a K-box.
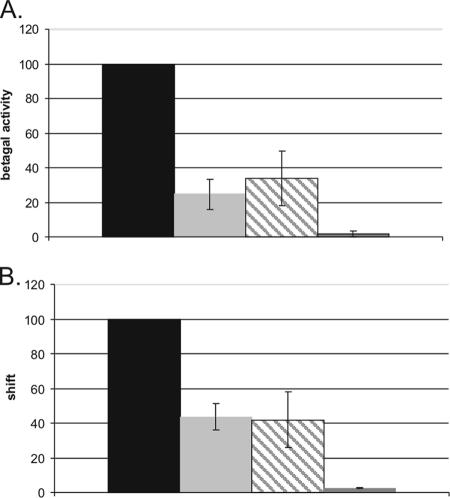
For further investigation of the importance of the consensus sequence of the AT-boxes, constructs with double mutations in a K-box were created. For this purpose, single mutations in AT-box 1 producing either a wild-type, an intermediate, or a low level of transcription activation were combined with the natural mutation found in the wild-type comG promoter K-box, i.e., A2-G in AT-box 2. Double mutants were tested for transcription activation and ComK binding and compared with the wild-type comG promoter in pG-wt. Mutants with mutations at A1 and T3 were the least affected, showing a relatively mild decrease in transcription to 60 to 80% of the wild-type level, while the observed transcription levels for mutants with mutations at positions T2 and T4 in AT-box 1, combined with the A2-G mutation in box 2, were severely reduced to at most 15% of the wild-type level (Fig. 5A). Similar effects could be seen on ComK binding, although these effects seem to be somewhat stronger than on transcription (Fig. 5B). This is especially clear for the mutant with the combination of A2-G in AT-box 2 and T3-G in AT-box 1, in which DNA binding affinity is only near 20% of the wild-type level, although transcription activation is still over 60% compared with that of the wild type. Apparently, a decrease in binding affinity does not necessarily result in a similar reduction in transcription activation.
FIG. 5.
Effect of double mutations on transcription activation and binding by ComK. (A) Five single AT-box mutants were combined with an A-G substitution at position 2 in AT-box 2, the mutation seen in the wild-type comG promoter. β-Galactosidase (betagal) assays were performed to determine the effects of those mutations on transcription activation by ComK in vivo. The activities determined 4 h into the stationary growth phase are shown. β-Galactosidase activities and standard deviations are determined as described for Fig. 4, in this case relative to the activity in the wild-type-comG promoter K-box. (B) To determine the effect of the mutations on DNA binding by ComK, EMSAs were performed for different concentrations of ComK. Quantifications were performed as described for Fig. 4, and values are relative to that determined for the wild-type comG promoter K-box, used as a control (standard deviations are included for each mutant). Results are shown for the binding reaction with 0.07 μM ComK. Wt indicates the wild-type comG promoter K-box (the sequence of which is depicted above the graph in panel A; this K-box was used as the control in this experiment). With Ax or Tx, x indicates the position that is replaced by a G in the A or T stretch, respectively, of AT-box 1 (right AT-box), which is in addition to the A2-G substitution in AT-box 2 (left AT-box).
Mutation of both T2s almost completely inactivates a K-box.
As mentioned in the introduction, the B. subtilis genome contains over 1,000 putative K-boxes, with a wide variety of deviations from the consensus sequence. Because of this natural variation, big effects of single mutations in a K-box were not expected. However, as shown in Fig. 4, a mutation at position T2 in AT-box 1 severely disturbs both transcription activation and DNA binding by ComK. For further investigation of the importance of this position for ComK-DNA interactions, K-boxes with mutated T2 positions in AT-box 1, AT-box 2, or both AT-boxes were tested for DNA binding and transcription activation by ComK. As is shown in Fig. 6, a T2-G mutation in AT-box 2 clearly reduces transcription activation and DNA binding by ComK, compared to wild-type or pG-perfect levels, but reduction is slightly less than with the same mutation in AT-box 1. A combination of mutations at both T2 positions, however, leads to an almost complete loss of transcription activation and DNA binding by ComK.
FIG. 6.
Importance of T2 positions in both AT-boxes for regulation by ComK. (A) β-Galactosidase (betagal) assays were performed to determine the effect on transcription activation by ComK of a single T-G substitution at position 2 in AT-box 1, in AT-box 2, and in both AT-boxes, using plasmid-borne reporters. β-Galactosidase activities are represented as percentages of the activity in a perfect K-box. The results are depicted for 4 h after entry into the stationary growth phase. Error bars were determined as described for Fig. 4 and 5. (B). EMSAs were performed to determine the effects of the mutations on DNA binding by ComK. Shifts were quantified and are represented as percentages of the shift determined for the perfect K-box. The averages and standard deviations were calculated and are depicted for the binding reaction with 0.07 μM ComK. Black bars, perfect K-box; light-gray bars, AT-box 1 with a T2-G substitution and a perfect AT-box 2; hatched bars, AT-box 2 with a T2-G substitution and a perfect AT-box 1; dark-gray bars, both AT-boxes with T2-G substitutions.
To corroborate these results, the assay was repeated using chromosomal integrations of the tested mutations, using the same strategy as with the single-base-pair mutations in the K-box consensus sequence (results not shown). The outcome of this experiment confirms the results obtained with the reporters on low-copy-number plasmids (Fig. 6A).
In silico analyses of ComK-regulated K-boxes.
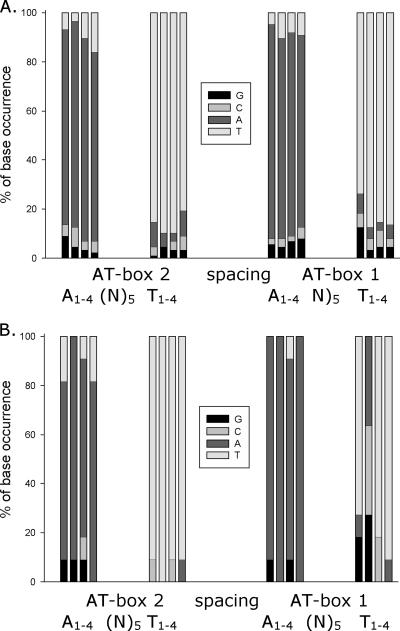
In 2002, three transcriptome studies of the ComK regulon were reported, providing an overview of natural transcription activation levels at K-boxes with various deviations from the consensus sequence (3, 11, 22). In that study, the ComK-regulated genes found in all three studies were grouped on the basis of their level of activation by ComK (strong to weak). Some ComK-dependent genes are organized in operons, meaning that the transcription of more genes depends on ComK binding to one K-box upstream of the entire operon. In this study, only unique K-boxes were included by taking into account only the most highly activated (n-fold) gene from an operon and assigning the transcription activation level for this gene to the K-box of the entire operon. The resulting list contained 88 ComK-regulated genes and was used as a reference provided by B. subtilis itself to determine the occurrence of mutations at each AT-box position. However, it should be mentioned that the possibility that some of the genes are regulated by additional factors (other than ComK) cannot be excluded. The ranking of the transcription activation levels, therefore, does not necessarily depend only on the occurrence of mutations in the K-box of a particular promoter but can also reflect influences of additional regulation. A weight matrix based on the sequences of all of these 88 ComK-regulated K-boxes shows a wide variety of mutations throughout the complete K-box (Fig. 7A), indicating that there are no positions where a mutation is never tolerated. This correlates with the previous knowledge that individual K-boxes with up to three deviations from the consensus sequence can still be good targets for regulation by ComK. Eleven out of these 88 ComK-activated genes were shown to contain a T-N substitution at position 2 in AT-box 1. A weight matrix based on these K-boxes clearly demonstrated that a concomitant mutation at the other T2 position in AT-box 2 is never seen among ComK-regulated boxes (Fig. 7B).
FIG. 7.
Weight matrices demonstrating natural variation in ComK-regulated K-boxes. (A) Weight matrix based on the sequences of 88 ComK-regulated K-boxes. On the x axis, the two AT-boxes, composing a K-box, are shown. In order to align the AT-boxes for the three classes of K-boxes, the spacer was in all cases set to the same length. For each position in the AT-boxes, which by consensus contain four A's and four T's, the percentage of occurrence of each of the four possible nucleotides among the 88 ComK-regulated K-boxes was calculated. This percentage is shown on the y axis. (B) Weight matrix based on the sequence of 11 out of the 88 ComK-regulated K-boxes, which contain a T-N substitution at position 2 in AT-box 1.
When, on the other hand, the non-ComK-regulated putative K-boxes with a mutation at the T2 position of AT-box 1 were used to build a weight matrix, mutations at the T2 position in the other AT-box could be observed (results not shown). The DNA of B. subtilis 168 contains 19 putative K-boxes with a T2-N mutation in both AT-boxes. However, the three transcriptome studies that have been published did not show ComK-dependent regulation of downstream genes from any of these K-boxes. These results show a very good correlation with the experimental data from this study, strongly suggesting that mutations occurring simultaneously at both T2 positions in one K-box result in the inactivation of this box.
DISCUSSION
The competence transcription factor ComK regulates gene transcription by binding to specific sequences, the so-called K-boxes, located upstream of promoters of regulated genes. Previous research determined a defined consensus sequence for ComK binding (9) but also demonstrated that a large degree of natural variation is allowed in ComK-regulated boxes. The most striking characteristic of K-boxes is the natural variation in spacer length, consisting of two, three, or four helical turns between the start positions of the two AT-boxes of a K-box.
Furthermore, K-boxes with up to three deviations from the consensus sequence have been shown to be activated by ComK, resulting in gene transcription. In the present study, an attempt was made to elucidate the role of the different elements of a B. subtilis K-box in regulation by ComK, which may explain why at least some of the unregulated putative K-boxes (92% of the total number of K-boxes) are not used by ComK to activate gene transcription.
It was demonstrated that the difference in the lengths of the spacers between the AT-boxes, as seen in the three classes of K-boxes, influences the levels of transcription activation and DNA binding by ComK (Fig. 2). In correlation with results of DNA arrays (11), it was shown that ComK activates transcription at a class III K-box to a lower level than at class I and II K-boxes, likely because binding to class III K-boxes requires higher ComK concentrations to achieve shifts similar to those obtained for the other two classes (Fig. 2B). A natural example of a class III K-box is found at the comK promoter. Of this K-box, it is known that efficient regulation by ComK requires the binding of DegU to prime ComK binding and transcription activation at the onset of competence development, when ComK concentrations in the cell are still low (10). It might be that additional regulators are also involved in other class III K-boxes to stimulate transcription activation by ComK.
Previous research demonstrated that binding of ComK results in DNA bending of the promoter region, as was shown at the comG and comF promoters, where bending of 60 to 70° was determined (9). DNA bending is known to be easier for the more-flexible AT-rich sequences than for GC combination sequences (17, 24), but surprisingly, increasing the GC content of the spacer from <40 to 60% did not significantly influence regulation by ComK, nor did the positioning of the GC base pairs throughout the spacer region. Apparently, the sequence of the spacer is not an important determinant for the level of regulation by ComK.
In contrast to the spacer, the consensus sequence of the AT-box repeats is more critical for determining whether or not a K-box has a high chance of being ComK regulated, as was determined by the introduction of an A-G or T-G mutation in one AT-box in a further idealized K-box background. In the setup of mutational scanning of a K-box, it should be noted that replacement of a T by a G might have a larger effect than replacement of an A by a G, since in the latter situation the base pair remains a purine, while mutation from a T to a G also implies a change from a pyrimidine to a purine. Indeed, our study reveals larger effects on T stretches than on A stretches (Fig. 4), so it should be noted that the latter effects might be an underestimate of the importance of the positions in the A stretch.
Using single-base-pair mutational scanning of a K-box, it was shown that, although large natural variations are allowed, a single replacement of the second thymine residue in AT-box 1 by guanine decreased transcription activation about threefold compared to that in a perfect K-box (Fig. 4). The reduction in transcription activation could be explained entirely by a decrease in DNA binding, which was reduced to a similar level, namely, 30% of the control K-box level. This is, however, not the case for all positions in the consensus sequence of a K-box, as for example in the case of a T3 or A3 mutation in AT-box 2 (Fig. 4). In both cases, binding is more affected than transcription activation. It might be that the level of ComK binding is reduced, but that it is still enough to drive transcription activation to the same level as in the control K-box or, for the A3-G mutant, to an even higher level. However, for most positions in the AT-box, binding and transcription activation by ComK are affected to similar extents.
Since a single mutation at the T2 position of a K-box was shown to most strongly reduce the activity of this box from regulation by ComK, additional experiments were performed to investigate the importance of the T2 positions. Transcription activation and DNA-binding assays demonstrated that introduction of a second T2 mutation in the K-box reduced transcription and DNA binding to below 5% of control levels, suggesting that a combination of mutations at both T2s is not allowed in ComK-regulated boxes (Fig. 6). This result was corroborated by in silico analyses demonstrating the absence of double T2 mutations among K-boxes upstream of ComK-regulated genes (Fig. 7). In contrast, for all other individual positions in an AT-box, mutations at the same positions in both boxes could be found among ComK-activated K-boxes. In addition to the base pair at the T2 position in AT-box 1, the base pairs at positions A2 and A4 in AT-box 2 are conserved when there is a T2 mutation in AT-box 2 (Fig. 7B). Whether this is of biological importance or just due to the relatively low number of boxes with a T2 mutation occurring in vivo among ComK-activated K-boxes cannot be concluded from this study.
A possible way of maintaining the activity of a K-box with a T2 mutation could be the introduction of compensating mutations in other base pairs in the AT-boxes. Often, protein-binding sites display direct or inverted symmetry in the recognition site, as was demonstrated for the E. coli cyclic AMP receptor protein, the arginine repressor ArgR, and the lac repressor LacI (4, 7, 23, 25). In the case of the K-box, the two AT-boxes form direct repeats separated by a spacer, and within each AT-box, an inverted repeat is formed by the A and T stretches. Potential candidates for compensating mutations would therefore be the very important T2 in the other AT-box or the A3 position in the same AT-box as the T2 mutation. However, in silico analyses did not clearly point out conserved mutations in AT-box base pairs, which could be introduced to maintain the activity of a K-box with a T2 mutation, as can be seen in Fig. 7B.
The most interesting question with respect to the importance of the T2 positions for regulation by ComK is what could be the reason for its dominating role. We demonstrated that a replacement of T2 by a guanine lowers the efficiency of ComK regulation by reducing ComK binding to the DNA. The extent of shift is not changed, so alteration of the T2 position does not affect, for example, the oligomerization of ComK on the DNA. Although the mechanism behind the reduced DNA binding by ComK at a K-box with a T2 mutation is unclear, it is likely that the T2 position is the residue that is in closest contact with or has the highest affinity for ComK in protein-DNA interactions. It is tempting to speculate that the positioning of the K-box along the DNA helix does not allow a guanine at the second position in the thymine stretches. It might be that a G/C base pair affects the minor groove of the DNA helix, thereby decreasing the binding efficiency of ComK at the K-box. This hypothesis is supported by research on the Arg-boxes bound by the arginine repressor protein in E. coli. Wang et al. (36) demonstrated that this repressor contacts the DNA at four major and two minor grooves of a long consensus sequence (Arg-box). The minor groove in the center of the Arg-boxes faces the repressor protein and was shown to contain almost exclusively A/T base pairs, suggesting that G/C combinations are not allowed in this region. They suggest that these base pairs would introduce an inhibitory group into the narrow minor groove, thereby inhibiting binding of the Arg repressor. Furthermore, they remark that there is growing evidence that the methyl groups of thymine containing base pairs are often essential for protein-DNA interactions. Although the present study describes a first, general overview of the important base pairs in a K-box, the three-dimensional structure of the protein-DNA complex should preferably be elucidated to unravel the exact nature of ComK-DNA interactions at the site of a K-box.
Acknowledgments
We thank Fabrizia Fusetti and Andy-Mark Thunnissen for helpful discussions and Aldert Zomer for help with the in silico analyses.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidham, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 3.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 4.Charlier, D., M. Roovers, F. van Vliet, A. Boyen, R. Cunin, Y. Nakamura, N. Glansdorff, and A. Pierard. 1992. Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression. J. Mol. Biol. 226:367-386. [DOI] [PubMed] [Google Scholar]
- 5.D'Souza, C., M. M. Nakano, and P. Zuber. 1994. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 91:9397-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubnau, D. 1993. Genetic exchange and homologous recombination, p. 555-584. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 7.Frank, D. E., R. M. Saecker, J. P. Bond, M. W. Capp, O. V. Tsodikov, S. E. Melcher, M. M. Levandoski, and M. T. Record, Jr. 1997. Thermodynamics of the interactions of lac repressor with variants of the symmetric lac operator: effects of converting a consensus site to a non-specific site. J. Mol. Biol. 267:1186-1206. [DOI] [PubMed] [Google Scholar]
- 8.Hamoen, L. W., H. Eshuis, J. Jongbloed, G. Venema, and D. van Sinderen. 1995. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 15:55-63. [DOI] [PubMed] [Google Scholar]
- 9.Hamoen, L. W., A. F. van Werkhoven, J. J. E. Bijlsma, D. Dubnau, and G. Venema. 1998. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 12:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamoen, L. W., A. F. van Werkhoven, G. Venema, and D. Dubnau. 2000. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:9246-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamoen, L. W., D. Kausche, M. A. Marahiel, D. van Sinderen, G. Venema, and P. Serror. 2003. The Bacillus subtilis transition state regulator AbrB binds to the −35 promoter region of comK. FEMS Microbiol. Lett. 218:299-304. [DOI] [PubMed] [Google Scholar]
- 13.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 14.Hoa, T. T., P. Tortosa, M. Albano, and D. Dubnau. 2002. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43:15-26. [DOI] [PubMed] [Google Scholar]
- 15.Innes, M. A., and D. H. Gelfand. 1990. Optimization of PCRs, p. 3-12. In M. A. Innes, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 16.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koo, H., H. Wu, and D. M. Crothers. 1986. DNA bending at adenine-thymine tracts. Nature 320:501-506. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Larsen, R., G. Buist, O. P. Kuipers, and J. Kok. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leenhouts, K. J., and G. Venema. 1993. Plasmids, a practical approach, p. 65-94. Oxford University Press, Oxford, United Kingdom.
- 22.Ogura, M., H. Yamaguchi, K. Kobayashi, N. Ogasawara, Y. Fujita, and T. Tanaka. 2002. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neill, M. C., K. Amass, and B. de Crombrugghe. 1981. Molecular model of the DNA interaction site for the cyclic AMP receptor protein. Proc. Natl. Acad. Sci. USA 78:2213-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez-Martín, J., F. Rojo, and V. de Lorenzo. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadler, J. R., H. Sasmor, and J. L. Betz. 1983. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl. Acad. Sci. USA 80:6785-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., F. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon, J., R. Magnuson, A. Srivastava, and A. D. Grossman. 1995. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 9:547-558. [DOI] [PubMed] [Google Scholar]
- 29.Susanna, K. A., A. F. van der Werff, C. D. den Hengst, B. Calles, M. Salas, G. Venema, L. W. Hamoen, and O. P. Kuipers. 2004. Mechanism of transcription activation at the comG promoter by the competence transcription factor ComK of Bacillus subtilis. J. Bacteriol. 186:1120-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susanna, K. A., F. Fusetti, A.-M. W. H. Thunnissen, L. W. Hamoen, and O. P. Kuipers. 2006. Functional analysis of the competence transcription factor ComK of Bacillus subtilis by characterization of truncation variants. Microbiology 152:473-483. [DOI] [PubMed] [Google Scholar]
- 31.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor (CTF), the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 34.Van Sinderen, D., and G. Venema. 1994. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J. Bacteriol. 176:5762-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venema, G., R. H. Pritchard, and T. Venema-Schroder. 1965. Fate of transforming deoxyribonucleic acid in Bacillus subtilis. J. Bacteriol. 89:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, H., N. Glansdorff, and D. Charlier. 1998. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J. Mol. Biol. 277:805-824. [DOI] [PubMed] [Google Scholar]