Abstract
Genetic differentiation by natural selection is readily observed among microbial populations, but a more comprehensive understanding of evolutionary forces, genetic causes, and resulting phenotypic advantages is not often sought. Recently, a surface population of Pseudomonas putida bacteria was shown to evolve rapidly by natural selection of better-adapted variants in a mixed-species biofilm consortium (S. K. Hansen, P. B. Rainey, J. A. Haagensen, and S. Molin, Nature 445:533-536, 2007). Adaptation was caused by mutations in a wapH homolog (PP4943) involved in core lipopolysaccharide biosynthesis. Here we investigate further the biofilm physiology and the phenotypic characteristics of the selected P. putida rough colony variants. The coexistence of the P. putida population in a mixed-species biofilm with Acinetobacter sp. strain C6 is dependent on the benzoate excreted from Acinetobacter during the catabolism of benzyl alcohol, the sole carbon source. Examination of biofilm development and the dynamics of the wild-type consortium revealed that the biofilm environment became oxygen limited, possibly with low oxygen concentrations around Acinetobacter microcolonies. In contrast to P. putida wild-type cells, which readily dispersed from the mixed-species biofilm in response to oxygen starvation, the rough variant cells displayed a nondispersal phenotype. However, in monospecies biofilms proliferating on benzoate, the rough variant (like the wild-type population) dispersed in response to oxygen starvation. A key factor explaining this conditional, nondispersal phenotype is likely to be the acquired ability of the rough variant to coaggregate specifically with Acinetobacter cells. We further show that the P. putida rough variant displayed enhanced production of a cellulose-like polymer as a consequence of the mutation in wapH. The resulting phenotypic characteristics of the P. putida rough variant explain its enhanced fitness and ability to form tight structural associations with Acinetobacter microcolonies.
It is frequently claimed that the vast majority of bacteria in natural settings live in surface-associated communities, also referred to as biofilms, and that such communities thrive on the presence of nutrients originating from the surfaces themselves or from the surrounding water (6, 12). Since most of these communities comprise multiple species, there will obviously be many instances of competition, commensalism, mutualism, synergy, parasitism, and other interactions between community members, and many of these will be driven by metabolic activities (7, 23, 32, 45, 49, 58). In structured communities, the heterogeneous distribution of biomass and of the various populations may allow the rapid development of nutrient gradients or opportunities for metabolic interactions, the consequence of which is that the actual spatial positioning of the various bacterial populations in relation to each other becomes an important ecological parameter (7, 19, 23, 46). In several documented cases, coaggregation of bacteria of different species seems to have evolved as an efficient strategy to optimize local opportunities (46). The best-studied case is that of the complex oral flora associated with dental plaque, where a very large number of species interact with each other to form a structured network of cells, which build up dental biofilms through numerous coaggregation events (30, 31). Some of these events seem to reflect metabolic interactions, thus documenting the assumptions indicated above. In the case of dental plaque, the microbial community is probably composed of the same groups of bacteria, which are naturally associated with the general oral flora, and many specific interactions, including coaggregation, have most likely evolved over a very long time. It is therefore not surprising that quite specific adherence factors have been identified as connecting cells of certain species to receptors present on surfaces of other specific cells (9, 10, 41). In other environments, where the conditions change more frequently, more stochastic interactions may be the rule, and in such cases specific coaggregation interactions may in fact be counterproductive. Instead, a diverse repertoire of structural interactions/coaggregations may be much more beneficial due to the flexibility provided by such ad hoc solutions. One such example of ad hoc association between two organisms has been characterized previously (7). A close association between environmental isolates of Pseudomonas putida R1 and Acinetobacter sp. strain C6 growing in flow chamber biofilms, with benzyl alcohol as the sole carbon and energy source, was observed. The driving force behind P. putida colonization of the Acinetobacter colonies seemed to be partial catabolism of benzyl alcohol by the latter organism resulting in excretion of benzoate, which was readily exploited by P. putida (37).
The flow chamber-based model system for studying biofilm formation and dynamics has the advantage of allowing online microscopic investigations of adaptive processes involving genetic changes (23). Evolved and wild-type (wt) genotypes can be reintroduced to the biofilm environment, and biofilm formation can be investigated under strictly controlled conditions (4). Eventually, such investigations may contribute to the understanding of the adaptation dynamics of more complex structured communities. Recently, we described the occurrence of evolution in a mixed-species biofilm consortium consisting of Acinetobacter sp. strain C6 and the laboratory strain Pseudomonas putida KT2440 growing on benzyl alcohol as the sole energy and carbon source (23). In this case, the P. putida strain was unable to degrade benzyl alcohol and was therefore totally dependent on the excreted benzoate. Genetic variants of P. putida appeared reproducibly, and the cause of this divergence was found (for a set of variants) to be mutations in a single gene (PP4943) homologous to the wapH gene of Pseudomonas aeruginosa PAO1 (35). These mutants had truncated core lipopolysaccharide (LPS) with the O antigen lacking (23) and a rough colony morphology, as often observed for core LPS variants (44). The P. putida rough colony variant was better adapted to the mixed-species biofilm environment, as shown by competitive fitness assays with the P. putida wt genotype. In addition, under conditions of very low levels of cross feeding, the derived variant was able to coexist with the Acinetobacter population, in striking contrast to the P. putida wt strain.
In the present study, we further investigate the structural interactions between P. putida KT2440 cells and Acinetobacter microcolonies in flow cell biofilms. We show that the consortium environment rapidly becomes oxygen limited, possibly creating conditions of low oxygen concentrations around Acinetobacter microcolonies. We further demonstrate that the P. putida wt population detaches from the biofilm in response to an oxygen downshift, indicating a possible explanation for the lacking structural interactions. In contrast, the P. putida rough variant displays a nondispersal phenotype in the mixed-species environment and forms coaggregates with Acinetobacter. Finally, we show that the P. putida rough variant has enhanced production of a cellulose-like polymer as a consequence of the core LPS mutation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The mixed-species biofilm consortium consisted of Acinetobacter sp. strain C6 (7) and derivatives of Pseudomonas putida KT2440 (17) without the TOL plasmid. The P. putida strains were the wt strain KT2440, with a gfp cassette inserted into the chromosome (SKH132), and a rough colony morphology variant (SKH208) derived from SKH132 by propagation for 10 days in a P. putida-Acinetobacter flow chamber biofilm (23). The rough variant was shown to have a spontaneous mutation in the wapH gene (PP4943) (TIGR Comprehensive Microbial Resource database [www.tigr.org]), which has a role in core LPS formation (23). In situ growth activity was visualized in the mixed-species biofilm population by using P. putida wt strain KT2440 with a mini-Tn7-Gmr-rrnBP1-gfp[AAV] cassette (34) inserted into the chromosome (AKN142). Acinetobacter sp. strain C6 was grown at 30°C in Luria-Bertani (LB) broth or on LB agar. P. putida strains were grown at 30°C in AB minimal medium (11) supplemented with citrate (40 mM in agar or 10 mM in broth). Biofilms were cultivated using FAB medium (AB minimal medium with 10 μM Fe-EDTA replacing FeCl3) containing benzyl alcohol (500 μM; Merck, Darmstadt, Germany) or benzoate (200 μM; Sigma Chemical Co., St. Louis, MO). FAB medium is composed of 1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM Fe-EDTA, 15 mM (NH4)2SO4, 33 mM Na2HPO4, 22 mM KH2PO4, and 51 mM NaCl. When required, antibiotics were added at final concentrations of 100 μg/ml of streptomycin, 10 μg/ml of gentamicin, and 10 μg/ml of kanamycin.
Cultivation of biofilms.
Biofilms were grown in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm. The flow system was assembled and prepared as described previously (8), with the modification of washing the system after sterilization with sterile milliQ water overnight. The substratum consisted of a microscope glass coverslip (st1; Knittel Gläser, Braunschweig, Germany). Each channel was supplied with a continuous flow of FAB medium containing the relevant carbon source. For propagation of mixed-species biofilm populations, flow cells were inoculated with a mixture of overnight cultures of Acinetobacter sp. strain C6 and P. putida KT2440 (wt or rough variant) diluted in a 0.9% NaCl solution. For monospecies biofilms, overnight cultures of the P. putida KT2440 wt or rough variant were used for inoculation. With arrested medium flow, the flow cells were turned upside down, and 250 μl of the diluted mixture was injected into each flow channel, using a small syringe. After 1 h, the flow cells were turned upside down, and the flow was resumed at a constant flow rate of 3.3 ml/h, using a Watson Marlow 205S peristaltic pump (Watson Marlow Inc., Wilmington, MA). After inoculation, each flow chamber contained ∼2 × 106 CFU of Acinetobacter and ∼2.5 × 105 CFU of P. putida (wt or rough variant) for mixed-species biofilms and ∼2.5 × 105 CFU of P. putida for monospecies biofilms. The mean flow velocity in the flow cells was 0.2 mm/s. Biofilms were grown at 24°C. When possible, Acinetobacter sp. strain C6 was visualized prior to image acquisition by staining the biofilm with a 0.1% solution of Syto62 (Molecular Probes Inc., Eugene, OR) in FAB medium containing 500 μM benzyl alcohol. The staining was allowed to progress for 15 min without arresting the flow to avoid biofilm detachment of the P. putida strain. Using this relatively short staining time, P. putida cells were stained at a relatively low level compared to Acinetobacter cells.
Microscopy and image analysis of biofilms.
All microscopic observations and image acquisitions were performed on a Zeiss LSM510 confocal laser scanning microscope (CSLM; Carl Zeiss, Jena, Germany) equipped with an argon-krypton laser and with detectors and filter sets for monitoring green fluorescent protein (GFP) and Syto62 and for the recording of reflection (light) images. Images were obtained using a 63×/1.4 Plan-APOChromat differential interference contrast objective or a 40×/1.3 Plan-Neofluor oil objective. Multichannel simulated fluorescence projection (SFP) images, vertical xz sections through the biofilms, and simulated three-dimensional (3D) images were generated by using the IMARIS software package (Bitplane). This software was used to remove the Syto62 signal from the GFP-fluorescent P. putida cells. Images were further processed for display by using Photoshop software (Adobe, Mountain View, CA).
Biofilm images of the mixed-species consortia were obtained to quantify biomass as described previously (23), using COMSTAT software. Twelve images from three independent biofilms were analyzed for each time point.
In order to quantify the degrees to which both the P. putida rough variant and the wt associated with the Acinetobacter microcolonies, specific 3D image algorithms were developed for calculating distributions of the distances from the surfaces of the P. putida cells to the surfaces of the Acinetobacter microcolonies/cells. In order to segment the biofilms from the background, the biofilm images of both Acinetobacter and P. putida were subjected to a threshold, using the Otsus method (40). In addition, morphological filtering (the so-called AreaOpen operation) was used to eliminate small colonies of background noise and interference noise between the channels. Additional image layers were then introduced by bilinear interpolation in order to obtain a voxel size with equidistant edges. Next, a 3D distance map defining the nearest distance from any voxel in the considered 3D mesh to the surfaces of P. putida cells was calculated by use of a previously described 3D Euclidean distance transform (60). The distance values belonging to voxels overlapping with the surface regions of the Acinetobacter biofilm were then collected to produce the distribution of distances between strains. A minimum of eight images from two independent biofilms were analyzed for each time point.
Oxygen upshift and downshift experiments.
A method was developed to control the oxygen concentration in the inflowing biofilm medium. Due to the high oxygen permeability of the silicone tubing feeding the flow channel, it was possible to control the oxygen concentration in the inflowing medium. This was done by passing the medium through 5 m of silicone tubing submerged in a water container purged with either nitrogen gas (99.8% N2; Hede Nielsen, Denmark) or oxygen gas (99.5%; Hede Nielsen, Denmark) prior to its entering the flow channel. Oxygen concentrations were measured using a microelectrode with a 0.5-mm tip (Unisense OX500) connected to an ampere meter with a built-in polarization source (Unisense PA2000). Before every set of measurements, a calibration curve was obtained by determining the value at zero oxygen (water vigorously purged with nitrogen gas) and the value for water saturated with air (which corresponds to approximately 250 μM oxygen at 24°C and 0.9% NaCl [18]).
Oxygen downshift experiments (one step down) were performed on 2-day-old biofilms consisting of mixed-species Acinetobacter and P. putida populations or a monospecies P. putida population grown as described above. To monitor downshift experiments online using scanning confocal microscopy, the flow cell was connected to the submerged silicone tubing by a glass tube to prevent influx of oxygen. The biofilm populations were fed with air-saturated biofilm medium until the desired time point for the downshift, shifted to the oxygen-stripped medium at time zero, and subsequently followed over time. In order to efficiently strip the oxygen from the biofilm medium, the silicone tubing had been submerged in N2-purged water additionally containing the reductant sodium ascorbate (0.1 M).
Oxygen upshift experiments were performed as described for the downshift experiments except that the water tank was purged with O2 gas, which resulted in an oxygen concentration of around 1 mM in the medium inflow.
Stepwise downshift experiments were performed on P. putida monospecies biofilms formed by the wt or the rough variant, propagated as described above. After inoculation, the silicone tubing, including the flow cells, was submerged and secured in a 15-liter water-filled tank. The oxygen concentration in the inflow medium was monitored by measuring the concentration of oxygen in the water container, which in turn was controlled by the purging rate of nitrogen gas (control experiments had shown a <2% difference in the concentrations in the inflow medium and the water surrounding the silicone tubing, which was mainly due to a difference in the saline concentration). The oxygen concentration in the water was adjusted to obtain an oxygen concentration of approximately 125 μM (50% of the normal level). After 18 h of incubation, the concentration was reduced to approximately 100 μM (40%), and after 24 h, it was reduced to approximately 75 μM (30%). After 32 h, the oxygen concentration was reduced to 35 μM (14%) and, finally, to near zero levels (<3 μM). During the experiment, the flow cells were briefly removed from the water tank for microscopic observations and image acquisition (brief upshifts had no significant effect on the biofilms).
Phenotypic characterization of the P. putida rough variant.
Swimming motility and chemotaxis (42) towards benzoate were investigated using 0.28% agar plates with AB medium supplemented with 1 mM benzoate (Sigma). Single colonies were inoculated, and plates were incubated for 24 h at 30°C.
P. putida wt and rough variant strains were investigated for pellicle formation in static broth cultures. A 100-ml Erlenmeyer flask containing 50 ml AB minimal medium supplemented with 40 mM glucose was inoculated with 1 ml overnight culture. Cultures were incubated without shaking at 30°C, and pellicle formation was observed after 2 days. The pellicle polysaccharide was stained with calcofluor white (Sigma) as described previously (36), except that the buffer (10 mM Tris buffer, pH 8, 0.9% NaCl) contained 1 μg/ml calcofluor white. The polysaccharide was examined using an Axioplan epifluorescence microscope (Carl Zeiss) with a 100× oil objective. The microscope was equipped with a 100-W mercury lamp and a DAPI (4′,6′-diamidino-2-phenylindole) filter. To confirm the presence of a cellulose-like polymer in the rough variant pellicle biofilm, pellicle material from static cultures was treated with cellulase (from Aspergillus niger; ICN Biomedicals Inc.) as described previously (51), with the addition of 5 μg/ml chloramphenicol to stop bacterial protein synthesis. Disintegration of pellicles was determined by visual inspection. Extracellular polysaccharide (EPS) formation in P. putida wt and variant colonies was examined on fresh LB agar plates with 25 μg/ml calcofluor white. Plates were incubated at 30°C for 5 days. The binding of calcofluor white to agar plate colonies was determined by fluorescence excitation with a 254-nm light source and photographed using a Canon digital camera.
Coaggregation of Acinetobacter and P. putida cells (GFP-tagged wt and rough variant cells) was investigated by mixing cells from stationary-phase cultures. The cell density (optical density at 600 nm) was adjusted to 1.5, and cells were subsequently mixed at a ratio of 1:1 in a total volume of 1 ml. Aggregation was allowed to proceed for 3 h before inspection. For microscopic visualization, cells were stained with 0.2% Syto62 for 30 min, and images were obtained using an LSM510 microscope as described above. Cellulase assays on coaggregated clumps were performed as described above for the rough variant pellicle biofilm material.
RESULTS
Biofilm development and in situ growth physiology.
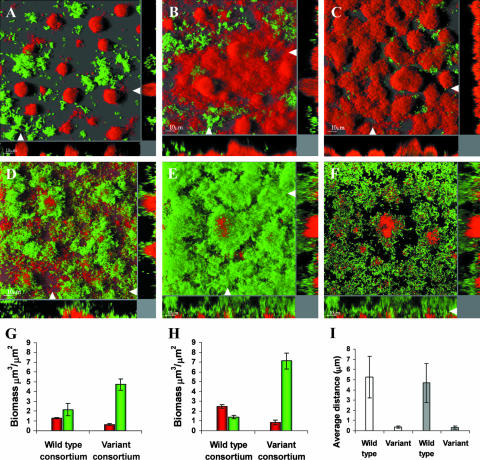
Biofilm development and structural coordination in mixed-species consortia of Acinetobacter sp. strain C6 and either P. putida wt KT2440 or the isolated rough variant were investigated by confocal microscopy in flow chambers irrigated with 500 μM of benzyl alcohol. Previous reports have shown that benzoate continuously leaks from the large Acinetobacter microcolonies (7, 37). P. putida KT2440 is totally dependent on the benzoate cross-feeding activity of Acinetobacter when the mixed-species consortium is propagated on benzyl alcohol, but despite the apparent advantage of associative behavior, it was obvious throughout the observed period of biofilm formation that the ancestral P. putida KT2440 strain was unable to enter close structural associations with Acinetobacter (Fig. 1A and B). Moreover, within 3 to 4 days, the Acinetobacter population became increasingly dominant in some areas (Fig. 1C). In contrast, the P. putida rough variant proliferated successfully in the presence of Acinetobacter while remaining tightly associated throughout the course of development (Fig. 1D and E). The P. putida wt biofilm structure was loose and protruding (no dense colonies), as previously described for P. putida OUS82 (54), with a flexible and elastic response to abrupt changes in flow velocity. In comparison, however, the rough variant developed a very inflexible biofilm structure. Rough variant cells covered almost the entire exposed surface of the Acinetobacter biofilm, and mixed microcolonies were regularly observed (Fig. 1F). The observed developments and dynamics of the wt and variant consortia were further supported by computer analysis of biofilm images, using COMSTAT software (26). Biomass quantification of the wt consortium thus revealed that the total P. putida wt population decreased from day 1 to day 3 while the Acinetobacter population nearly doubled (Fig. 1G and H). Quantification of the rough variant consortium demonstrated a successfully reproducing rough variant population with a previously described (23) exploitative (negative) effect on the Acinetobacter population, in contrast to the wt consortium (Fig. 1G and H). The associative patterns of the wt and variant consortia on day 1 and day 3 were analyzed using a novel COMSTAT extension (see Materials and Methods) that calculated the average distance (μm) from the surfaces of Acinetobacter microcolonies and cells to the nearest P. putida cells. These data showed that rough variant cells of P. putida were associated closely with Acinetobacter microcolonies, whereas the nearest P. putida wt cells, on average, were located at a short distance (Fig. 1I). The differences in biofilm behavior and productivity between P. putida wt and variant populations could not be explained by differences in the ability to proliferate on benzoate, as growth rates of shaken batch cultures could not be distinguished data not shown).
FIG. 1.
Structural relationships and development of wt and evolved consortium biofilms. Biofilms of Acinetobacter sp. strain C6 and P. putida wt (A to C) or a P. putida rough variant (D to F) were grown in flow chambers supplemented with 500 μM benzyl alcohol as the sole carbon source. P. putida cells (green) had a gfp expression cassette inserted into the chromosome. Acinetobacter sp. strain C6 cells (red) were visualized using Syto62. CSLM micrographs were obtained for biofilms grown for 1 day (A and D) and 3 days (B, C, E, and F). The main frames (A to E) are horizontal shadow projection (SFP) images, and side panels are xz sections in the positions indicated with white arrows. The main frame in panel F is a single image slide from the image shown in panel E showing a high association between microcolonies of Acinetobacter and the P. putida rough variant. Biomass (μm3/μm2) distributions were determined on day 1 (G) and day 3 (H) by image quantification of biofilms consisting of the wt consortium (P. putida wt and Acinetobacter) and the variant consortium (P. putida rough variant and Acinetobacter), respectively. (Day 3 data were reproduced from Nature [23] with permission of the publisher.) Red, biomass of the Acinetobacter population; green, biomass of the P. putida population. Values are means ± standard deviations. (I) The structural association between P. putida and Acinetobacter was quantified by image analysis of the mixed-species biofilms (see Materials and Methods for details). The average distances from the surfaces of Acinetobacter microcolonies to the nearest P. putida cells were determined for the wt consortium and the rough variant consortium on day 1 (white bars) and on day 3 (gray bars). Values are means ± standard deviations.
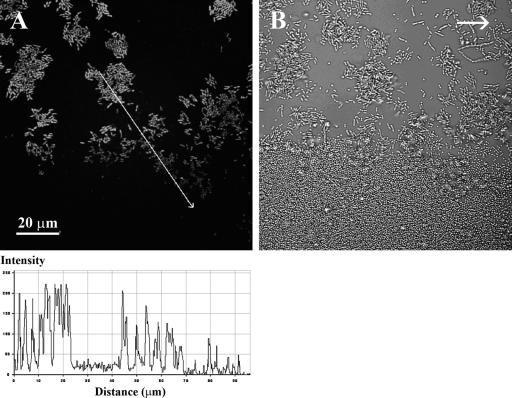
In order to understand the inability of the ancestral P. putida population to efficiently colonize Acinetobacter microcolonies, the development and dynamics of in situ growth activity in the mixed-species biofilm were investigated. The P. putida KT2440 wt strain used in this study had a growth activity monitor cassette with a gene encoding an unstable version of GFP downstream of a ribosomal promoter inserted into the chromosome. This unstable version of GFP has a half-life of about 1 h in P. putida cells (34). With this insertion, growing cells will appear fluorescent green, whereas nongrowing cells will quickly lose fluorescence due to degradation of GFP in cells with no net protein synthesis (52). This construct allowed online studies of the growth activity distribution of P. putida cells growing in the biofilm consortium. Microscopic investigations revealed both temporary and structure-related dynamic changes in growth physiology. Although generally not associating with Acinetobacter microcolonies, the P. putida cells were fluorescent green, indicating active growth, throughout the first 24 to 36 h (data not shown). At this time, it was observed that very large Acinetobacter microcolonies seemed to exert a growth rate-decreasing activity in a gradient mode on P. putida cells situated at the substratum inside or in very close proximity to Acinetobacter microcolonies (Fig. 2). Gradually, within 48 h, the entire P. putida population had a significantly decreased growth activity (low GFP fluorescence), possibly caused by nutrient limitation. Aerobic degradation of aromatics requires molecular oxygen, and nutrient gradients are likely to appear due to the structured heterogeneous environment of the biofilm (5, 13, 57, 59). It was therefore considered possible that oxygen limitation could develop, which led us to investigate if oxygen limitations could explain part of the observed biofilm development and dynamics.
FIG. 2.
Gradients in growth activity of P. putida cells at the bottom edge of large Acinetobacter sp. strain C6 microcolonies. To visualize in situ growth activity, the P. putida wt strain had a mini-Tn7-Gmr-rrnBP1-gfp[AAV] cassette inserted into the chromosome. Mixed-species biofilms of Acinetobacter and P. putida were grown in flow chambers for 36 h. (A) Image of GFP fluorescence of P. putida cells (substratum layer) obtained using CSLM. The gradient in fluorescence intensity (bottom) was quantified along the line indicated with the white arrow. (B) Light reflection image captured in the same viewing field, showing both P. putida and Acinetobacter cells. The typical cell morphology of Acinetobacter is coccoid, and that of P. putida is rod-shaped, but the nonfluorescent rod-shaped cells in panel B are stressed or undivided cells of Acinetobacter, which are frequently seen for this strain when it is proliferated on a surface. The arrow in panel B shows the direction of flow.
Investigation of oxygen limitations.
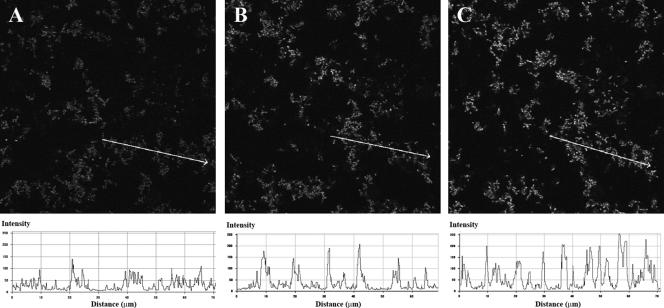
The inflowing biofilm medium contained approximately 250 μM oxygen (medium saturated with air at 24°C) and 500 μM benzyl alcohol. To determine the oxygen consumption of biofilms, replicate mixed-species populations of Acinetobacter and P. putida wt were propagated in flow chambers, and the total oxygen consumption was measured. Measurements were obtained using an oxygen microelectrode with a 0.5-mm tip and a T-connector device applied to the system just before and after each flow channel, including a control without a biofilm population. The oxygen concentration dropped from approximately 250 μM in the influent biofilm medium to 8 μM ± 4 μM (mean ± standard deviation) on day 1 and <2 μM on day 2 in the effluent medium, as determined for three independent experiments. It should be noted that the determined oxygen levels reflect the activities of both the resident biofilm population and the detached planktonic cells. The results thus showed a rapid decrease in total oxygen concentration, but this does not necessarily imply that oxygen became the limiting nutrient in situ in the mixed-species consortium. To this end, the mixed-species consortium with the P. putida wt strain harboring the growth activity reporter cassette was propagated. On day 2, when the general growth activity of the P. putida wt population was quite low, an oxygen upshift (see Materials and Methods) was performed by increasing the oxygen concentration in the inflowing medium from 250 μM to around 1 mM oxygen. Solely by raising the oxygen level, we found that the growth activity of P. putida was enhanced within 1 h following the upshift (Fig. 3). These results strongly suggest that oxygen did in fact become a limiting nutrient, and taken together with the high level of oxygen consumption, it is possible that the temporary and structural heterogeneity in growth activity observed for P. putida was due to oxygen limitation. The growth rate-decreasing activity exerted by Acinetobacter on P. putida cells associated with the very large Acinetobacter microcolonies could thus simply be the result of oxygen limitation due to oxygen consumption by the Acinetobacter cells.
FIG. 3.
Oxygen upshift causes increased growth activity of P. putida wt cells grown in mixed biofilms with Acinetobacter sp. strain C6 cells. P. putida wt cells containing a mini-Tn7-Gmr-rrnBP1-gfp[AAV] cassette were grown in flow cells containing Acinetobacter. On day 2, the oxygen concentration in the inflowing medium was increased approximately fourfold, as described in Materials and Methods. CSLM micrographs of the GFP fluorescence were captured in the same viewing field before (A) and 30 min (B) and 60 min (C) after the oxygen upshift. Increased growth activity was observed in three independent experiments. Images were recorded as single horizontal scans. The gradient in fluorescence intensity shown below each image was quantified along the line indicated with the arrow, using LSM510 CSLM software. Acinetobacter cells are not visible.
Biofilm dispersal in response to oxygen starvation.
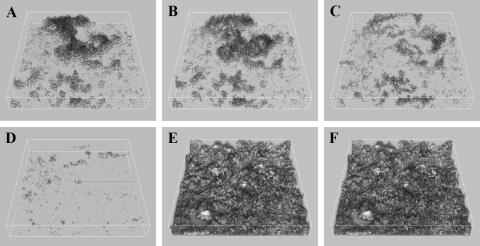
Biofilm dispersal in response to nutrient starvation has been reported for several biofilm populations (1, 14, 20, 53), and this phenomenon is recognized as an important part of surface colonization strategies (22). For Pseudomonas putida strain OUS82, biofilm dispersal in response to carbon starvation was demonstrated (20), and for Shewanella oneidensis biofilms, removal of oxygen resulted in biofilm detachment (53). These findings made us investigate if the ancestral P. putida KT2440 biofilm would detach in response to oxygen starvation and whether the P. putida rough variant would display a different biofilm dispersal response. In order to investigate this possibility, mixed populations of Acinetobacter sp. strain C6 and the P. putida wt strain or rough variant were propagated in flow chambers with benzyl alcohol as the sole carbon source. On day 2, an oxygen downshift was performed by decreasing the oxygen concentration in the inflowing biofilm medium from 250 μM to near zero levels (<2 μM). Biofilms were studied online using confocal microscopy and followed over time (Fig. 4). Within minutes, the first signs of biofilm detachment were observed for the P. putida wt strain (Fig. 4B), and the majority of the biofilm had detached after only 25 min (Fig. 4D). In contrast, the P. putida rough variant was unresponsive to the oxygen downshift in the consortium environment, and no biofilm dispersal was observed (Fig. 4E and F), even after several hours of oxygen starvation. Investigations of two separately evolved, isolated rough variants showed that they also did not disperse the biofilm in response to an oxygen downshift (data not shown). Biofilm dispersal of the P. putida wt was not observed in response to carbon starvation in the consortium environment following 5 h of starvation (data not shown). These findings suggest that the inability of P. putida KT2440 to successfully compete in the mixed-species biofilm environment and to efficiently colonize Acinetobacter microcolonies may be due at least in part to the inherent biofilm dispersal in response to oxygen starvation. This suggestion is supported by the findings that the repeatedly selected rough variant efficiently colonized Acinetobacter microcolonies and, furthermore, showed a nondispersal biofilm phenotype in the consortium environment.
FIG. 4.
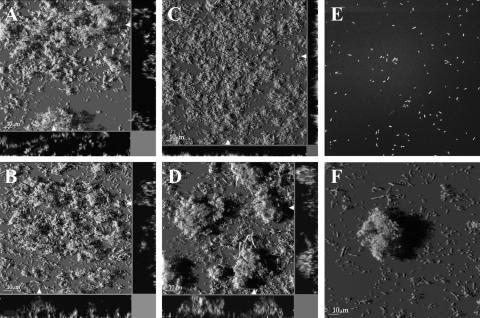
Different biofilm dispersal phenotypes of P. putida wt and the rough variant in response to oxygen downshift. Acinetobacter cells were proliferated in flow chambers with GFP-tagged cells of P. putida wt and the rough variant. On day 2, an oxygen downshift was performed online as described in Materials and Methods. CSLM micrographs of the P. putida wt biofilm were captured in the same viewing field before (A) and 5 min (B), 9 min (C), and 25 min (D) after the oxygen downshift. CSLM micrographs of the P. putida rough variant biofilm were likewise captured in the same viewing field before (E) and 25 min after (F) the oxygen downshift. CSLM micrographs are presented as simulated three-dimensional images. Acinetobacter cells are not visible.
P. putida rough variant biofilm physiology.
In a further phenotypic characterization of the P. putida rough variant, the oxygen downshift experiment was carried out with monospecies biofilms (in the absence of Acinetobacter) of P. putida wt and the rough variant. Biofilm populations were propagated on 200 μM benzoate (since P. putida does not grow on benzyl alcohol) and followed microscopically for 2 days before the oxygen downshift was performed. Both strains showed a complete dispersal response to oxygen starvation (data not shown). In fact, the P. putida rough variant displayed a biofilm structure very similar to the loose and protruding structure of the wt biofilm described above (Fig. 5A and B). The addition of benzyl alcohol, in addition to benzoate, to the inflowing biofilm medium did not significantly influence the biofilm formation of the two strains (data not shown).
FIG. 5.
Structures and phenotypes of P. putida wt and rough variant monospecies biofilms during a stepwise oxygen downshift. Biofilms were proliferated in flow chambers supplemented with benzoate as the sole carbon source. Green fluorescence from GFP-tagged P. putida cells was recorded using CSLM imaging. Monospecies biofilms of P. putida wt (A) and the rough variant (B) displayed a similar structure (day 2) when biofilms were proliferated with standard oxygen concentrations (approximately 250 μM in the inflowing medium). A graduated (stepwise) oxygen downshift was performed on monospecies biofilms as described in Materials and Methods. After 32 h, the inflowing medium contained approximately 75 μM oxygen, resulting in a very flat and thin biofilm of the P. putida wt population (C), in contrast to the thick microcolonies observed for the rough variant (D). After 48 h and prolonged starvation (down to <3 μM oxygen), both P. putida wt (E) and the rough variant (F) showed significant dispersal of the biofilm. The main frames are horizontal shadow projection (SFP) images, and side panels are xz sections in the positions indicated with white arrows.
In an attempt to imitate the oxygen concentration dynamics in the mixed-species consortium, the oxygen downshift experiment was modified to gradually decrease the oxygen concentration in the inflowing biofilm medium. Monospecies populations of P. putida wt and the rough variant were inoculated in flow chambers irrigated with 1 mM benzoate, after which the oxygen concentration was gradually decreased (stepwise) over a 2-day period. Microscopic observations showed (after 32 h) that the P. putida rough variant population formed thick microcolonies in a biofilm medium containing around 30% of the normal oxygen level (approximately 75 μM), in contrast to the rather flat, thin wt biofilm (Fig. 5C and D). However, under conditions of prolonged starvation (48 h), both the wt and variant populations significantly dispersed the biofilm (Fig. 5E and F). These results show that although the P. putida rough variant displayed a transient improvement in biofilm persistence, conditions of decreased oxygen concentrations alone (absence of both benzyl alcohol and Acinetobacter) did not give rise to the nondispersal, low-oxygen-persistence phenotype.
These data show that the low-oxygen-persistence physiology of the rough variant is not expressed under all conditions. Since the rough variants evolved specifically in the presence of Acinetobacter sp. strain C6, we investigated if the sole presence of Acinetobacter cells in the flow chamber with the rough variant would cause the nondispersal, low-oxygen-persistence phenotype of the variant independent of the nutrient conditions. This was done by proliferating populations of the rough variant and Acinetobacter in the flow chamber on benzoate instead of benzyl alcohol. Flow chambers were supplied with standard air-saturated medium inflow. In this scenario, both strains were competing for the primary carbon source, and hence there was no apparent advantage for the rough variant to associate with Acinetobacter sp. strain C6. Not surprisingly, a large part of the rough variant population was found dissociated from the Acinetobacter microcolonies, and this part dispersed after the oxygen downshift, as observed for the P. putida wt population (data not shown). However, the part that was found associated with Acinetobacter microcolonies showed the characteristic nondispersal phenotype, indicating that proliferation associated with Acinetobacter microcolonies may change the rough variant phenotype.
Coaggregate formation.
The P. putida rough variant lacks the LPS O antigen as a consequence of the wapH mutation, which is very likely to change the physiochemical characteristics of the cell surface (47). This led us to investigate if the mutation had caused a change in surface compatibility between the rough variant and Acinetobacter cells, leading to increased coadhesion. Stationary-phase cultures were mixed at a ratio of 1:1 and allowed to aggregate for 3 h. At this time, coaggregation of Acinetobacter and P. putida rough variant cells formed large visible clumps in the mixture (Fig. 6A), whereas no significant clumping was observed for the mixture of Acinetobacter and P. putida wt (Fig. 6B). Rough variant cells grown on FAB minimal medium containing benzoate coaggregated into visual clumps with Acinetobacter cells grown in FAB medium containing benzyl alcohol; however, larger clumps formed at a higher rate when the rough variant was mixed with Acinetobacter cells grown in LB medium.
FIG. 6.
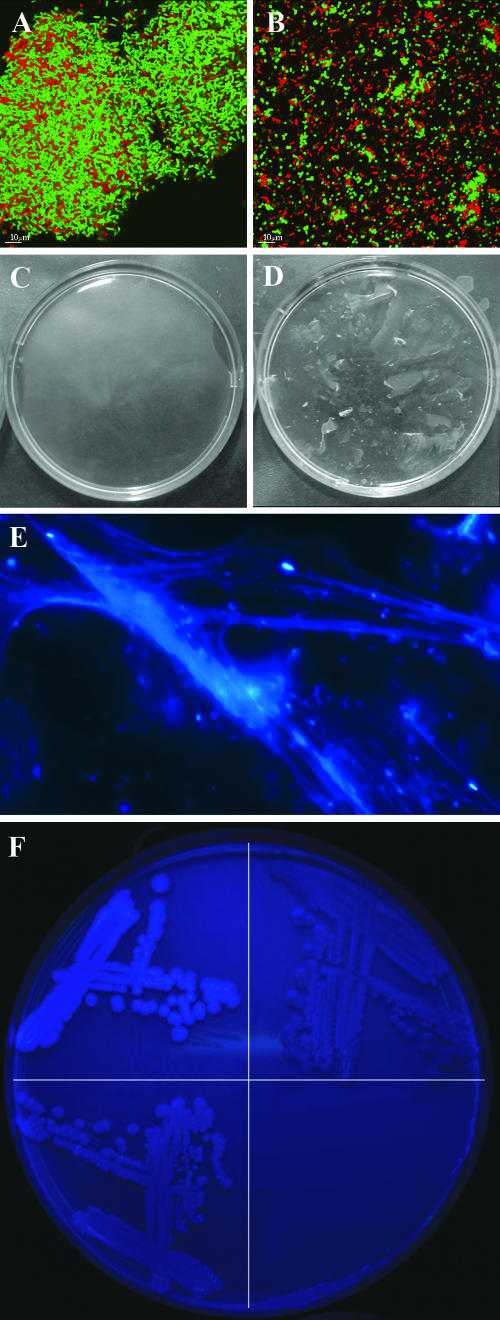
Coaggregation with Acinetobacter, biofilm pellicle formation, and polysaccharide formation observed for the P. putida rough variant are caused by the wapH mutation. (A and B) A stationary-phase cell culture of P. putida (green) grown on FAB medium with 5 mM benzoate was mixed with a stationary-phase cell culture of Acinetobacter sp. strain C6 (red) grown on LB. CSLM micrographs were obtained of coaggregates of Acinetobacter and P. putida rough variant cells (A) or Acinetobacter and P. putida wt cells (B) after 3 h of incubation. (C and D) Rough variant biofilm pellicles were transferred to a petri dish and incubated at 37°C overnight in the presence (C) and absence (D) of cellulase enzyme (from Aspergillus niger), as described in Materials and Methods. Dissolution was observed only after incubation with the cellulase enzyme. (E) Polysaccharide fibers of condensed biofilm pellicle material stained with calcofluor white. (F) Colonies of P. putida with a constructed mutation in the wapH gene (SNZ83) (top left), P. putida KT2440 wt (top right), and P. putida (SNZ83) complemented with a plasmid containing the wapH gene (bottom right) were grown on LB agar containing 25 μg/ml calcofluor white for 5 days.
EPS formation in P. putida rough variants.
The P. putida rough variant used in this study has a mutation in wapH (PP4943), a P. aeruginosa PAO1 wapH homolog (33, 35) involved in outer core LPS formation, conferring the rough colony morphology and cell shape (23). The conditionally inflexible biofilm structure observed for the variant suggested that cell surface properties and/or extracellular matrix materials were involved in changing the biofilm structure. Rough or wrinkled colony variants of other bacteria have previously been found to overproduce cellulose or cellulose-like polymers, making them capable of forming a pellicle in standing cultures (48, 50, 61). Interestingly, we found that the derived rough variant, but not the wt, was able to produce an elastic gel-like pellicle in standing broth cultures containing LB broth or glucose minimal medium (Fig. 6D). The pellicle was visible from day 2, progressively thickening over the following days. Epifluorescence microscopy of condensed pellicle material showed that the variant EPS material bound calcofluor white, an indicator of cellulose(-like) polymers that specifically binds to (1-3)- and (1-4)-β-d-glucopyranosyl units (Fig. 6E). In order to test if the robustness of the biofilm was secured by the calcofluor white-binding exopolysaccharide, standing broth cultures of the rough variant were incubated for 5 days, after which the pellicles were treated with cellulase enzyme. After overnight incubation at 37°C, the treated pellicles had disintegrated (Fig. 6C), whereas untreated pellicles remained intact (Fig. 6D). To compare cellulose(-like) production in the wt versus variant strains and to determine if the wapH mutation alone was causing the polysaccharide production, we used the untagged P. putida KT2440 wt strain and a constructed rough variant thereof (SNZ83) (see reference 23 for construction details) with an introduced nonpolar mutation in the wapH gene. Pellicle formation was also observed in SNZ83. Colonies grown on agar plates containing the calcofluor white stain clearly showed an enhanced production of the cellulose-like polymer in the constructed rough variant (Fig. 6F, top left quadrant). Additionally, the calcofluor white-stainable material was not expressed in P. putida wt at any recordable levels, in accordance with previous findings (55) (Fig. 6F, top right quadrant). The level of calcofluor white staining in P. putida SNZ83 harboring the wapH gene on plasmid pBBR4943-1 (23) was similar to the wt level, suggesting that production of the exopolysaccharide is a direct consequence of the mutation in wapH (Fig. 6F, bottom left quadrant).
In order to investigate the role of cellulose production in stabilizing the coaggregates of Acinetobacter and P. putida rough variant cells, aggregates were treated with cellulase as described for the pellicle biofilm (see above). However, no signs of dissolution were observed after overnight incubation (data not shown). Attempts to dissolve the mixed-species biofilm of Acinetobacter and the P. putida rough variant with cellulase treatment also failed (data not shown).
DISCUSSION
When P. putida KT2440 and Acinetobacter sp. strain C6 are propagated in mixed-species biofilms on benzyl alcohol, the P. putida population is totally dependent on benzoate cross-feeding from Acinetobacter cells. In the course of biofilm development, the reproducible appearance of better-adapted rough colony morphology variants of P. putida was observed as described previously (23). The rough variant formed tight structural associations with the Acinetobacter population and efficiently colonized the Acinetobacter biofilm surface, which was confirmed by image analysis. The ability to interact structurally is very likely part of the selective advantage of this variant due to the dependence on cross-fed benzoate. In contrast, the P. putida wt strain clearly lacked the capacity to form such tight structural associations with Acinetobacter microcolonies and to persist successfully over time. These observations raise the question of which specific phenotypic properties are needed for the completely changed persistence phenotype of P. putida KT2440.
One possible explanation for the observed phenotypic differences could be a change in chemotactic behavior, given that P. putida is chemotactic towards benzoate, the cross-fed substrate (24, 25). Chemotaxis and motility have been shown to be very important for biofilm structure and coordination in surface populations (28, 29, 39). Our own investigations have documented that both P. putida wt and rough variant cells are motile and show a positive chemotactic response to increasing concentrations of benzoate (data not shown). Despite a small reduction in swimming speed of the rough variant, the differences in motility behavior cannot explain the observed difference in structural association in the mixed-species biofilms.
The P. putida wt population was unable to establish a tight association with the Acinetobacter microcolonies, and furthermore, a decline in population size was observed over time. The underlying theory behind this failure involves the oxygen starvation response. It is hypothesized that as they are confronted with reductions in oxygen levels, P. putida cells respond by leaving the biofilm rather than taking up competition for oxygen. The exact molecular basis for biofilm dispersal in P. putida KT2440 is unknown, but in P. putida OUS82, proteins containing the GGDEF and EAL domains have been shown to be involved in biofilm formation and dispersal (21). The biofilm dispersal mechanism was observed for P. putida wt biofilms in response to both a rapid and a more gradual oxygen downshift. The findings that the P. putida wt biomass decreased from day 1 to day 3, that the total oxygen consumption reached almost exhaustive levels within 1 to 2 days, that the growth activity of wt P. putida decreased first locally around large Acinetobacter microcolonies (Fig. 2) and then globally within 2 days, and that the growth rate-limiting nutrient at this point was in fact oxygen are all in accordance with our hypothesis. Benzoate limitation was unlikely to be the cause of the local growth activity decrease observed, because previous investigations have shown that benzoate is continuously leaking from the large microcolonies (37) and accumulating in the effluent medium (7) under similar conditions. Upshifting the oxygen concentration increased the growth activity of P. putida cells, showing that there was no lack of carbon source under the prevailing conditions (Fig. 3). Also, we found that benzoate was not toxic to P. putida at relevant concentrations (data not shown). Since the P. putida wt population disperses in response to oxygen starvation, a niche is open to any new variant that would be able to overcome this problem.
A complete change in dispersal response was observed for the P. putida rough variant: when mixed-species biofilms of the rough variant and Acinetobacter sp. strain C6 were exposed to low-oxygen conditions, no dispersal of the variant population was observed. Explaining the lack of an oxygen starvation response of the rough variant genotype in the mixed-species biofilms is complicated by the finding that these cells in fact do possess the normal biofilm dispersal response if confronted with a sudden depletion of oxygen in the absence of Acinetobacter, so the rough variant is not merely a biofilm dispersal mutant. In addition to the normal dispersal response observed for P. putida rough variant monospecies biofilms was the finding of loose and flexible biofilm structures similar to those observed for the wt. Hence, the rough variant flow chamber population displayed behavioral and structural similarity with the wt population in the absence of Acinetobacter cells, in support of our previous results (23) showing that rough variants did not evolve in monospecies biofilms of P. putida wt and that the rough variants did not have any fitness advantage relative to the ancestral P. putida cells in this environment.
One of the real challenges in this investigation was to explain how a single mutation resulting in a changed LPS phenotype gives rise to the observed conditional nondispersal phenotype. We therefore investigated various factors in the mixed-species biofilm environment in an attempt to resolve the specific conditions leading to the characteristic conditional phenotype of the P. putida rough variant. As discussed above, it was clear that the presence of the Acinetobacter population in the biofilm was needed for the nondispersal phenotype to occur. One effect of the Acinetobacter population in the mixed-species biofilms was a gradual reduction in oxygen concentration due to extensive oxygen consumption from metabolizing the carbon source, benzyl alcohol. However, reduced oxygen conditions alone did not produce the complete nondispersal phenotype, as a gradual reduction in the externally applied oxygen concentration resulted in a transiently thicker monospecies biofilm of the rough variant compared to that of the wt, and prolonged starvation caused both strains to disperse significantly.
The lack of the O antigen in the rough variant is very likely to change the physiochemical characteristics of the cell surface (47), which could have a direct effect on the cell-to-cell interaction with Acinetobacter cells by changing the surface compatibility. It was previously shown that core or rough LPS variant derivatives of P. aeruginosa were more hydrophobic and showed changed biofilm-forming properties (16, 47). Experiments with batch cultures revealed that cells of Acinetobacter and the P. putida rough variant, but not wt P. putida, form extensive coaggregates when mixed together. The mechanism of coadherence is unknown, but this feature seems to be a key factor in understanding the conditional behavior of the rough variant: it is able to stick firmly and specifically to the Acinetobacter microcolonies or cells in the mixed-species biofilms and thereby ensure close access to the excreted benzoate. In this way, the Acinetobacter population may function as an anchor point for the rough variant, an opportunity not present in monospecies biofilms. However, just serving as an anchor point was not sufficient for the Acinetobacter population to cause the complete nondispersal phenotype of the rough variant population. This was demonstrated by propagating the mixed-species rough variant biofilm on a different nutrient source, i.e., benzoate, instead of benzyl alcohol. The part of the rough variant population that was found associated with Acinetobacter microcolonies when propagated on benzoate showed the characteristic nondispersal phenotype. However, other niches contained rough variant microcolonies with a phenotype similar to that of the wt; these microcolonies would detach when the oxygen concentration was shifted down. This suggests that proliferating in the environment in tight association with Acinetobacter microcolonies could change the rough variant physiology, although this was neither advantageous nor necessary under conditions where both strains were able to degrade the primary carbon source, benzoate.
We have previously shown that the rough variant has a parasitic or negative effect on the Acinetobacter population when they proliferate together in mixed-species biofilms (23). The results from this work point to intense oxygen competition in the biofilm environment, which offers a plausible explanation for the parasitic effect, as follows: the rough variant grows as a mantle on the Acinetobacter microcolonies (Fig. 1) and therefore acts as a living shield by preventing Acinetobacter cells from efficiently reaching the important nutrient oxygen. In the wt situation, the P. putida population responds by leaving when competition for oxygen becomes too intense.
We have shown that both benzyl alcohol and the Acinetobacter population are necessary but that neither one is sufficient for the complete nondispersal phenotype of the rough variant to occur. Acinetobacter cells may serve as an attachment point for the P. putida rough variant cells, but other adherence factors or matrix components seem to be needed to stabilize the variant biofilm, as not all variant cells can be attached directly to the Acinetobacter cells. It is very likely that growing in the stressful environment of the Acinetobacter microcolonies may induce the rough variant population to produce some extracellular polymeric substances. Some of the stress factors could be the very low concentration of oxygen, surface contact with Acinetobacter cells, or the generally poor nutrient environment. It is highly possible that the cellulose-like polymer that has been observed in the rough variant could be induced in the mixed-species biofilm and may be responsible, to some degree, for the nondispersal phenotype. However, it is clear that the cellulose-like polymer was not the only factor causing the nondispersal biofilm phenotype, as cellulase enzyme treatment of mixed-species biofilms with Acinetobacter did not disintegrate the biofilm.
Evidence of enhanced formation of a cellulose-like polymer in the rough variant was found on agar plates and in standing broth cultures producing a biofilm pellicle that could be disintegrated by cellulase enzyme treatment. Sequence analysis has revealed a cellulose operon in the P. putida genome (15, 38), and the EPS material could therefore be cellulose. In agreement with our results, no pellicle formation was observed for P. putida KT2440 wt in standing cultures of KB medium, but interestingly, the cellulose machinery was demonstrated to be intact and functioning (55). Additionally, cellulose production seems to be fairly common among bacteria and pseudomonads in relation to biofilm formation (55). The enhanced production of the cellulose-like polymer was shown to be the result of one mutation in wapH, a gene involved in core LPS production. The link between the mutation and the enhanced EPS production in P. putida is not clear, but a similar phenotype was found for the root-colonizing bacterium Azospirillum brasilense. In this case, a deletion causing a modification in the LPS core structure resulted in enhanced production of a calcofluor white-stainable polysaccharide (27). Deep rough LPS variant phenotypes of Escherichia coli have been shown to overproduce colanic acid EPS. These mutants are unable to cross-link the LPS core part (43), but the homologous genes involved in inner core LPS formation in P. aeruginosa were shown to be essential (56).
Our investigations show that biofilms may readily develop into very complex and heterogeneous environments, with structural niches, microenvironments, and gradients of nutrients, a finding that is consistent with the prevailing view of the biofilm environment (2). Nutrient (or oxygen) starvation causes subpopulation stress (3). The rapid and repeated emergence of rough colony variants is the result of natural selection (23) in response to the physical/chemical environment afforded by the presence of benzyl alcohol-degrading Acinetobacter cells; the shortcomings of the wt genotype are quickly overcome by adaptive mutations. The present results confirm our previous suggestion (23) that the rough variant phenotype is adapted to the very specific environment from which it was derived, and there is no reason to assume that the mutation has any selective advantage under other environmental conditions. The specific adaptation could be caused by one single mutation in wapH altering entirely the interspecies interaction, biofilm structure, and phenotype, causing coaggregation and polysaccharide formation.
Acknowledgments
Support from the Danish Research Councils to S.M. is gratefully acknowledged.
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 2.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13:16-19. [DOI] [PubMed] [Google Scholar]
- 3.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 4.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 48:2659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell, D., G. M. Wolfaardt, D. R. Korber, and J. R. Lawrence. 1997. Do bacterial communities transcend Darwinism? Adv. Microb. Ecol. 15:105-191. [Google Scholar]
- 7.Christensen, B. B., J. A. Haagensen, A. Heydorn, and S. Molin. 2002. Metabolic commensalism and competition in a two-species microbial consortium. Appl. Environ. Microbiol. 68:2495-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 9.Cisar, J. O., S. H. Curl, P. E. Kolenbrander, and A. E. Vatter. 1983. Specific absence of type 2 fimbriae on a coaggregation-defective mutant of Actinomyces viscosus T14V. Infect. Immun. 40:759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 65:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, J. D., and O. Maaløe. 1967. DNA replication and the cell cycle in Escherichia coli cells. J. Mol. Biol. 23:293-300. [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.DeBeer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structures on oxygen distribution and mass-transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 14.Delaquis, P. J., D. E. Caldwell, J. R. Lawrence, and A. R. McCurdy. 1989. Detachment of Pseudomonas fluorescens from biofilm on glass surfaces in response to nutrient stress. Microb. Ecol. 18:199-210. [DOI] [PubMed] [Google Scholar]
- 15.Dos Santos, V. A., S. Heim, E. R. Moore, M. Stratz, and K. N. Timmis. 2004. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6:1264-1286. [DOI] [PubMed] [Google Scholar]
- 16.Flemming, C. A., R. J. Palmer, A. A. Arrage, H. C. Van de Mei, and D. C. White. 1998. Cell surface physicochemistry alters biofilm development of Pseudomonas aeruginosa lipopolysaccharide mutants. Biofouling 13:213-231. [Google Scholar]
- 17.Franklin, F. C., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, H. E., and L. I. Gordon. 1992. Oxygen solubility in seawater: better fitting equations. Limnol. Oceanogr. 37:1307-1312. [Google Scholar]
- 19.Gieseke, A., L. Bjerrum, M. Wagner, and R. Amann. 2003. Structure and activity of multiple nitrifying bacterial populations co-existing in a biofilm. Environ. Microbiol. 5:355-369. [DOI] [PubMed] [Google Scholar]
- 20.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894-906. [DOI] [PubMed] [Google Scholar]
- 21.Gjermansen, M., P. Ragas, and T. Tolker-Nielsen. 2006. Proteins with GGDEF and EAL domains regulate Pseudomonas putida biofilm formation and dispersal. FEMS Microbiol. Lett. 265:215-224. [DOI] [PubMed] [Google Scholar]
- 22.Hall-Stoodley, L., and P. Stoodley. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7-10. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, S. K., P. B. Rainey, J. A. Haagensen, and S. Molin. 2007. Evolution of species interactions in a biofilm community. Nature 445:533-536. [DOI] [PubMed] [Google Scholar]
- 24.Harwood, C. S., and L. N. Ornston. 1984. TOL plasmid can prevent induction of chemotactic responses to aromatic acids. J. Bacteriol. 160:797-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harwood, C. S., M. Rivelli, and L. N. Ornston. 1984. Aromatic acids are chemoattractants for Pseudomonas putida. J. Bacteriol. 160:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 27.Jofre, E., A. Lagares, and G. Mori. 2004. Disruption of dTDP-rhamnose biosynthesis modifies lipopolysaccharide core, exopolysaccharide production, and root colonization in Azospirillum brasilense. FEMS Microbiol. Lett. 231:267-275. [DOI] [PubMed] [Google Scholar]
- 28.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 29.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 30.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 31.Kolenbrander, P. E., P. G. Egland, P. I. Diaz, and R. J. Palmer, Jr. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13:11-15. [DOI] [PubMed] [Google Scholar]
- 32.Komlos, J., A. B. Cunningham, A. K. Camper, and R. R. Sharp. 2005. Interaction of Klebsiella oxytoca and Burkholderia cepacia in dual-species batch cultures and biofilms as a function of growth rate and substrate concentration. Microb. Ecol. 49:114-125. [DOI] [PubMed] [Google Scholar]
- 33.Lam, J. S., M. Matewish, and K. K. H. Poon. 2004. Lipopolysaccharides of Pseudomonas aeruginosa, p. 3-51. In J. Ramos (ed.), Pseudomonas, vol. 3. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 34.Lambertsen, L., C. Sternberg, and S. Molin. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6:726-732. [DOI] [PubMed] [Google Scholar]
- 35.Matewish, J. M. 2004. The functional role of lipopolysaccharide in the cell envelope and surface proteins of Pseudomonas aeruginosa. Ph.D. thesis. University of Guelph, Guelph, Ontario, Canada.
- 36.Matthysse, A. G., K. V. Holmes, and R. H. Gurlitz. 1981. Elaboration of cellulose fibrils by Agrobacterium tumefaciens during attachment to carrot cells. J. Bacteriol. 145:583-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Møller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, L. P. Chris, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, A. T., T. Tolker-Nielsen, K. B. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 40.Otsu, A. 1979. Threshold selection method from gray level histograms. Trans. Syst. Man Cybernet. 9:62-66. [Google Scholar]
- 41.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parales, R. E. 2004. Nitrobenzoates and aminobenzoates are chemoattractants for Pseudomonas strains. Appl. Environ. Microbiol. 70:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raetz, C. R. H. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 45.Rao, D., J. S. Webb, and S. Kjelleberg. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 47.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 49.Schink, B. 2002. Synergistic interactions in the microbial world. Antonie Leeuwenhoek 81:257-261. [DOI] [PubMed] [Google Scholar]
- 50.Spiers, A. J., J. Bohannon, S. M. Gehrig, and P. B. Rainey. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 51.Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano, and P. B. Rainey. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sternberg, C., B. B. Christensen, T. Johansen, N. A. Toftgaard, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thormann, K. M., R. M. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ude, S., D. L. Arnold, C. D. Moon, T. Timms-Wilson, and A. J. Spiers. 2006. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 8:1997-2011. [DOI] [PubMed] [Google Scholar]
- 56.Walsh, A. G., M. J. Matewish, L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Lipopolysaccharide core phosphates are required for viability and intrinsic drug resistance in Pseudomonas aeruginosa. Mol. Microbiol. 35:718-727. [DOI] [PubMed] [Google Scholar]
- 57.Werner, E., F. Roe, A. Bugnicourt, M. J. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, S. J. Caldwell, and D. E. Caldwell. 1994. Multicellular organization in a degradative biofilm community. Appl. Environ. Microbiol. 60:434-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, K. D., P. S. Stewart, F. Xia, C. T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zampirolli, F., and R. Lotufo. 2001. Fast multidimensional parallel Euclidean distance transform based on mathematical morphology, p. 100-105. In D. Borges and T. Wu (ed.), XIV Brazilian Symposium on Computer Graphics and Image Processing. IEEE Press.
- 61.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]