Adenosylcobalamin (AdoCbl, also known as [aka] coenzyme B12) is the most complex coenzyme known, and its biosynthesis requires a great deal of genetic information (Fig. 1) (61). The core of AdoCbl is a cobalt-containing cyclic tetrapyrrolidine/pyrroline known as the corrin ring. The latter is synthesized only by prokaryotes via either of two pathways, the aerobic or anaerobic pathway, which differ in several important ways, but most notably in the timing of cobalt insertion (early in the anaerobic pathway, late in the aerobic pathway) and the type of cobaltochelatase used by each pathway (reviewed in references 5, 49, and 61). Unlike other members of the metal-containing cyclic tetrapyrroles, AdoCbl has upper and lower ligands, and in prokaryotes such as Salmonella enterica serovar Typhimurium (hereafter serovar Typhimurium), these ligands are attached to the corrin ring under aerobic or anaerobic conditions (21, 28).
FIG. 1.
Structure of AdoCbl, aka coenzyme B12. The indicated gene functions involved in the synthesis of different parts of AdoCbl are those found in serovar Typhimurium. AdoCba's differ in their structures by the nature of the lower-ligand base (reviewed in reference 44). (A) Cbi, synthesized by CbiABCDETFGHJKLMNQOP. (B) Cobyric acid, synthesized by CbiACDETFGHJKLMNQOP. (C) 1-Aminopropanol-O-2-phosphate, synthesized by CobD. (D) Nucleotide loop, assembled by CobUTSC. Me, methyl; FMN, flavin mononucleotide.
Recently, C. A. Roessner and A. I. Scott published a minireview on the topic of anaerobic de novo corrin ring biosynthesis which fine-tuned our understanding of the assembly of the core of Cbl (46). It is only fitting to offer the readership of the journal a minireview of what has been learned about other important aspects of coenzyme B12 synthesis in bacteria and archaea.
Unfortunately, the nomenclature for coenzyme B12 is complex. The rules for naming complete molecules and precursors have been published by the Commission on Biochemical Nomenclature (CBN) of the International Union of Pure and Applied Chemistry (IUPAC) and revised by the International Union of Biochemistry (IUB) (2, 3). For the reader's benefit, I define below terminology used in this minireview.
Corrinoids.
Molecules containing the cobalt-containing cyclic tetrapyrrole known as the corrin ring.
Incomplete corrinoids.
Corrinoids lacking the lower (Coα) axial ligand base.
Cobamide (Cba).
A complete corrinoid, that is, a corrinoid containing the nucleotide loop. The name of the Cba depends on the nature of the lower ligand.
Cobalamin (Cbl).
The Cba whose lower (Coα) ligand base is 5,6-dimethylbenzimidazole (DMB).
Adenosylcobamide (AdoCba).
The coenzymic form of a Cba whose lower (Coα) ligand is not DMB.
PseudoAdoCba.
The coenzymic form of AdoCba with adenine as the lower (Coα) ligand.
Vitamin B12, or cyanocobalamin.
Cbl with a cyano group as the upper (Coβ) ligand.
Coenzyme B12 or AdoCbl.
Cbl with 5′-deoxyadenosine (Ado) as the upper (Coβ) ligand and DMB as the lower (Coα) ligand.
α-5,6-Dimethylbenzimidazole-ribosyl-5′-phosphate (α-ribazole-P).
The nucleotide loop consisting of the DMB attached to ribose-5′-P. The N-glycosidic bond in this nucleotide is in the alpha configuration.
Here I review significant differences in the strategies used by archaea and bacteria to salvage the precursor cobinamide (Cbi) from the environment. Cbi is an incomplete corrinoid lacking the structure known as the nucleotide loop and the 5′-deoxyadenosine (Ado) ligand (Fig. 1). Cbi must be found in diverse ecosystems, given the fact that bacteria and archaea have evolved different enzymes to convert Cbi to AdoCba. Cbi salvaging is important to all cells, because this capability saves them the great deal of energy they spend synthesizing the machinery to make the corrin ring. I also discuss recent discoveries that fill important gaps in the knowledge of how prokaryotes synthesize this important coenzyme.
CORRINOID TRANSPORT ACROSS MEMBRANES
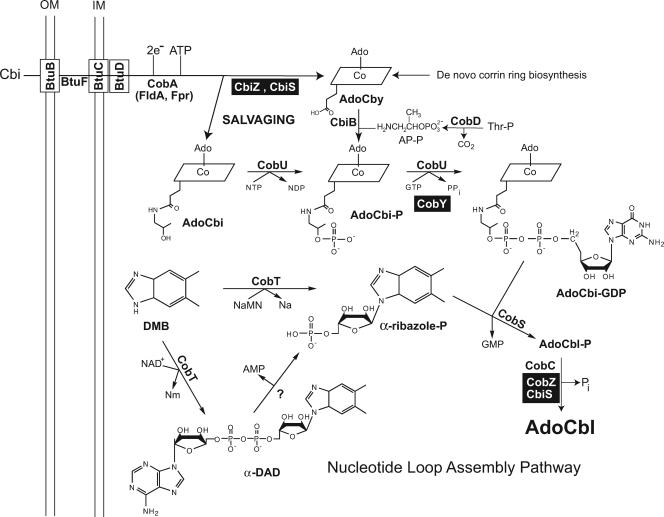
The salvaging of exogenous corrinoids (e.g., Cbi) starts with their transport into the cell. The highly specific BtuBFCD system found in gram-negative bacteria (e.g., serovar Typhimurium and Escherichia coli) efficiently translocates exogenous corrinoids present in the environment in fM concentrations (1). The BtuB protein is a Ca-dependent transporter located in the outer membrane (9, 18, 27) that delivers the corrinoid to the periplasmic corrinoid-binding BtuF protein (7, 13, 60). BtuB function requires energy input, which is provided through interactions with TonB, a protein anchored to the inner membrane whose function is also critical to iron acquisition (22, 42). Once bound with the corrinoid, BtuF delivers Cbi to the ABC transporter BtuCD located in the inner membrane (8, 20), which translocates it using an ATP-dependent mechanism (Fig. 2).
FIG. 2.
The late steps of AdoCbl biosynthesis in bacteria and archaea. Shown in black boxes are archaeal enzymes that are nonorthologous replacements of the indicated bacterial enzyme. Abbreviations: Pi, orthophosphate; Na, nicotinate; Nm, nicotinamide; AdoCbl-P, AdoCbl-5′-phosphate.
Corrinoid transport across archaeal membranes has been reported in Halobacterium sp. strain NRC-1 (65). Because Halobacterium lacks an outer membrane, orthologs of the btuB gene are not present in the genome of this archaeon. All the other components of the Btu system, namely, BtuFCD, are functional in Halobacterium (65).
UPPER LIGAND ATTACHMENT IS THE FIRST STEP IN Cbi SALVAGING
Upon entry into the cell, Cbi is adenosylated by an ATP:co(I)rrinoid adenosyltransferase enzyme; otherwise it cannot be converted into AdoCbl or be used to activate gene expression (12, 21, 50). In serovar Typhimurium and E. coli, the cobA and btuR genes, respectively, encode the housekeeping corrinoid adenosyltransferase. Although the gene nomenclature is different, the primary amino acid sequences of the CobA and BtuR proteins are 89% identical and the proteins have the same enzymatic activity (35, 54). The CobA-type corrinoid adenosyltransferase of Pseudomonas denitrificans is the only other bacterial enzyme that has been studied in some detail (19). The CobA enzyme of serovar Typhimurium is the paradigm for this type of corrinoid adenosyltransferase enzyme, and a great deal of structural, functional, and mechanistic information has been published (10, 23-25, 51, 53).
To date, only one CobA-type archaeal enzyme has been studied in vivo and in vitro. Although there are important differences between the bacterial and archaeal enzymes in regard to ATP binding, the by-product of the reaction (triphosphate) is the same in both cases (11).
There are Cbi-salvaging prokaryotes that lack CobA (e.g., Listeria monocytogenes, Lactobacillus reuteri, and others) (45). Instead, these bacteria use enzymes encoded by orthologs of the pduO gene of serovar Typhimurium, where it was first discovered as part of a 21-gene operon encoding functions required for the catabolism of 1,2-propanediol (6).
PduO-type enzymes (including the human enzyme) have been characterized in vitro and in vivo, and crystal structures with and without ATP bound to them have been reported (29, 30, 33, 34, 47, 48, 52). A striking feature of PduO-type enzymes is that, unlike CobA-type enzymes, they lack a P loop for ATP binding and use a novel motif to bind ATP (48, 52).
BACTERIA AND ARCHAEA USE DIFFERENT PATHWAYS TO SALVAGE Cbi
There are several important differences in the pathways used by bacteria and archaea to convert Cbi to AdoCbl (Fig. 2).
In bacteria, AdoCbi is the substrate of a bifunctional enzyme with kinase and guanylyltransferase activities. This enzyme goes by the name of CobU (in serovar Typhimurium), or CobP (in P. denitrificans) (4, 39); CobU and CobP are homologs. From structural studies of the CobU enzyme of serovar Typhimurium, we know the enzyme works as a trimer; although some aspects of the catalytic mechanism are known, our understanding of how the enzyme works is incomplete. The CobU enzyme phosphorylates the hydroxyl group of the aminopropanol (AP) moiety of AdoCbi, yielding AdoCbi-P, which is used by the enzyme to generate AdoCbi-GDP by transferring the guanylate moiety of GTP to it (39, 57).
Archaea use a different strategy to salvage Cbi from their environments, because archaea lack CobU. Instead they use CobY, a nonorthologous replacement for CobU that has GTP:AdoCbi-P guanylyltransferase activity but lacks AdoCbi kinase activity (56, 64). Archaea solve the problem of not having an AdoCbi kinase enzyme by removing the AP moiety of AdoCbi first, to yield adenosylcobyric acid, which is condensed with 1-aminopropanol-O-2-phosphate by the AdoCbi-P synthase (CbiB) enzyme to yield AdoCbi-P (Fig. 2) (43; C. L. Zayas and J. C. Escalante-Semerena, unpublished results).
In archaea, the cbiZ gene encodes the amidohydrolase that converts AdoCbi to adenosylcobyric acid (66). The enzymatic activity of CbiZ has been studied in vitro, and its involvement in AdoCbl synthesis has been demonstrated in vivo in Halobacterium sp. strain NRC-1 (62, 66). Hyperthermophilic archaea (e.g., Methanopyrus kandleri AV19, Aeropyrum pernyx, and Pyrobaculum aerophilum) synthesize CbiZ fused to CobZ, the nonorthologous replacement of the AdoCbl-5′-P CobC phosphatase of bacteria (68). The CbiZ-CobZ fusion is known as CbiS (63). The reasons for fusing CbiZ to CobZ are not clear. The latter enzymes do not catalyze sequential reactions of the pathway (Fig. 1), so traffic of intermediates between these proteins is not expected. Interestingly, the CbiZ and CobZ portions of CbiS can be separated and are functional in vivo and in vitro (63). Whether CbiS evolved as a means to stabilize CbiZ, CobZ, or both at very high temperatures needs further investigation.
SYNTHESIS AND ACTIVATION OF THE LOWER LIGAND BASE
The lower ligand base of AdoCbl is DMB. There are two known, distinct pathways for DMB synthesis. The chief difference between the two is the involvement of oxygen (reviewed in reference 44). Very recently, results of studies of the BluB protein isolated from Rhodospirillum rubrum and Sinorhizobium meliloti showed that BluB is necessary and sufficient to convert reduced flavin mononucleotide into DMB via an O2-dependent mechanism (26, 55). The studies of the S. meliloti BluB enzyme also reported the crystal structure of the protein at a resolution of 2.9 Å with substrates in the active site. These findings are a major advance in the field, because the identities of the enzymes involved in the synthesis of DMB in the presence of O2 remained unknown for a long time. The detailed mechanistic analysis of the reaction should follow in the not-so-distant future.
In some bacteria (e.g., Streptomyces, Thermobifida, and others), the BluB protein is fused to the CobT enzyme that activates DMB into its unique α-nucleotide, known as α-ribazole-P. Detailed analyses of these BluB-CobT fusion proteins would provide insights into metabolite channeling.
The pathway for DMB synthesis under anoxic conditions was delineated by tracer analyses performed in Eubacterium limosum (reviewed in reference 44), but the enzymes catalyzing these reactions have not been identified. The anaerobic pathway of DMB synthesis is not present in serovar Typhimurium, since under anoxic conditions, this bacterium does not synthesize DMB; instead it makes AdoCba's that have, as the lower ligand, either adenine (aka pseudoAdoCba or pseudoAdoB12) or 2-methyladenine [aka 2-methyladenylyl-(Coα)-AdoCba] (31, 32). To date, work on lower ligand synthesis in archaea has not been reported.
In serovar Typhimurium and E. coli, activation of the lower ligand base is performed by the nicotinate mononucleotide (NaMN) (NAD+):DMB phosphoribosyl (ADP-ribosyl) transferase (CobT) enzyme (Fig. 2). The phosphoribosyltransferase activity of CobT has been studied in P. denitrificans (the protein is known as CobU in this bacterium) and serovar Typhimurium (14, 58, 59). Structural analyses of the CobT protein in its apo form and bound to substrates provided insights into the mechanism of catalysis and the specificity of the enzyme for its base substrate (15-17). The ADP-ribosyltransferase activity of CobT was recently described, uncovering the previously unknown intermediate of the pathway known as α-DAD (α-5,6-dimethylbenzimidazole adenine dinucleotide) (37). The enzyme responsible for cleaving α-DAD into AMP and α-ribazole-P has not been identified. The two routes of α-ribazole-P synthesis do not exclude each other. The argument for α-DAD being the preferred route in serovar Typhimurium is based on reports in the literature which show that NaMN is not present in sufficiently high amounts for the CobT enzyme to support AdoCbl synthesis (37). Prokaryotes that have elevated levels of NaMN are probably making α-ribazole-P directly, not via the α-DAD route.
Our understanding of the pathways responsible for the synthesis of other lower-ligand bases found in different complete corrinoids (aka Cba's) is limited (44).
THE LAST TWO STEPS OF THE AdoCbl BIOSYNTHETIC PATHWAY
The penultimate step of the AdoCbl biosynthetic pathway is catalyzed by the AdoCbl-5′-P (CobS) enzyme, an integral membrane protein (36, 38, 40). This activity was also characterized in P. denitrificans, where this enzyme is known as CobV (14). Until recently, it was unclear whether CobS catalyzed the last or penultimate step of the pathway. The results of in vitro studies did not unequivocally place this enzyme in the pathway (14, 38). The answer to this question was obtained through phenotypic analyses of mutants defective in CobC phosphatase function (41) (Fig. 2). These studies provided the needed in vivo support to the assignment of CobS as the penultimate step and CobC as the last step of the pathway (67).
Archaea synthesize CobZ, a nonorthologous replacement of the bacterial CobC enzyme (68), and as mentioned above, in some extremely thermophilic archaea, CobZ is fused to the CbiZ amidohydrolase enzyme (62, 63). In other archaea (e.g., Halobacterium) the nonorthologous replacement for CobC has not been identified.
WHAT IS NEXT IN AdoCbl RESEARCH?
There are many open questions of interest regarding the late steps of the AdoCbl biosynthetic pathway. Chief among them are those concerning the locations of the AdoCbi-P synthase (CbiB) and AdoCbl-5′-P synthase (CobS) enzymes at the cell membrane. What physiological need is met at this location? Do the proteins involved in the nucleotide loop assembly pathway form a complex? We need to know more about the structures of these proteins so we can better understand their function. Because AdoCbl synthesis is unique to prokaryotes, the enzymes involved in making this important coenzyme would be good targets for the development of antimicrobials that would not affect the host.
Acknowledgments
This work was supported by Public Health Service grant R01-GM40313 to J.C.E.-S.
I am indebted to Ivan Rayment and coworkers (Department of Biochemistry, University of Wisconsin—Madison) and to Thomas Brunold and coworkers (Department of Chemistry, University of Wisconsin—Madison) for longstanding, fruitful collaborations. Without these collaborations, our understanding of the nucleotide loop assembly and adenosylation pathways would be very limited. Special thanks go to past and present graduate and undergraduate students in my laboratory who contributed so much to our understanding of how this magnificent molecule is assembled.
Footnotes
Published ahead of print on 4 May 2007.
REFERENCES
- 1.Bassford, P. J., Jr., and R. J. Kadner. 1977. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J. Bacteriol. 132:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biochemistry. 1974. The nomenclature of corrinoids (1973 recommendations). IUPAC-IUB Commission on Biochemical Nomenclature. Biochemistry 13:1555-1560. [DOI] [PubMed] [Google Scholar]
- 3.Biochimica et Biophysica Acta. 1966. Tentative rules. Nomenclature of corrinoids. Biochim. Biophys. Acta 117:285-288. [PubMed] [Google Scholar]
- 4.Blanche, F., L. Debussche, A. Famechon, D. Thibaut, B. Cameron, and J. Crouzet. 1991. A bifunctional protein from Pseudomonas denitrificans carries cobinamide kinase and cobinamide phosphate guanylyltransferase activities. J. Bacteriol. 173:6052-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanche, F., D. Thibaut, L. Debussche, R. Hertle, F. Zipfel, and G. Muller. 1993. Parallels and decisive differences in vitamin B12 biosynthesis. Angew. Chem. Int. Ed. Engl. Suppl. 32:1651-1653. [Google Scholar]
- 6.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borths, E. L., K. P. Locher, A. T. Lee, and D. C. Rees. 2002. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. USA 99:16642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borths, E. L., B. Poolman, R. N. Hvorup, K. P. Locher, and D. C. Rees. 2005. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 44:16301-16319. [DOI] [PubMed] [Google Scholar]
- 9.Bradbeer, C., P. R. Reynolds, G. M. Bauler, and M. T. Fernandez. 1986. A requirement for calcium in the transport of cobalamin across the outer membrane of Escherichia coli. J. Biol. Chem. 261:2520-2523. [PubMed] [Google Scholar]
- 10.Buan, N. R., and J. C. Escalante-Semerena. 2005. Computer-assisted docking of flavodoxin with the ATP:co(I)rrinoid adenosyltransferase (CobA) enzyme reveals residues critical for protein-protein interactions but not for catalysis. J. Biol. Chem. 280:40948-40956. [DOI] [PubMed] [Google Scholar]
- 11.Buan, N. R., K. Rehfeld, and J. C. Escalante-Semerena. 2006. Studies of the CobA-type ATP:co(I)rrinoid adenosyltransferase enzyme of Methanosarcina mazei strain Gö1. J. Bacteriol. 188:3543-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buan, N. R., S. J. Suh, and J. C. Escalante-Semerena. 2004. The eutT gene of Salmonella enterica encodes an oxygen-labile, metal-containing ATP:corrinoid adenosyltransferase enzyme. J. Bacteriol. 186:5708-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadieux, N., C. Bradbeer, E. Reeger-Schneider, W. Koster, A. K. Mohanty, M. C. Wiener, and R. J. Kadner. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cameron, B., F. Blanche, M. C. Rouyez, D. Bisch, A. Famechon, M. Couder, L. Cauchois, D. Thibaut, L. Debussche, and J. Crouzet. 1991. Genetic analysis, nucleotide sequence, and products of two Pseudomonas denitrificans cob genes encoding nicotinate-nucleotide:dimethylbenzimidazole phosphoribosyltransferase and cobalamin (5′-phosphate) synthase. J. Bacteriol. 173:6066-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheong, C. G., J. C. Escalante-Semerena, and I. Rayment. 2002. Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica. J. Biol. Chem. 277:41120-41127. [DOI] [PubMed] [Google Scholar]
- 16.Cheong, C. G., J. C. Escalante-Semerena, and I. Rayment. 2001. Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica. J. Biol. Chem. 276:37612-37620. [DOI] [PubMed] [Google Scholar]
- 17.Cheong, C. G., J. C. Escalante-Semerena, and I. Rayment. 1999. The three-dimensional structures of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella typhimurium complexed with 5,6-dimethybenzimidazole and its reaction products determined to 1.9Å resolution. Biochemistry 38:16125-16135. [DOI] [PubMed] [Google Scholar]
- 18.Cherezov, V., E. Yamashita, W. Liu, M. Zhalnina, W. A. Cramer, and M. Caffrey. 2006. In meso structure of the cobalamin transporter, BtuB, at 1.95 Å resolution. J. Mol. Biol. 364:716-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debussche, L., M. Couder, D. Thibaut, B. Cameron, J. Crouzet, and F. Blanche. 1991. Purification and partial characterization of cob(I)alamin adenosyltransferase from Pseudomonas denitrificans. J. Bacteriol. 173:6300-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeVeaux, L. C., and R. J. Kadner. 1985. Transport of vitamin B12 in Escherichia coli: cloning of the btuCD region. J. Bacteriol. 162:888-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escalante-Semerena, J. C., S. J. Suh, and J. R. Roth. 1990. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J. Bacteriol. 172:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson, A. D., C. A. Amezcua, N. M. Halabi, Y. Chelliah, M. K. Rosen, R. Ranganathan, and J. Deisenhofer. 2007. Signal transduction pathway of TonB-dependent transporters. Proc. Natl. Acad. Sci. USA 104:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fonseca, M. V., N. R. Buan, A. R. Horswill, I. Rayment, and J. C. Escalante-Semerena. 2002. The ATP:co(I)rrinoid adenosyltransferase (CobA) enzyme of Salmonella enterica requires the 2′-OH group of ATP for function and yields inorganic triphosphate as its reaction byproduct. J. Biol. Chem. 277:33127-33131. [DOI] [PubMed] [Google Scholar]
- 24.Fonseca, M. V., and J. C. Escalante-Semerena. 2001. An in vitro reducing system for the enzymic conversion of cobalamin to adenosylcobalamin. J. Biol. Chem. 276:32101-32108. [DOI] [PubMed] [Google Scholar]
- 25.Fonseca, M. V., and J. C. Escalante-Semerena. 2000. Reduction of cob(III)alamin to cob(II)alamin in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:4304-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray, M. J., and J. C. Escalante-Semerena. 2007. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc. Natl. Acad. Sci. USA 104:2921-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heller, K., B. J. Mann, and R. J. Kadner. 1985. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J. Bacteriol. 161:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeter, R. M., B. M. Olivera, and J. R. Roth. 1984. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J. Bacteriol. 159:206-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, C. L., M. L. Buszko, and T. A. Bobik. 2004. Purification and initial characterization of the Salmonella enterica PduO ATP:cob(I)alamin adenosyltransferase. J. Bacteriol. 186:7881-7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, C. L., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keck, B., M. Munder, and P. Renz. 1998. Biosynthesis of cobalamin in Salmonella typhimurium: transformation of riboflavin into the 5,6-dimethylbenzimidazole moiety. Arch. Microbiol. 171:66-68. [DOI] [PubMed] [Google Scholar]
- 32.Keck, B., and P. Renz. 2000. Salmonella typhimurium forms adenylcobamide and 2-methyladenylcobamide, but no detectable cobalamin during strictly anaerobic growth. Arch. Microbiol. 173:76-77. [DOI] [PubMed] [Google Scholar]
- 33.Leal, N. A., H. Olteanu, R. Banerjee, and T. A. Bobik. 2004. Human ATP:cob(I)alamin adenosyltransferase and its interaction with methionine synthase reductase. J. Biol. Chem. 279:47536-47542. [DOI] [PubMed] [Google Scholar]
- 34.Leal, N. A., S. D. Park, P. E. Kima, and T. A. Bobik. 2003. Identification of the human and bovine ATP:cob(I)alamin adenosyltransferase cDNAs based on complementation of a bacterial mutant. J. Biol. Chem. 278:9227-9234. [DOI] [PubMed] [Google Scholar]
- 35.Lundrigan, M. D., and R. J. Kadner. 1989. Altered cobalamin metabolism in Escherichia coli btuR mutants affects btuB gene regulation. J. Bacteriol. 171:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maggio-Hall, L. A., K. R. Claas, and J. C. Escalante-Semerena. 2004. The last step in coenzyme B12 synthesis is localized to the cell membrane in bacteria and archaea. Microbiology 150:1385-1395. [DOI] [PubMed] [Google Scholar]
- 37.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 2003. Alpha-5,6-dimethylbenzimidazole adenine dinucleotide (α-DAD), a putative new intermediate of coenzyme B12 biosynthesis in Salmonella typhimurium. Microbiology 149:983-990. [DOI] [PubMed] [Google Scholar]
- 38.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 1999. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc. Natl. Acad. Sci. USA 96:11798-11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole, G. A., and J. C. Escalante-Semerena. 1995. Purification and characterization of the bifunctional CobU enzyme of Salmonella typhimurium LT2. Evidence for a CobU-GMP intermediate. J. Biol. Chem. 270:23560-23569. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole, G. A., M. R. Rondon, and J. C. Escalante-Semerena. 1993. Analysis of mutants defective in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 175:3317-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Toole, G. A., J. R. Trzebiatowski, and J. C. Escalante-Semerena. 1994. The cobC gene of Salmonella typhimurium codes for a novel phosphatase involved in the assembly of the nucleotide loop of cobalamin. J. Biol. Chem. 269:26503-26511. [PubMed] [Google Scholar]
- 42.Postle, K., and R. A. Larsen. 17 January 2007, posting date. TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals. doi: 10.1007/s10534-006-9071-6. [Epub ahead of print.] [DOI] [PubMed]
- 43.Raux, E., A. Lanois, F. Levillayer, M. J. Warren, E. Brody, A. Rambach, and C. Thermes. 1996. Salmonella typhimurium cobalamin (vitamin B12) biosynthetic genes: functional studies in S. typhimurium and Escherichia coli. J. Bacteriol. 178:753-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renz, P. 1999. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of other bases found in natural corrinoids, p. 557-575. In R. Banerjee (ed.), Chemistry and biochemistry of B12. John Wiley & Sons, Inc., New York, NY.
- 45.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148-41159. [DOI] [PubMed] [Google Scholar]
- 46.Roessner, C. A., and A. I. Scott. 2006. Fine-tuning our knowledge of the anaerobic route to cobalamin (vitamin B12). J. Bacteriol. 188:7331-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanishvili, R., M. Pennycooke, J. Gu, X. Xu, A. Joachimiak, A. M. Edwards, and D. Christendat. 2004. Crystal structure of the hypothetical protein TA1238 from Thermoplasma acidophilum: a new type of helical super-bundle. J. Struct. Funct. Genomics 5:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert, H. L., and C. P. Hill. 2006. Structure of ATP-bound human ATP:cobalamin adenosyltransferase. Biochemistry 45:15188-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott, A. I., and C. A. Roessner. 2002. Biosynthesis of cobalamin (vitamin B12). Biochem. Soc. Trans. 30:613-620. [DOI] [PubMed] [Google Scholar]
- 50.Sheppard, D. E., J. T. Penrod, T. Bobik, E. Kofoid, and J. R. Roth. 2004. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 186:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stich, T. A., N. R. Buan, J. C. Escalante-Semerena, and T. C. Brunold. 2005. Spectroscopic and computational studies of the ATP:corrinoid adenosyltransferase (CobA) from Salmonella enterica: insights into the mechanism of adenosylcobalamin biosynthesis. J. Am. Chem. Soc. 127:8710-8719. [DOI] [PubMed] [Google Scholar]
- 52.St. Maurice, M., P. E. Mera, M. P. Taranto, F. Sesma, J. C. Escalante-Semerena, and I. Rayment. 2007. Structural characterization of the active site of the PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri. J. Biol. Chem. 282:2596-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suh, S., and J. C. Escalante-Semerena. 1995. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J. Bacteriol. 177:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suh, S. J., and J. C. Escalante-Semerena. 1993. Cloning, sequencing and overexpression of cobA which encodes ATP:corrinoid adenosyltransferase in Salmonella typhimurium. Gene 129:93-97. [DOI] [PubMed] [Google Scholar]
- 55.Taga, M. E., N. A. Larsen, A. R. Howard-Jones, C. T. Walsh, and G. C. Walker. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, M. G., and J. C. Escalante-Semerena. 2000. Identification of an alternative nucleoside triphosphate:5′-deoxyadenosylcobinamide phosphate nucleotidyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 182:4227-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, M. G., T. B. Thompson, I. Rayment, and J. C. Escalante-Semerena. 2000. Analysis of the adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) enzyme of Salmonella typhimurium LT2. Identification of residue H46 as the site of guanylylation. J. Biol. Chem. 275:27376-27386. [DOI] [PubMed] [Google Scholar]
- 58.Trzebiatowski, J. R., and J. C. Escalante-Semerena. 1997. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J. Biol. Chem. 272:17662-17667. [DOI] [PubMed] [Google Scholar]
- 59.Trzebiatowski, J. R., G. A. O'Toole, and J. C. Escalante-Semerena. 1994. The cobT gene of Salmonella typhimurium encodes the NaMN:5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-α-d-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 176:3568-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Bibber, M., C. Bradbeer, N. Clark, and J. R. Roth. 1999. A new class of cobalamin transport mutants (btuF) provides genetic evidence for a periplasmic binding protein in Salmonella typhimurium. J. Bacteriol. 181:5539-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warren, M. J., E. Raux, H. L. Schubert, and J. C. Escalante-Semerena. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19:390-412. [DOI] [PubMed] [Google Scholar]
- 62.Woodson, J. D., and J. C. Escalante-Semerena. 2004. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12 precursor cobinamide in archaea. Proc. Natl. Acad. Sci. USA 101:3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woodson, J. D., and J. C. Escalante-Semerena. 2006. The cbiS gene of the archaeon Methanopyrus kandleri AV19 encodes a bifunctional enzyme with adenosylcobinamide amidohydrolase and α-ribazole-phosphate phosphatase activities. J. Bacteriol. 188:4227-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woodson, J. D., R. F. Peck, M. P. Krebs, and J. C. Escalante-Semerena. 2003. The cobY gene of the archaeon Halobacterium sp. strain NRC-1 is required for de novo cobamide synthesis. J. Bacteriol. 185:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woodson, J. D., A. A. Reynolds, and J. C. Escalante-Semerena. 2005. ABC transporter for corrinoids in Halobacterium sp. strain NRC-1. J. Bacteriol. 187:5901-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodson, J. D., C. L. Zayas, and J. C. Escalante-Semerena. 2003. A new pathway for salvaging the coenzyme B12 precursor cobinamide in archaea requires cobinamide-phosphate synthase (CbiB) enzyme activity. J. Bacteriol. 185:7193-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zayas, C. L., and J. C. Escalante-Semerena. 2007. Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: evidence that dephosphorylation of adenosylcobalamin-5′-phosphate by the CobC phosphatase is the last step of the pathway. J. Bacteriol. 189:2210-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zayas, C. L., J. D. Woodson, and J. C. Escalante-Semerena. 2006. The cobZ gene of Methanosarcina mazei Gö1 encodes the nonorthologous replacement of the α-ribazole-5′-phosphate phosphatase (CobC) enzyme of Salmonella enterica. J. Bacteriol. 188:2740-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]