Abstract
Background
The lemurs of Madagascar provide an excellent mammalian radiation to explore mechanisms and processes favouring species diversity and evolution. Species diversity, in particular of nocturnal species, increased considerably during the last decade. However, the factors contributing to this high diversity are not well understood. We tested predictions derived from two existing biogeographic models by exploring the genetic and morphological divergence among populations of a widely distributed lemur genus, the sportive lemur (Lepilemur ssp.) along a 560 km long transect from western to northern Madagascar.
Results
By using the phylogenetic analyses of mtDNA sequence data, molecular diagnostic sites and phenotypic morphometric traits, we uncovered two previously undetected species whose distributions contradict the two existing biogeographic models. Brief species descriptions are provided and a new biogeographic model is proposed (the ”large river model“).
Conclusion
According to the ”large river model“, large rivers in north and northwestern Madagascar acted as geographical barriers for gene flow and facilitated speciation events on a much smaller spatial scale than previously thought. Thereby, this study does not only show that species diversity in nocturnal Malagasy primates is continuously underestimated but aims to emphasize the need for conservation actions if those species with small ranges shall not face extinction in the near future.
Background
Malagasy lemurs constitute one of six major radiations of extant primates [1]. Lemurs show a remarkable species diversity, both numerically and in terms of adaptations making them an excellent mammalian radiation to explore mechanisms and processes underlying speciation and evolution. During the last decade, species diversity in lemurs increased from 33 to currently 74 [2,3]. In relation to the small surface area of Madagascar, diversity of species within this primate radiation is quite high. Individual lemur species tend to have small geographic ranges in comparison to other primates. Because of such limited geographic ranges and the high rate of deforestation, the need for conservation action including genetic monitoring and effective management policies is particularly urgent [2,4]. Two major models have been proposed to explain diversity of Malagasy mammals.
The "Martin model" divided northern and northwestern Madagascar into four biogeographical zones (circles in Fig. 1) [5,6]. The western zone (W1) covers the area between the two major rivers Tsiribihina and Betsiboka. The northwestern zone 1 covers the area between the two major rivers Betsiboka and Maevarano (NW), the northwestern zone 2 the area between the rivers Maevarano and Mahavavy (X). The northern zone (N) covers the area between the rivers Mahavavy and Fanambana. These riverine barriers were hypothesized to form geographical boundaries to gene flow and consequently favour allopatric speciation. This model of speciation within Madagascar was refined [6] and it was shown that it is compatible with a reconstruction of speciation within the families Lemuridae, Cheirogaleidae and Indridae [7].
Figure 1.
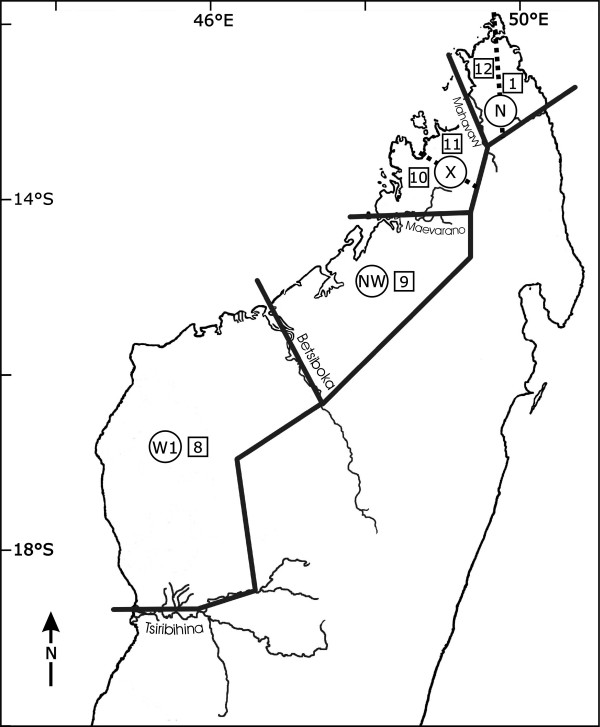
Zonation of northwestern Madagascar described by Martin (1972), zones marked with letters, and by Wilmé et al. (2006), zones marked with numbers.
The recent "Wilmé model" explained the process of explosive speciation on the island using a mechanistic model [8]. Madagascar's rivers and associated watersheds with sources at relatively low elevations suggested to be zones of isolation that led to the evolution of locally endemic taxa, whereas those at higher elevations were proposed to have functioned as zones of retreat and dispersion and contain a lower level of microendemism. Wilmé et al. (2006) divided northern and northwestern Madagascar into six centres of endemism (squares in Fig. 1). The western zone (zone 8) covers the area between the two major rivers Tsiribihina and Betsiboka, corresponding to Martin's W1. One large northwestern zone, zone 9, corresponded to Martin's NW. Two smaller northwestern zones, zone 10, between the two rivers Maevarano and Sambirano, and zone 11 between the two rivers Sambirano and Mahavavy were suggested. In addition, two northern zones, one (zone 12), between the river Mahavavy and the continental divide between eastern and western draining watersheds, and another (zone 1), between the continental divide and the river Bemarivo, divided the N-zone of Martin into two partitions.
The geographical settings in northwestern and northern Madagascar are perfect to test if allopatric speciation of a widely distributed lemur genus follows one of the models. Each model predicts a different minimum number of species in this region and divergent distributions. Whereas the "Martin model" predicts four species, the "Wilmé model" proposes six species.
Sportive lemurs (Lepilemur spp.) are an excellent lemur group to test these two models of mammalian distribution in Madagascar, because they occur in almost all forested regions on the island. They are cat-sized vertical clingers and leapers with powerful hind legs. They are nocturnal and totally arboreal. They live in dispersed pairs and have an elaborated vocal repertoire [9-11]. Because differences in pelage colouration and other external characteristics between species are inconspicous, their early classification [12,13] based on morphological features was disputed until comprehensive cytogenetic approaches and molecular studies allowed the recognition of twelve species [11,14-18].
The aim of this study is to test the predictions from the models with the largest available genetic and morphological data set of a larger-sized lemur. We sequenced three mitochondrial genes of particular diagnostic importance for phylogeography (D-loop, Cytochrome B and NADH-dehydrogenase subunit 4) of individuals captured in 14 different localities that covered a 560 km transect and the area between eight large rivers (Inter-River-Systems, IRS) from western to northern Madagascar. In addition, morphometric data were analysed in order to explore, to which extent genetic differentiation coincides with morphological diversification. As in similar studies [e.g. [19,20]] we favour the phylogenetic species concept [21,22], where fixed molecular differences among parapatric populations indicate the existence of species barriers.
Results
Phylogenetic relationships
The 48 sequences available for the D-loop (43 own sequences + five reference sequences), after having cut out the hypervariable part, varied from 388 to 390 bp in length. 128 characters were constant, 201 variable characters are parsimony-uninformative and 66 were parsimony-informative. There were 17 different haplotypes. The 72 sequences available for the partial cytochrome B (43 own sequences + 29 reference sequences) were 352 bp long, with no indels. 211 characters were constant, 17 variable characters are parsimony-uninformative and 124 were parsimony-informative. There were 32 different haplotypes. The 50 sequences available for the partial ND4 (43 own sequences + seven reference sequences) varied from 630 to 631 bp in length. 408 characters were constant, 72 variable characters were parsimony-uninformative and 153 were parsimony-informative characters. There were 19 different haplotypes. Table 1 shows the best-fit models for the three loci selected by the hierarchical likelihood ratio test (hLRT) implemented in Modeltest 3.5.mac. Based on the single-gene-trees, derived for the new and reference sequences, the samples in this study could be classified as follows: The individuals found in IRS 0 clustered with L. aeeclis, the individuals found in IRS I with L. edwardsi, the individuals found in IRS IV with L. sahamalazensis, the individuals found in IRS V and VI with L. dorsalis, the individuals found in IRS VII with L. ankaranensis, the individuals from Kirindy with L. ruficaudatus and the individuals from Mantadia with L. mustelinus. The individuals from IRS II and III did not cluster with any of the reference sequences. No sampled individual clustered with the reference sequences of L. leucopus, L. microdon, L. randrianasoli or L. septentrionalis.
Table 1.
Best-fit mutation model for the three mitochondrial loci and the concatenated sequence selected by the hierarchical likelihood ratio test (hLRT) implemented in Modeltest 3.5.mac.
| Locus | Method | Model | Base | Nst | Alpha | Pinvar | TRatio |
| D-loop | ML, NJ | HKY+G | 0.3066 0.2151 0.1828 0.2955 | 2 | 0.1752 | 0 | 3.2075 |
| Cyt B | ML, NJ | HKY+I+G | 0.3092 0.3229 0.1219 0.2460 | 2 | 3.5808 | 0.5469 | 12.5738 |
| ND4 | ML, NJ | HKY+G | 0.3372 0.2697 0.1127 0.2804 | 2 | 0.2736 | 0 | 8.1268 |
| concatenated | ML, NJ | HKY+I+G | 0.3124 0.2661 0.1377 0.2838 | 2 | 0.8801 | 0.3833 | 6.1953 |
Nst: number of substitution types
Pinvar: assumed proportion of invariable sites
Alpha: shape parameter
TRatio: transition/transversion ratio
In order to reconstruct the phylogenetic relationships within the genus Lepilemur, we combined these three loci to one concatenated sequence, 1380 bp in length. 768 characters were constant, 333 variable characters were parsimony-uninformative, and 279 were parsimony-informative. There were 21 different haplotypes. The best-fit model selected by hLRT in Modeltest 3.5.mac was the HKY+I+G model (Table 1). Figure 2 shows the Neighbour-Joining tree based on the concatenated sequence. All populations within each IRS clustered together, so that each IRS (including the populations Kirindy (West) and Mantadia (East)) built separate terminal clades, supported by high bootstrap values (Fig. 2). The phylogram consists of four major clades, a western, a northwestern, a northern clade, and the clade of L. mustelinus. L. mustelinus branched off first, followed by the western clade that consisted of IRS 0 and the individuals found in Kirindy (West) (bootstrap values between 93 and 96). The northern clade consisted of IRS IV, V, VI and VII (bootstrap values of 100), and the northwestern clade of IRS I, II and III (bootstrap values of 100). All so far recognized species formed distinct terminal clades with moderate (L. ankaranensis, L. dorsalis) to large (L. mustelinus, L. ruficaudatus, L. aeeclis) branch lengths. Branch lengths among IRS I, II and III in the northwestern clade were in the same scale as these between L. ankaranensis and L. dorsalis.
Figure 2.
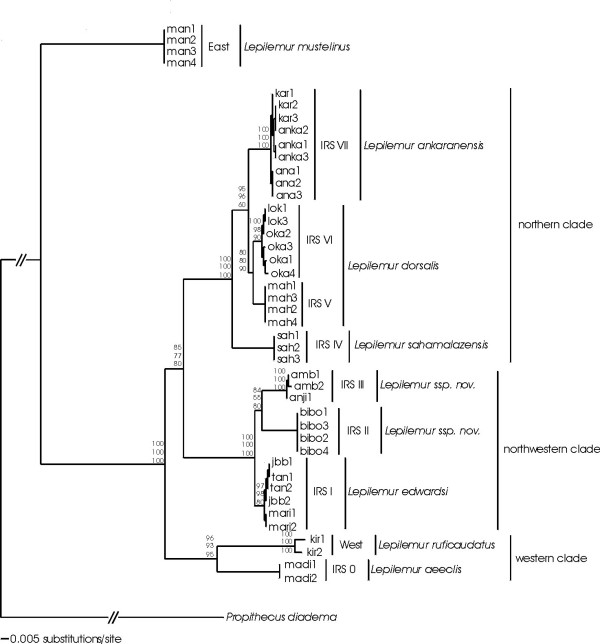
Neighbour-Joining tree based on the concatenated sequences of the three loci. The branch lengths indicate the number of substitutions, the numbers at the nodes indicate bootstrap values for internal branches (top: NJ, middle: MP, bottom: ML).
The absolute pairwise distances within an IRS ranged from zero to seven characters (Fig. 3). The absolute pairwise distances among IRSs (including Kirindy (West) and Mantadia (East)) varied from 18 to 199 characters. The largest absolute pairwise distance (199 characters) existed between L. aeeclis and L. mustelinus. The smallest absolute pairwise distance (18 to 23 characters) among IRSs existed between IRS V and IRS VI, both of them were previously supposed to give home to L. dorsalis. The relative genetic distance between these two IRSs can be defined as intermediate between the intra-IRS differences (0–7 bp) and the interspecific differences (32–199 bp). This level of differentiation could indicate the presence of two subspecies of L. dorsalis. In accordance with the deep phylogenetic splits in the Lepilemur tree among the western, northwestern, and northern clade, absolute pairwise distances were always largest when crossing borders among neighbouring biogeographic zones (L. aeeclis to L. edwardsi and IRS III to L. sahamalazensis, Fig. 3). When examining the absolute pairwise distances among the IRSs within the northwestern clade, they were the same size or even larger than between L. dorsalis and L. ankaranensis, which are accepted species (Fig. 3).
Figure 3.
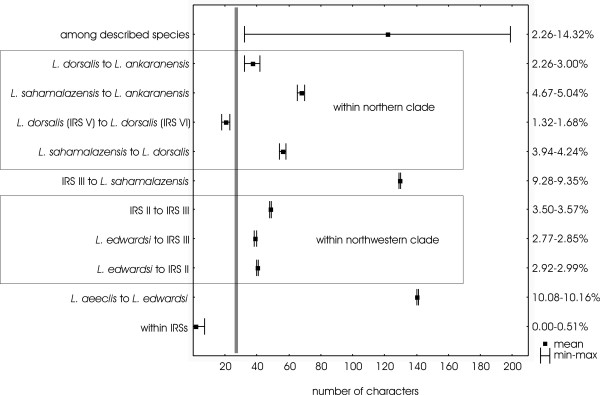
Absolute pairwise distances (minimum-maximum and mean) within IRSs, between neighbouring IRSs/species and among the seven already described species.
Additional files 1, 2, 3 show the molecular diagnostic sites for each terminal clade in each of the three genes. Recognized species had total number of one (L. dorsalis) to 73 (L. mustelinus) sites that allowed to identify them unmistakably. The two terminal clades containing the individuals of IRS II and III had a total of eleven and seven diagnostic sites, respectively. The absolute pairwise distances as well as the analysis of the diagnostic sites indicate the presence of two new Lepilemur species in northwestern Madagascar, one in IRS II and one in IRS III. Consequently, the geographic range of L. edwardsi is much smaller than previously assumed, and limited exclusively to IRS I. By mapping each of these species with respect to their geographical setting (IRS), it can be concluded that all large rivers act as genetic barriers in this genus (Fig. 4).
Figure 4.
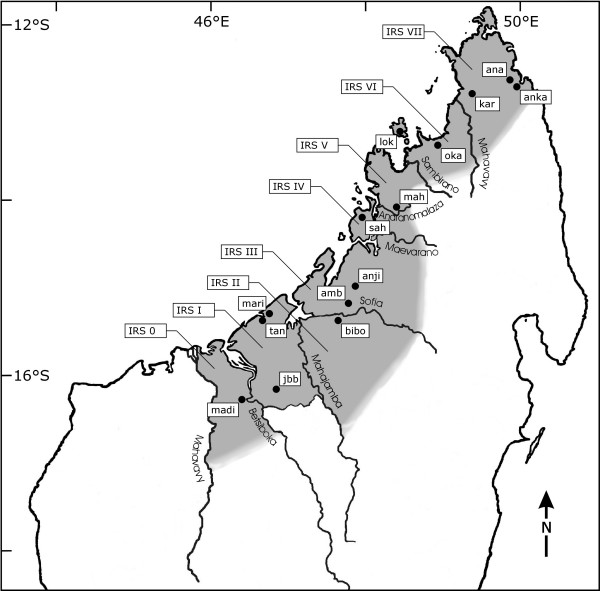
Map of study sites, large rivers and the zonation of the eight Inter-River-Systems (IRSs).
Morphometry
The means and standard deviations of all morphometric variables for the nine phylogenetically defined species are provided in Table 2. All variables showed significant differences for species in the ANOVA. Post-hoc tests revealed that ear length and intraorbital distance was significantly different in 15 of the 28 possible pairs of species and thereby the two most distinct variables, followed by snout length (14/28), weight (12/28), head width and lower leg length (10/28), tail circumference (9/28), 3rd toe length (8/28), interorbital distance and hind foot length (6/28) and tail length (4/28). Six variables showed tendencies (0.05 ≤ p < 0.1) in one to two possible pairs of species. The Post-hoc tests revealed significant differences between the northwestern and northern clade and between the northern clade and L. mustelinus. Moreover, it could distinguish between all neighbouring species (established and proposed), except between L. aeeclis from the western clade and L. edwardsi from the northwestern clade (Table 2).
Table 2.
Descriptive statistics (mean ± SD) for 10 morphometric variables from each species.
|
L. aeeclis (n = 5) |
L. edwardsi (n = 11) |
L. sp. nov. in IRS II (n = 6) |
L. sp. nov. in IRS III (n = 8) |
L. sahamalazensis (n = 7) |
L. dorsalis (n = 30) |
L. ankaranensis (n = 26) |
L. mustelinus (n = 7) |
Results of ANOVA | |||||||||||||||||
| Variable | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | df | F | p | ||||||
| Ear length [mm] | 30.72 | 0.83 | 31.75 | 1.93 | 33.60 | 0.99 | 33.41 | 2.10 | ** | 26.73 | 1.79 | 25.59 | 1.52 | ** | 28.79 | 1.71 | ** | 31.93 | 3.11 | 7 | 37.081 | 0.000 | |||
| Head width [mm] | 36.38 | 0.83 | 34.93 | 1.36 | * | 37.88 | 2.24 | 37.61 | 2.29 | ** | 34.00 | 1.37 | 34.69 | 1.72 | 34.12 | 1.97 | ** | 38.36 | 2.33 | 7 | 9.105 | 0.000 | |||
| Snout length [mm] | 15.50 | 1.13 | 17.52 | 2.25 | ** | 21.00 | 0.83 | 19.34 | 1.45 | ** | 15.10 | 1.05 | 14.74 | 1.69 | 13.97 | 1.67 | ** | 18.66 | 2.14 | 7 | 23.894 | 0.000 | |||
| Intraorbital distance [mm] | 13.60 | 0.65 | 14.51 | 0.79 | 15.40 | 1.04 | 15.29 | 1.12 | ** | 13.31 | 0.96 | ** | 12.08 | 0.89 | 12.34 | 0.76 | 12.79 | 1.17 | 7 | 25.088 | 0.000 | ||||
| Interorbital distance [mm] | 36.98 | 0.80 | 37.80 | 1.33 | 36.05 | 1.92 | 36.88 | 0.82 | 34.96 | 1.39 | ** | 36.82 | 1.45 | ** | 35.48 | 1.13 | ** | 38.41 | 1.77 | 7 | 7.470 | 0.000 | |||
| Lower leg length [mm] | 96.30 | 1.37 | 96.22 | 5.37 | 101.37 | 3.63 | 105.44 | 2.91 | 99.27 | 1.40 | 98.05 | 5.88 | 99.74 | 3.69 | ** | 113.30 | 3.94 | 7 | 12.874 | 0.000 | |||||
| Hind foot length [mm] | 48.14 | 2.98 | 51.96 | 2.68 | 50.23 | 1.43 | 51.76 | 3.45 | 49.00 | 2.01 | 49.83 | 2.31 | 48.90 | 2.51 | ** | 54.94 | 1.45 | 7 | 6.953 | 0.000 | |||||
| 3rd toe length [mm] | 21.16 | 0.90 | 22.24 | 1.47 | 22.37 | 1.62 | 23.75 | 1.60 | 21.70 | 0.82 | 20.61 | 2.38 | 20.80 | 1.13 | ** | 25.76 | 2.91 | 7 | 9.127 | 0.000 | |||||
| Tail length [mm] | 260.00 | 16.58 | 279.73 | 14.64 | ** | 253.00 | 13.58 | ** | 280.63 | 15.24 | 257.57 | 11.87 | 263.40 | 15.53 | 267.88 | 17.73 | 252.57 | 16.16 | 7 | 4.006 | 0.001 | ||||
| Tail circumference [mm] | 34.80 | 0.84 | 35.73 | 3.26 | 40.33 | 3.27 | * | 35.25 | 3.58 | 33.14 | 4.38 | 34.40 | 2.19 | 34.42 | 1.60 | ** | 42.43 | 6.83 | 7 | 9.046 | 0.000 | ||||
| Body mass [g] | 795.20 | 80.79 | 934.73 | 109.06 | 938.50 | 116.15 | 939.50 | 96.97 | ** | 673.57 | 120.13 | 713.07 | 93.24 | 706.31 | 61.71 | ** | 964.57 | 96.27 | 7 | 20.680 | 0.000 | ||||
Two asterisks indicate significant differences (p≤0.05) and one asterisk indicate a statistical trend (0.05≤p<0.1) between the neighbouring species/columns.
The discriminant function analysis used five variables for model calculation, ear length, snout length, lower leg length, interorbital distance and intraorbital distance. Two functions were computed explaining a significant part of the morphometric variability between the six established and two proposed species (Wilk's λ = 0.006; F(35,120) = 8.355; p < 0.000). Table 3 shows the classification matrix, with correct classification in 82.5% of the cross-validated cases. The differences between the classification accuracy of each species ranged from 60% to 100%. The individuals of L. aeeclis, L. sp. nova in IRS III and L. mustelinus were correctly classified in 100% of the cases. The individuals of L. sp. nova in IRS II and L. ankaranensis were correctly classified in 80%, and the individuals of L. edwardsi, L. sahamalazensis and L. dorsalis had the smallest percentage of correct classifications (60%). All misclassifications occurred within each major clade, indicating again cryptic speciation within the genus Lepilemur.
Table 3.
Classification matrix of the discriminant function analysis.
| % correct | L. aeeclis | L. edwardsi | L. sp. nov. in IRS II | L. sp. nov. in IRS III | L. sahamalazensis | L. dorsalis | L. ankaranensis | L. mustelinus | |
| L. aeeclis | 100.0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. edwardsi | 80.0 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. sp. nov. in IRS II | 80.0 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 0 |
| L. sp. nov. in IRS III | 100.0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| L. sahamalazensis | 60.0 | 0 | 0 | 0 | 0 | 3 | 1 | 1 | 0 |
| L. dorsalis | 60.0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 1 |
| L. ankaranensis | 80.0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 |
| L. mustelinus | 100.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| total | 82.5 | 6 | 4 | 4 | 6 | 3 | 5 | 6 | 6 |
Discussion
Revised phylogeny of the genus Lepilemur
Molecular methods, such as DNA sequencing provide powerful tools to understand diversity and phylogeny [7,17,23-30]. This could be confirmed by our study in sportive lemurs. The phylogenetic trees distinguished all previously described species. Moreover, it provided evidence for two previously unknown species in northwestern Madagascar. The absolute pairwise distances between all species were in the range of those observed in other lemur genera such as Mirza [31], Microcebus [31-33], Hapalemur [34,35] and Propithecus [7,36]. The two new taxa occurred in a single IRS (II and III) each. Their phylogenetic position in the tree, the genetic distances and the number of diagnostic sites, suggest a separation at the species level. Similar conclusions were drawn with comparable approaches in other taxa [17,20,27,31,33].
Besides the molecular evidence, the discriminant function analysis of the morphometric data further supported the species status of the two new Lepilemur taxa (IRS II and III) in northwestern Madagascar. Between 80% (IRS II) and 100% (IRS III) of the animals were correctly classified into their IRS of origin. The ANOVA of the morphometric data detected significant differences between the northwestern and northern clade and between the northern clade and L. mustelinus. Moreover, it could differentiate between the neighbouring species (established and proposed) within the northwestern and northern clade, but it could not distinguish between L. edwardsi and L. aeeclis that are geographically separated by the river Betsiboka. The differentiation between the northwestern and northern clade, and between the northern clade and L. mustelinus is stronger than between the species within these major clades. Although the lack of differentiation between the western and northwestern clade may also partly be due to a sample size effect, it may also suggest cryptic speciation events in the genus Lepilemur not only within major clades as it is known in other taxa [37-39], but also between major clades.
The revised phylogeny of the genus Lepilemur is based on the combination of molecular differences (genetic distances and diagnostic sites) and morphometric traits. Diagnostic sites are also routinely used in DNA barcoding, which is becoming an increasingly important tool in species identification [40]. Although DNA barcoding requires a large and nearly complete database of sequences to which individuals can be compared [41,42], the diagnostic sites we identified can be seen as first step towards such a database in Lepilemur.
Description of two new species
Lepilemur otto sp. nov
Holotype
Individual 02y04bibo, adult male captured in Ambodimahabibo on 1st August 2004 by M. Craul (Fig. 5, 6, 7).
Figure 5.

Lepilemur otto, portrait of individual 02y04bibo (photograph by M. Craul).
Figure 6.

Lepilemur otto, body of individual 02y04bibo (photograph by M. Craul).
Figure 7.

Lepilemur otto, back of individual 02y04bibo (photograph by M. Craul).
Material
Tissue and hair samples, morphometric measurements as well as photographs of 02y04bibo are stored at the Institute of Zoology of the University of Veterinary Medicine Hannover, Hannover, Germany.
Type locality
Madagascar: Province de Mahajanga, Ambodimahabibo (15°29'54,2"S, 47°28'47,2"E).
Paratype
Individuals 01y04bibo, 03y04bibo and 04y04bibo were captured in Ambodimahabibo by M. Craul in 2004. Tissue and hair samples, morphometric measurements as well as photographs of each paratype are stored at the Institute of Zoology of the University of Veterinary Medicine Hannover, Hannover, Germany.
Description
The dorsal pelage, including shoulders and the upper and lower arms, is predominantly grey-brown. A dark diffuse line runs from the middle of the upper skull down the spine, ending in the middle or at the lower part of the back, but is never present on the tail. The ventral pelage is generally grey to creamy. The coloration of the tail is grey-brown to deep brown, sometimes with a white tail tip. The face and forehead are essentially grey.
Diagnosis
The sequenced mtDNA of Lepilemur otto has eleven diagnostic sites, eight in the ND4 (positions 42 = G, 57 = T, 123 = G, 255 = A, 306 = C, 630 = A, 631 = T, 632 = :C; see additional file 2: Diagnostic sites of the ND4 region for each terminal clade.), and three in the D-loop (positions 20 = C, 22 = A, 23 = T; see additional file 3: Diagnostic sites of the D-loop region for each terminal clade.). L. otto differs from its closest relative, L. edwardsi, in 2.92–2.99% and from its sister taxon L. manasamody in 3.50–3.57% in the sequenced mtDNA, respectively. The few morphometric data, which are available at the moment indicate that L. otto has a significant longer snout than the neighbouring species south of the Mahajamba River, L. edwardsi. The tail is significant short compared to the neighbouring species north of the Sofia River, L. manasamody and to L. edwardsi. L. otto shows a tendency to have a wider head than L. edwardsi and a bigger tail circumference than L. manasamody.
Distribution
The known distribution range of Lepilemur otto is so far limited to the sample site of Ambodimahabibo. This site is situated in the IRS II, which is limited by the Mahajamba River in the west and the Sofia River in the north. Intensive surveys are now required in this vastly deforested area to obtain additional information about the location and viability of other remaining populations, so that conservation measures can be proposed.
Etymology
The name Lepilemur otto was chosen to acknowledge the donation of Dr. Michael Otto for the purpose of research and conservation of Malagasy lemurs.
Vernacular name
Otto's sportive lemur or Lépilemur de Otto.
Lepilemur manasamody sp. nov
Holotype
Individual 16y03amb, adult female captured in Ambongabe on 20th September 2003 by M. Craul (Fig. 8, 9, 10).
Figure 8.

Lepilemur manasamody, portrait of individual 16y03amb (photograph by M. Craul).
Figure 9.

Lepilemur manasamody, body of individual 16y03amb (photograph by M. Craul).
Figure 10.

Lepilemur manasamody, back of individual 16y03amb (photograph by M. Craul).
Material
Tissue and hair samples, morphometric measurements as well as photographs of 16y03amb are stored at the Institute of Zoology of the University of Veterinary Medicine Hannover, Hannover, Germany.
Type locality
Madagascar: Province de Mahajanga, Ambongabe (15°19'38.3"S, 46°40'44.4"E) and Anjiamangirana I (15°09'24.6"S, 47°44'06.2"E).
Paratype
Individuals 14y03amb and 15y03amb were captured in Ambongabe and individuals 07y03anji, 08y03anji and 09y03anji in Anjiamangirana I by M. Craul in 2003. Tissue and hair samples, morphometric measurements as well as photographs of all paratypes are stored at the Institute of Zoology of the University of Veterinary Medicine Hannover, Hannover, Germany.
Description
The dorsal pelage is predominantly grey-brown, including shoulders, the upper and lower arms. The ventral pelage is generally grey to creamy. The face and forehead are essentially grey. From the middle of the upper skull, a dark diffuse line runs down the spine, ending in the middle of lower part of the back. This line is never present on the tail. The tail is grey-brown to deep brown, sometimes with a white tail tip.
Diagnosis
The sequenced mtDNA of Lepilemur manasamody has seven diagnostic sites, two of them in the Cytochrome B (positions 86 = G, 140 = G; see additional file 1: Diagnostic sites of the Cytochrome B region for each terminal clade.), three in the ND4 (positions 171 = T, 201 = G, 333 = A; see additional file 2: Diagnostic sites of the ND4 region for each terminal clade.), and two in the D-loop (positions 75 = G, 156 = G; see additional file 3: Diagnostic sites of the D-loop region for each terminal clade.). L. manasamody differs from its sister taxa L. otto in 3.50–3.57% and from L. edwardsi in 2.77–2.92% in the sequenced mtDNA, respectively. The few morphometric data, which are available at the moment, indicate that L. manasamody has a significantly longer tail than L. otto. L. manasamody has significantly longer ears and a longer snout, a significantly wider head and bigger intraorbital distance and is heavier than the neighbouring species to the north, L. sahamalazensis. It also shows a tendency to have a smaller tail circumference than L. otto.
Distribution
The known distribution range of Lepilemur manasamody is so far limited to the sample sites of Ambongabe and Anjiamangirana I. Both sites are situated in the IRS III, which is limited by the Sofia River in the south and the Maevarano River in the north. Intensive surveys are now required to obtain additional information about the location and viability of the remaining populations, so that conservation measures can be proposed.
Etymology
The name Lepilemur manasamody was chosen after the forest region Manasamody, west of Anjiamangirana I between the Sofia and Maevarano River.
Vernacular name
Manasamody sportive lemur or Lépilemur de Manasamody.
Conclusion
Our results showed that all species, except for L. mustelinus from the East, grouped in three major clades (western, northwestern and northern). Taking into account the species diversity within each major clade, however, we can define seven biogeographic zones in northern and northwestern Madagascar. When compared to the predictions derived from the "Martin model" and the "Wilmé model", we find several inconsistencies to our data.
The "Martin model" defined four biogeographic zones from western to northern Madagascar (W1, NW, X and N). They corresponded well to the three deep phylogenetic splits, that gave rise to the western, northwestern and northern clade in our study. However, the species diversity within each major clade could not be explained by this model.
The "Wilmé model" defined six biogeographic zones from western to northern Madagascar (numbers 8, 9, 10, 11, 12 and 1). This model may also explain the deep splits between the three major clades, but it proposed two more splits. One between IRS V and VI, divided by the Sambirano River. This split could be confirmed by our study, although it seems not to be a species barrier for sportive lemurs. The level of absolute pairwise distances is intermediate and may rather suggest a variation on a subspecies level. Very recently however, Rabarivola et al. (2006) proposed species status for the individuals in IRS V based on cytogenetics [18]. They collected samples in IRS V at a locality further north of Mahilaka and the number of chromosomes differed between individuals from IRS V (2N = 24) and the neighbouring Lepilemur sahamalazensis (2N = 26) and Lepilemur dorsalis (2N = 26). The second additional split indicated by the "Wilmé model" is that between zone 12 and 1. This split could not be confirmed by our study, since all individuals found in IRS VII (corresponding to Wilmé's zones 12 and 1) clustered together and belonged to the species L. ankaranensis. One major discrepancy exists between our findings and the "Wilmé model". The "Wilmé model" predicts one centre of endemism in northwestern Madagascar (zone 9), which should correspond to one Lepilemur species in that area. Our study provided evidence, however, for three species of sportive lemurs between the Betsiboka and Maevarano River, each restricted to one of the three IRSs. Thus, we showed that each IRS is represented as a separate terminal clade in the phylogenetic trees, building distinct phylogenetic units. At least six of the seven large rivers act as species barriers for Lepilemur. Therefore, we propose a new model, the "large river model" to explain the biogeography of this larger-sized nocturnal lemur genus. Large rivers acted as insurmountable barriers for gene flow, leading to cryptic speciation within larger biogeographic units. Except for IRSs V and VI, the genetic distances among all IRSs reach species level.
The deep splits between the major clades may indicate initial colonization events, with the Betsiboka and Maevarano River playing a major role in long-term and continuous isolation of western, northwestern and northern Madagascar. The splits within each major clade however, indicate younger cryptic speciation events. Populations, initially belonging to one founder species, entered the IRSs I-III and VI-VII respectively, and were subsequently separated from each other by the rivers Mahajamba and Sofia, and the Andranomalaza, respectively. Quaternary paleoclimatic variation may have played another important role in shaping biogeography and speciation events on Madagascar. The climate during periods of glaciation was cooler and drier than today [8,43,44]. Rivers with year-round water course could have acted as retreats/refugia in times of aridification. All seven large rivers in northwestern and northern Madagascar should have belonged to this category, since the genetic isolation of the IRSs would otherwise not have persisted over time and signs of repeated introgression should be detectable. Subsequent recolonization of the IRSs should thereby have originated from small and isolated refugia, which further promoted genetic differentiation between the IRSs.
In conclusion, we presented evidence for an unexpected species diversity of sportive lemurs in northwestern and northern Madagascar. Current biogeographic models were not sufficient to explain the underlying processes of speciation. We therefore suggest a new model of biogeographical zonation, the "large river model". In this model, biogeographic zones are separated and maintained over time by all large rivers with permanent water bodies that may have provided retreat zones during periods of aridification and may have harboured founder populations for subsequent recolonization. The importance of large rivers as biogeographic barriers was previously emphazized for mouse lemurs [3], but also for neotropical primates [45,46]. Further studies are now needed to test the relevance of this model for other terrestrial taxa, such as the insectivores, rodents, or other lemurs.
Methods
Fieldwork
A total of 157 Lepilemur individuals were captured at 14 different localities along a 560 km transect from western to northern Madagascar (Fig. 4, Table 4). This region is divided by eight large rivers (over 50 km wide 20 km inlands) into eight Inter-River-Systems (IRS 0 to IRS VII, Fig. 4). Six localities were sampled by Mathias Craul (MC) and eight localities were sampled by Solofo Rasoloharijaona (SR) and Blanchard Randrianambinina (BR). At each site we performed daily and nightly surveys to capture the animals. At daytime we used a net to capture the animals out of their sleeping holes and briefly anesthetised them with Ketasel-5 (Selectavet). At night time we anesthetised the animals using a blowpipe (TELINJECT B22T) with Ketasel-5 (Selectavet). Each captured sportive lemur was then characterised with regard to sex, skin colour, reproductive status (testis size or form of vulva), 13 external morphometric measures (ear length, ear width, head length, head width, snout length, interorbital distance, intraorbital distance, lower leg length, hind foot length, 3rd toe length, body length, tail length, tail circumference) and body mass [10,47]). In addition, a small biopsy from one or both pinnae was taken as tissue samples. Tissue samples were stored in Queen's lysis buffer [48] for later DNA extraction and genetic analyses.
Table 4.
Details of study sites.
| Locality | Abbreviation | Coordinates | Origin |
| Madirovalo | madi | 16°22'45.6"S, 46°29'01.9"E | IRS 0 |
| Ampijoroa | jbb | 16°17'S, 46°48'E | IRS I |
| Mariarano | mari | 15°28'50.3"S, 46°41'19.0"E | IRS I |
| Tananvaovao | tan | 15°28'15.5"S, 46°39'59.4"E | IRS I |
| Ambodimahabibo | bibo | 15°29'54.2"S, 47°28'47.2"E | IRS II |
| Ambongabe | amb | 15°19'38.3"S, 47°40'44.4"E | IRS III |
| Anjiamangirana I | anji | 15°09'24.6"S, 47°44'06.2"E | IRS III |
| Ankarafa | sah | 14°22'47.8"S, 47°45'26.3"E | IRS IV |
| Mahilaka | mah | 14°17'12.0"S, 48°12'12.0"E | IRS V |
| Lokobe | lok | 13°23'23.9"S, 48°20'31.0"E | IRS VI |
| Manehoka | oka | 13°25'49.0"S, 48°47'51.0"E | IRS VI |
| Ankavana | anka | 12°46'55.7"S, 49°22'27.4"E | IRS VII |
| Ankarana | kar | 12°58'05.0"S, 49°08'18.0"E | IRS VII |
| Analabe | ana | 12°45'13.8"S, 49°30'03.9"E | IRS VII |
| Kirindy | kir | 20°03'S, 44°37'E | West |
| Mantadia | man | 18°47'S, 48°25'E | East |
Molecular methods and analyses
DNA from the tissue of 37 individuals was isolated with the DNeasy Tissue Kit (Qiagen), or extracted using a standard proteinase K digestion followed by a Phenol/Chloroform protocol [49] and stored at -20°C. In addition, we analysed the DNA of two individuals of Lepilemur ruficaudatus (Kirindy forest, western Madagascar) provided by Yves Rumpler and of four individuals from Mantadia (eastern Madagascar) sampled previously by SR and BR. We sequenced the mitochondrial genes D-loop, cytochrome B and NADH-dehydrogenase subunit 4 (ND4), because reference sequences from all eleven recognized species were available for these particular markers. The complete D-loop was amplified with the oligonucleotide primers DLp-1.5: 5'-GCA CCC AAA GCT GAR RTT CTA-3' and DLp-5: 5'-CCA TCG WGA TGT CTT ATT TAA GRG GAA-3' [19]. Standard PCRs were carried out in a 25 μl reaction with a final concentration of 1 μM for each primer, 1.5 mM for MgCl2, 0.2 mM for each dNTP, 1 × NH4 reaction buffer (50 mM Tris-HCl pH 8.8, 16 mM (NH4)2 SO4, 0.1% Tween® 20), 1.25 units of Taq DNA polymerase, and 1 μl of DNA. Successful amplifications were obtained using the following protocol: 35 cycles of denaturing at 94°C for 60 seconds, primer annealing at 47°C for 60 seconds and extension at 72°C for 90 seconds. The partial Cytochrome B was amplified with the oligonucleotide primers L14841: 5'-AAA AAG CTT CCA TCC AAC ATC TCA GCA TGA TGA AA-3' and H15149: 5'-AAA CTG CAG CCC CTC AGA ATG ATA TTT GTC CTC A-3' [50]. Standard PCRs were carried out in a 25 μl reaction with a final concentration of 1 μM for each primer, 1.5 mM for MgCl2, 0.2 mM for each dNTP, 1 × NH4 reaction buffer (50 mM Tris-HCl pH 8.8, 16 mM (NH4)2 SO4), 1.25 units of Taq DNA polymerase, and 1 μl of DNA. Successful amplifications were obtained using the following protocol: 35 cycles of denaturing at 94°C for 60 seconds, primer annealing at 47°C for 60 seconds and extension at 72°C for 90 seconds. The partial NADH-dehydrogenase subunit 4 was amplified with the oligonucleotide primers LepiP1: 5'-TTG ATG TAG TAT GAC TRT TCC-3' and LepiR1: 5'-GCC AAA CCG ATG GCT GCT TCA CAG GCT GCA AG-3' [51]. Standard PCRs were carried out in a 25 μl reaction with a final concentration of 1 μM for each primer, 1.5 mM for MgCl2, 0.2 mM for each dNTP, 1 × NH4 reaction buffer (50 mM Tris-HCl pH 8.8, 16 mM (NH4)2 SO4), 1.25 units of Taq DNA polymerase, and 1 μl of DNA. Successful amplifications were obtained using the following protocol: 40 cycles of denaturing 95°C for 30 seconds, primer annealing at 60°C for 60 seconds and extension at 72°C for 60 seconds. The PCR products were cleaned with the Invisorb Spin PCRapid Kit (Invitek) or Quick-Clean (Bioline) and checked for successful amplification by running an aliquot on a 1.5% agarose gel, stained with 1.3 × 10-4 mg/ml ethidium bromide. After cleaning the PCR products, cycle sequencing reactions were carried out using DYEnamic™ ET dye terminator kit (Amersham Biosciences) and the primers indicated above. After a second cleaning with ammonium acetate, provided with the DYEnamic™ ET dye terminator kit, the PCR products were sequenced on a MegaBACE™ 1000 DNA Sequencing System (Amersham Biosciences). The respective sequences were deposited in GenBank (Table 5).
Table 5.
Locality, origin, sample type and GenBank accession number of analysed individuals for genetic studies.
| Species | Locality | Abbreviation | Origin | Sample type | D-loop | Cyt B | ND4 |
| L. ruficaudatus | Kirindy | kir1 | west | DNA | EF686766 | EF686723 | EF686680 |
| L. ruficaudatus | Kirindy | kir2 | west | DNA | EF686767 | EF686724 | EF686681 |
| L. ruficaudatus | Kirindy | west | sequence | DQ109013–DQ109015, DQ109017 | AF224596 | ||
| L. randrianasoli | west | sequence | AY321456 | ||||
| L. randrianasoli | Andramasay | west | sequence | AY441463, DQ109019, DQ234891–DQ234894 | |||
| L. randrianasoli | Ambalarano | west | sequence | DQ234890 | |||
| L. aeeclis | Madirovalo | madi1 | IRS 0 | tissue | EF686768 | EF686725 | EF686682 |
| L. aeeclis | Madirovalo | madi2 | IRS 0 | tissue | EF686769 | EF686726 | EF686683 |
| L. aeeclis | Anjamena | IRS 0 | sequence | AF224593 | |||
| L. aeeclis | Antafia-Anjahamena | IRS 0 | sequence | DQ108999–DQ109003, DQ234899 | |||
| L. edwardsi | Ampijoroa | jbb1 | IRS I | tissue | EF686756 | EF686713 | EF686670 |
| L. edwardsi | Ampijoroa | jbb2 | IRS I | tissue | EF686757 | EF686714 | EF686671 |
| L. edwardsi | Mariarano | mari1 | IRS I | tissue | EF686760 | EF686717 | EF686674 |
| L. edwardsi | Mariarano | mari2 | IRS I | tissue | EF686761 | EF686718 | EF686675 |
| L. edwardsi | Tananvaovao | tan1 | IRS I | tissue | EF686758 | EF686715 | EF686672 |
| L. edwardsi | Tananvaovao | tan2 | IRS I | tissue | EF686759 | EF686716 | EF686673 |
| L. edwardsi | Ampijoroa | IRS I | sequence | DQ109006 | AF224595 | ||
| L. edwardsi | Andofombombe | IRS I | sequence | DQ109004, DQ109005, DQ234888 | |||
| L. sp. nov. IRS II | Ambodimahabibo | bibo1 | IRS II | tissue | EF686762 | EF686719 | EF686676 |
| L. sp. nov. IRS II | Ambodimahabibo | bibo2 | IRS II | tissue | EF686763 | EF686720 | EF686677 |
| L. sp. nov. IRS II | Ambodimahabibo | bibo3 | IRS II | tissue | EF686764 | EF686721 | EF686678 |
| L. sp. nov. IRS II | Ambodimahabibo | bibo4 | IRS II | tissue | EF686765 | EF686722 | EF686679 |
| L. sp. nov. IRS III | Ambongabe | amb1 | IRS III | tissue | EF686753 | EF686710 | EF686667 |
| L. sp. nov. IRS III | Ambongabe | amb2 | IRS III | tissue | EF686754 | EF686711 | EF686668 |
| L. sp. nov. IRS III | Anjiamangirana I | anji1 | IRS III | tissue | EF686755 | EF686712 | EF686669 |
| L. sahamalazensis | Ankarafa | sah1 | IRS IV | tissue | EF686750 | EF686707 | EF686664 |
| L. sahamalazensis | Ankarafa | sah2 | IRS IV | tissue | EF686751 | EF686708 | EF686665 |
| L. sahamalazensis | Ankarafa | sah3 | IRS IV | tissue | EF686752 | EF686709 | EF686666 |
| L. sahamalazensis | Sahamalaza | IRS IV | sequence | DQ108990–DQ108992, DQ234882, DQ234883 | |||
| L. dorsalis | Mahilaka | mah1 | IRS V | tissue | EF686746 | EF686703 | EF686660 |
| L. dorsalis | Mahilaka | mah2 | IRS V | tissue | EF686747 | EF686704 | EF686661 |
| L. dorsalis | Mahilaka | mah3 | IRS V | tissue | EF686748 | EF686705 | EF686662 |
| L. dorsalis | Ambanja | IRS V | sequence | DQ108995–DQ108997, DQ234886, DQ234887 | |||
| L. dorsalis | Mahilaka | mah4 | IRS V | tissue | EF686749 | EF686706 | EF686663 |
| L. dorsalis | Lokobe | lok1 | IRS VI | tissue | EF686740 | EF686697 | EF686654 |
| L. dorsalis | Lokobe | lok3 | IRS VI | tissue | EF686741 | EF686698 | EF686655 |
| L. dorsalis | Manehoka | oka1 | IRS VI | tissue | EF686744 | EF686701 | EF686658 |
| L. dorsalis | Manehoka | oka2 | IRS VI | tissue | EF686742 | EF686699 | EF686656 |
| L. dorsalis | Manehoka | oka3 | IRS VI | tissue | EF686743 | EF686700 | EF686657 |
| L. dorsalis | Manehoka | oka4 | IRS VI | tissue | EF686745 | EF686702 | EF686659 |
| L. dorsalis | Nosy Be | IRS VI | sequence | AY441464, DQ108993, DQ108994, DQ108998, DQ234885 | |||
| L. ankaranensis | Ankavana | anka1 | IRS VII | tissue | EF686735 | EF686692 | EF686649 |
| L. ankaranensis | Ankavana | anka2 | IRS VII | tissue | EF686734 | EF686691 | EF686648 |
| L. ankaranensis | Ankavana | anka3 | IRS VII | tissue | EF686736 | EF686693 | EF686650 |
| L. ankaranensis | Ankarana | kar1 | IRS VII | tissue | EF686731 | EF686688 | EF686645 |
| L. ankaranensis | Ankarana | kar2 | IRS VII | tissue | EF686732 | EF686689 | EF686646 |
| L. ankaranensis | Ankarana | kar3 | IRS VII | tissue | EF686733 | EF686690 | EF686647 |
| L. ankaranensis | Analabe | ana1 | IRS VII | tissue | EF686737 | EF686694 | EF686651 |
| L. ankaranensis | Analabe | ana2 | IRS VII | tissue | EF686738 | EF686695 | EF686652 |
| L. ankaranensis | Analabe | ana3 | IRS VII | tissue | EF686739 | EF686696 | EF686653 |
| L. ankaranensis | Ankarana | IRS VII | sequence | DQ109028–DQ109032 | AF304597 | ||
| L. ankaranensis | Analamera | IRS VII | sequence | DQ109022–DQ109024, DQ234884 | |||
| L. ankaranensis | Andrafiamena | IRS VII | sequence | DQ109025, DQ109027, DQ234881 | |||
| L. septentrionalis | Sahafary | IRS VII | sequence | AJ304651 | DQ109020, DQ109021, DQ234900 | ||
| L. mustelinus | Mantadia | man1 | east | tissue | EF686727 | EF686684 | EF686641 |
| L. mustelinus | Mantadia | man2 | east | tissue | EF686728 | EF686685 | EF686642 |
| L. mustelinus | Mantadia | man3 | east | tissue | EF686729 | EF686686 | EF686643 |
| L. mustelinus | Mantadia | man4 | east | tissue | EF686730 | EF686687 | EF686644 |
| L. mustelinus | Behasina | east | sequence | DQ109033 | |||
| L. mustelinus | near Mantadia | east | sequence | DQ109034 | |||
| L. microdon | Vohiparara | east | sequence | DQ109008 | |||
| L. microdon | Antarando | east | sequence | DQ109009, DQ109010 | |||
| L. microdon | Ambatolampy | east | sequence | DQ234889 | |||
| L. leucopus | south | sequence | DQ109007 | ||||
| P. diadema | sequence | AF354743 | AY441452 | AF224599 |
For a comprehensive phylogenetic analysis of the sequence data, we expanded our data set with reference sequences from all eleven recognized species available from GenBank (Table 5). As outgroup for phylogenetic tree reconstructions, we selected Propithecus diadema. Sequences were aligned using the program CLUSTALX [52] and checked by eye. Tree reconstructions of each single gene were carried out to phylogenetically classify the sampled individuals within the genus Lepilemur. Because of the lack of reference sequences of single individuals for all three genes, further phylogenetic tree reconstructions based on all three genes were performed only with our own data set consisting of 43 sequences. Phylogenetic tree reconstructions were carried out with the maximum-parsimony (MP), neighbour-joining (NJ) and maximum-likelihood (ML) algorithms as implemented in PAUP4.0b10 [53]. Throughout the analyses, all characters were treated as unordered and equally weighted. Gaps were considered as missing data in NJ and ML, but were treated as fifth character in MP analysis. The NJ and ML trees were constructed using the best-fit model selected by the hierarchical likelihood ratio test (hLRT) in Modeltest 3.5.mac [54]. Relative support of internal nodes was provided by bootstrap analyses with 1,000 replications for MP and NJ and 100 replications for ML. Absolute pairwise distances were calculated using PAUP4.0b10 [50] and ARLEQUIN 1.1 to describe the variation among taxa. To determine fixed molecular differences among terminal clades (indicating barriers for gene flow), diagnostic sites for each terminal clade to all others were identified using the program MEGA 3.1 [55].
Statistical analyses of morphometric data
Quantitative analyses of morphometric data were carried out with two different sample sizes. The ANOVA was conducted with 100 individuals. After removing two variables that differed among researchers (1-way ANOVA, STATISTICA 6.0, Statsoft, Inc.), the 11 remaining variables were tested for normality using the Kolmogorov-Smirnov test (Statistica 6.0, StatSoft, Inc.) at a level of p ≤ 0.05. All were normally distributed. A MANOVA revealed no differences in sex. The variables were then tested for correlation. All variables had an r < 0.75 and were therefore defined as sufficiently independent to be used in a discriminant function analysis [56]. This analysis was limited to five adult individuals per species (established and proposed) in order to equilibrate the samples. The discriminant function analysis tested only for species differences and for differences between the IRSs. A stepwise forward method (statistic: Wilk's λ) with the criteria Ftoenter = 3.84 and Ftoremove = 2.71 and a tolerance level of p ≤ 0.01 was used to calculate the discriminant function model. The computed discriminant functions were used to classify cases with regard to their group membership. All cases were cross-validated by the "leave-one-out" method, where each case in the analysis is classified by the functions derived from all cases other than that case. The discriminant function analysis was carried out with the program SPSS 13.0 (SPSS, Inc.).
Footnote
During the review process of this paper, Louis Jr. et al. (2006) described a new sportive lemur species in IRS III [57]. It was named Lepilemur grewcocki. This might be a synonym to L. manasamody, as our sampling sites of this species were in the same IRS. However, a joint phylogenetic analysis is still needed to verify the identity of both forms.
Authors' contributions
MC participated in the design of the study, conducted part of the field work and all lab work, performed the comparative genetic and morphometric analyses and wrote the MS.
EZ initiated, financed and designed the study. She organized and conceptualized the field work, supervised data analyses and critically revised the MS several times and approved its final version.
SR conducted part of the field work including capturing, measuring and sampling L. ankaranensis, L. dorsalis, L. mustelinus and L. aeeclis.
BR conducted part of the field work including capturing, measuring and sampling L. ankaranensis, L. dorsalis, L. mustelinus and L. aeeclis.
UR designed the study. She organized and conceptualized the field work, supervised data analyses and critically revised the MS several times and approved its final version.
Supplementary Material
Diagnostic sites of the Cytochrome B region for each terminal clade. Dashes (-) indicate deletions. Points (.) indicate identical bases.
Diagnostic sites of the ND4 region for each terminal clade. Dashes (-) indicate deletions. Points (.) indicate identical bases.
Diagnostic sites of the D-loop region for each terminal clade. Dashes (-) indicate deletions. Points (.) indicate identical bases.
Acknowledgments
Acknowledgements
We like to thank the late B. Rakotosamimanana, as well as C. Andrianarivo, O. Ramilijaona, D. Rakotondravony and the following institutions for their authorization and support of this study: Faculty of Sciences, University of Antananarivo, the Association Nationale pour la Gestion des Aires Protégées (ANGAP), the Ministère des Eaux et Forêts and CAFF/CORE (Commision Tripartite), Madagascar. Many thanks go to G. Olivieri, K. Guschanski, C. Schwitzer, N. Schwitzer, R. Rakotondravony, A. Rabesandratana, J.-A. Rakotonirina, T. Razafimamonjy, B. Rakoloarinelina and J. Rajanary for their assistance in the field, to H. Breitrück and S. Sippel for their help in the lab and to Y. Rumpler for providing DNA of Lepilemur ruficaudatus. The project was funded by the Volkswagen Foundation (VW I, Aktenzeichen I/76828, Zimmermann), and the "Gesellschaft der Freunde der Tierärztlichen Hochschule Hannover e.V.". We dedicate this paper to the late Berthe Rakotosamimanana.
Contributor Information
Mathias Craul, Email: mathias.craul@tiho-hannover.de.
Elke Zimmermann, Email: elke.zimmermann@tiho-hannover.de.
Solofonirina Rasoloharijaona, Email: solofo_r01@yahoo.fr.
Blanchard Randrianambinina, Email: blanrand@yahoo.fr.
Ute Radespiel, Email: ute.radespiel@tiho-hannover.de.
References
- Martin RD. Primate Origins and Evolution: A Phylogenetic Reconstruction. London/New Jersey: Chapman Hall/Princeton University Press; 1990. [Google Scholar]
- Mittermeier RA, Konstant WR, Hawkins F, Louis EE, Langrand O, Ratsimbazafy J, Rasoloarion R, Ganzhorn JU, Rajaobelina S, Tattersall I, Meyers DM. Lemurs of Madagascar. 2. Washington, DC: Conservation International; 2006. [Google Scholar]
- Olivieri G, Zimmermann E, Randrianambinina B, Rasoloharijaona S, Rakotondravony D, Guschanski K, Radespiel U. The ever-increasing diversity in mouse lemurs: three new species in north and northwestern Madagascar. Mol Phylogenet Evol. 2006 doi: 10.1016/j.ympev.2006.10.026. doi: 10.1016/j.ympev.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Ganzhorn JU, Langrand O, Wright PC, O'Connor S, Rakotosamimanana B, Feistner ATC, Rumpler Y. The state of lemur conservation in Madagascar. Prim Conserv. 1996;17:70–86. [Google Scholar]
- Martin RD. Adaptive radiation and behaviour of the Malagasy lemurs. Philos Trans R Soc Lond B Biol Sci. 1972;264:295–352. doi: 10.1098/rstb.1972.0013. [DOI] [PubMed] [Google Scholar]
- Martin RD, Alterman L, Doyle GA, Izard MK. Creatures of the dark: the nocturnal prosimians. New York: Plenum Press; 1995. Prosimians: from obscurity to extinction? pp. 535–563. [Google Scholar]
- Pastorini J, Thalmann U, Martin RD. A molecular approach to comparative phylogeography of extant Malagasy lemurs. Proc Natl Acad Sci USA. 2003;100:5879–5884. doi: 10.1073/pnas.1031673100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmé L, Goodman S, Ganzhorn J. Biogeographic evolution of Madagascar's microendemic biota. Science. 2006;312:1063–1065. doi: 10.1126/science.1122806. [DOI] [PubMed] [Google Scholar]
- Rasoloharijaona S, Rakotosamimanana B, Randrianambinina B, Zimmermann E. Pair-specific usage of sleeping sites and their implications for social organization in a nocturnal Malagasy primate, the Milne Edwards' sportive lemur (Lepilemur edwardsi) Am J Phys Anthropol. 2003;122:251–258. doi: 10.1002/ajpa.10281. [DOI] [PubMed] [Google Scholar]
- Rasoloharijaona S, Randrianambinina B, Braune P, Zimmermann E. Loud calling, spacing, and cohesiveness in a nocturnal primate, the Milne Edwards' sportive lemur (Lepilemur edwardsi) Am J Phys Anthropol. 2006;129:591–600. doi: 10.1002/ajpa.20342. [DOI] [PubMed] [Google Scholar]
- Thalmann U, Ganzhorn J. Lepilemur, Sportive Lemur. In: Goodmann SM, Benstead JP, editor. The Natural History of Madagascar. Chicago: The University of Chicago Press; 2003. pp. 1336–1340. [Google Scholar]
- Petit G. Le genre Lepidolemur et sa répartition géographique. C R Soc Biogéogr. 1933;10:33–37. [Google Scholar]
- Petter JJ, Petter-Rousseaux A. Remarque sur la systématique du genre Lepilemur. Mammalia. 1960;24:76–86. [Google Scholar]
- Petter JJ, Albignac R, Rumpler Y. Faune de Madagascar. Vol. 44. Paris: C.N.R.S./O.R.S.T.O.M; 1977. Mammifères lémuriens (Primates prosimiens) pp. 1–513. [Google Scholar]
- Rumpler Y, Albignac R. Chromosomes studies of the Lepilemur, an endemic Malagasy genus of lemurs: Contribution of the cytogenetics to their taxonomy. J Hum Evol. 1978;7:191–196. doi: 10.1016/S0047-2484(78)80110-9. [DOI] [Google Scholar]
- Tattersall I. The primates of Madagascar. New York: Columbia University Press; 1982. [Google Scholar]
- Andriaholinirina N, Fausser JL, Roos C, Zinner D, Thalmann U, Rabarivola C, Ravoarimanana I, Ganzhorn JU, Meier B, Hilgartner R, Walter L, Zaramody A, Langer C, Hahn T, Zimmermann E, Radespiel U, Craul M, Tomiuk J, Tattersall I, Rumpler Y. Molecular phylogeny and taxonomic revision of the sportive lemurs (Lepilemur, Primates) BMC Evol Biol. 2006;6:17. doi: 10.1186/1471-2148-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabarivola C, Zaramody A, Fausser JL, Andriaholinirina N, Roos C. Cytogenetic and molecular characteristics of a new species of sportive lemur from Northern Madagascar. Lemur News. 2006;11:45–49. [Google Scholar]
- Wyner YM, Amato G, Desalle R. Captive breeding, reintroduction, and the conservation genetics of black and white ruffed lemurs, Varecia variegata variegata. Mol Ecol. 1999;8:107–115. doi: 10.1046/j.1365-294X.1999.00815.x. [DOI] [PubMed] [Google Scholar]
- Ravaoarimanana IB, Tiedemann R, Montagnon D, Rumpler Y. Molecular and cytogenetic evidence for cryptic speciation within a rare endemic Malagasy lemur, the Northern Sportive Lemur (Lepilemur septentrionalis) Mol Phylogenet Evol. 2004;31:440–448. doi: 10.1016/j.ympev.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Cracraft J. Species concepts and speciation analysis. Current Ornithology. 1983;1:159–187. [Google Scholar]
- Davis JI, Nixon KC. Populations, genetic variation and the delimitation of phylogenetic species. Syst Biol. 1992;41:421–435. doi: 10.2307/2992584. [DOI] [Google Scholar]
- Yoder AD, Olson LE, Hanley C, Heckman KL, Rasoloarison R, Russell AL, Ranivo J, Soarimalala V, Karanth KP, Raselimanana AP, Goodman SM. A multidimensional approach for detecting species patterns in Malagasy vertebrates. Proc Natl Acad Sci USA. 2005;102:6587–6594. doi: 10.1073/pnas.0502092102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences M, Vieites DR, Glaw F, Brinkmann H, Kosuch J, Veith M, Meyer A. Multiple overseas dispersal in amphibians. Proc Biol Sci. 2003;270:2435–2442. doi: 10.1098/rspb.2003.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Burns MM, Zehr S, Delefosse T, Veron G, Goodman SM, Flynn JJ. Single origin of Malagasy carnivora from an African ancestor. Nature. 2003;421:734–737. doi: 10.1038/nature01303. [DOI] [PubMed] [Google Scholar]
- Jansa SA, Barker FK, Heaney LR. The pattern and timing of diversification of Philippine endemic rodents: evidence from mitochondrial and nuclear gene sequences. Syst Biol. 2006;55:73–88. doi: 10.1080/10635150500431254. [DOI] [PubMed] [Google Scholar]
- Asher RJ, Hofreiter M. Tenrec phylogeny and the noninvasive extraction of nuclear DNA. Syst Biol. 2006;55:181–194. doi: 10.1080/10635150500433649. [DOI] [PubMed] [Google Scholar]
- Farias IP, Orti G, Sampaio I, Schneider H, Meyer A. Mitochondrial DNA phylogeny of the family Cichlidae: monophyly and fast molecular evolution of the neotropical assemblage. J Mol Evol. 1999;48:703–711. doi: 10.1007/PL00006514. [DOI] [PubMed] [Google Scholar]
- Nagy ZT, Joger U, Wink M, Glaw F, Vences M. Multiple colonization of Madagascar and Socotra by colubrid snakes: evidence from nuclear and mitochondrial gene phylogenies. Proc Biol Sci. 2003;270:2613–2621. doi: 10.1098/rspb.2003.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Lalueza-Fox C, Anderson S, Rambaut A, Austin J, Ward R. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature. 2001;409:704–707. doi: 10.1038/35055536. [DOI] [PubMed] [Google Scholar]
- Kappeler PM, Rasoloarison RM, Razafimanantsoa L, Walter L, Roos C. Morphology, Behaviour and Molecular Evolution of Giant Mouse Lemurs (Mirza spp.) Gray, with Description of a new Species. Primate Report. 1870;71:3–26. [Google Scholar]
- Pastorini J, Martin RD, Ehresmann P, Zimmermann E, Forstner MRJ. Molecular phylogeny of the lemur family Cheirogaleidae (Primates) based on mitochondrial DNA sequences. Mol Phylogenet Evol. 2001;19:45–56. doi: 10.1006/mpev.2000.0904. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Rasoloarison RM, Goodman SM, Irwin JA, Atsalis S, Ravosa MJ, Ganzhorn JU. Remarkable species diversity in Malagasy mouse lemurs (primates, Microcebus) Proc Natl Acad Sci USA. 2000;97:11325–11330. doi: 10.1073/pnas.200121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausser JL, Prosper P, Donati G, Ramanamanjato JB, Rumpler Y. Phylogenetic relationships between Hapalemur species and subspecies based on mitochondrial DNA sequences. BMC Evol Biol. 2002;2:4. doi: 10.1186/1471-2148-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorini J, Forstner MRJ, Martin RD. Phylogenetic relationships among Lemuridae (Primates): evidence from mtDNA. J Hum Evol. 2002;43:463–478. [PubMed] [Google Scholar]
- Mayor MI, Sommer JA, Houck ML, Zaonarivelo JR, Wright PC, Ingram C, Engel SR, Louis EE. Specific status of Propithecus spp. Int J Primatol. 2004;25:875–900. doi: 10.1023/B:IJOP.0000029127.31190.e9. [DOI] [Google Scholar]
- Camargo A, de Sá RO, Heyer WR. Phylogenetic analyses of mtDNA sequences reveal three cryptic lineages in the widespread neotropical frog Leptodactylus fuscus (Schneider, 1799) (Anura, Leptodactylidae) Biol J Linnean Soc. 2006;87:325–341. doi: 10.1111/j.1095-8312.2006.00581.x. [DOI] [Google Scholar]
- Goetze E. Cryptic speciation on the high seas; global phylogenetics of the copepod family Eucalanidae. Proc Biol Sci. 2003;270:2321–2331. doi: 10.1098/rspb.2003.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggio AJ, Perkins SL. Molecular phylogeny of North American long-eared bats (Vespertilionidae: Corynorhinus); inter- and intraspecific relationships inferred from mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 2005;37:762–775. doi: 10.1016/j.ympev.2005.03.029. [DOI] [PubMed] [Google Scholar]
- DeSalle R. Species Discovery versus Species Identification in DNA Barcoding Efforts: Response to Rubinoff. Cons Biol. 2006;20:1545–1547. doi: 10.1111/j.1523-1739.2006.00543.x. [DOI] [PubMed] [Google Scholar]
- Moritz C, Cicero C. DNA barcoding: promise and pitfalls. PloS Biol. 2004;2:1529–1531. doi: 10.1371/journal.pbio.0020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will KW, Rubinoff D. Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics. 2004;20:47–55. doi: 10.1111/j.1096-0031.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- Haffer J. Speciation in Amazonian Forest Birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- de Wit MJ. MADAGASCAR: Heads It's a Continent, Tails It's an Island. Annual Review of Earth and Planetary Sciences. 2003;31:213–248. doi: 10.1146/annurev.earth.31.100901.141337. [DOI] [Google Scholar]
- Ayres JM, Clutton-Brock TH. River boundaries and species range size in Amazonian primates. American Naturalist. 1992;140:531–537. doi: 10.1086/285427. [DOI] [PubMed] [Google Scholar]
- Lehman SM. Distribution and Diversity of Primates in Guyana: Species-Area Relationships and Riverine Barriers. Int J Primatol. 2004;25:73–95. doi: 10.1023/B:IJOP.0000014646.82182.51. [DOI] [Google Scholar]
- Zimmermann E, Cepok S, Rakotoarison N, Zietemann V, Radespiel U. Sympatric mouse lemurs in north-west Madagascar: a new rufous mouse lemur species (Microcebus ravelobensis) Folia Primatol. 1998;69:106–114. doi: 10.1159/000021571. [DOI] [PubMed] [Google Scholar]
- Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Can J Zool. 1991;69:82–90. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor: Cold Spring Harbor press; 1989. [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorini J. PhD thesis. University of Zürich; 2000. Molecular Systematics of Lemurs. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 40 b2. Sunderland, MA, Sinauer Associates; 1999. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Braune P, Schmidt S, Zimmermann E. Spacing and group coordination in a nocturnal primate, the golden brown mouse lemur (Microcebus ravelobensis): the role of olfactory and acoustic signals. Behav Ecol Sociobiol. 2005;58:587–596. doi: 10.1007/s00265-005-0944-4. [DOI] [Google Scholar]
- Louis EE, Jr, Engberg SE, Lei R, Geng H, Sommer JA, Randriamampionona R, Randriamanana JC, Zaonarivelo JR, Andriantompohavana R, Randria G, Prosper , Ramaromilanto B, Rakotoarisoa G, Rooney A, Brenneman RA. Special Publications. Vol. 49. Museum of Texas Tech University; 2006. Molecular and Morphological Analyses of the Sportive Lemurs (Family Megaladapidae: Genus Lepilemur) Reveals 11 Previously Unrecognized Species; pp. 1–47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagnostic sites of the Cytochrome B region for each terminal clade. Dashes (-) indicate deletions. Points (.) indicate identical bases.
Diagnostic sites of the ND4 region for each terminal clade. Dashes (-) indicate deletions. Points (.) indicate identical bases.
Diagnostic sites of the D-loop region for each terminal clade. Dashes (-) indicate deletions. Points (.) indicate identical bases.


