Abstract
Background
In the human pathogen Bacillus cereus, the expression of most extracellular virulence factors is controlled by the transcriptional activator PlcR. Among these virulence factors, cereolysin O (Clo) is an haemolysin belonging to the cholesterol-dependant cytolysins, a protein family extensively studied in Gram-positive bacteria.
Results
In the genomes of bacteria belonging to the B. cereus group, including Bacillus anthracis and Bacillus thuringiensis, a small gene encoding a 26-amino acid peptide was present in multicopy. One copy was always found upstream from the gene encoding Clo. In B. cereus ATCC 14579, the small gene and the clo gene are co-transcribed. Transcriptional fusions showed that the three paralogues identified in this strain were expressed in a PlcR-dependent manner. We propose to name these peptides Spp for small PlcR-regulated peptides. We show that a synthetic peptide corresponding to the deduced product of the spp genes displayed antibacterial activity.
Conclusion
The co-expression of spp, a small PlcR-regulated multicopy gene with clo suggests a yet unidentified relationship between Spp and the cholesterol-dependent cytolysin in bacteria belonging to the B.cereus group.
Background
Bacillus cereus is an opportunistic pathogen of humans, causing local and systemic infections, and is a frequent cause of food poisoning. This species belongs to the B. cereus group, which includes the closely related species Bacillus anthracis, Bacillus thuringiensis, Bacillus weihenstephanensis,Bacillus mycoides and Bacillus pseudomycoides [1,2]. B. cereus produces several secreted proteins, including enterotoxins, cytolysins, phospholipases and proteases that may contribute to B. cereus pathogenicity. The expression of most of these virulence factors is controlled by the pleiotropic transcriptional activator PlcR [3,4]. This global regulator has been shown to contribute to B. cereus virulence in mice and insects [5] and in rabbit endophthalmitis [6]. Expression of the PlcR regulon is activated at the onset of the stationary phase of growth [7]. This activation results from cell-cell communication under the control of PapR, a small peptide that is exported, processed, and re-imported into bacterial cells in its mature form, presumably a pentapeptide, by the oligopeptide permease [8,9].
Haemolysins of the cholesterol-dependent cytolysins (CDC) family (also known as thiol-activated cytolysins) have been identified in several genera of Gram-positive bacteria [10]. These pore-forming toxins appear to play a significant role in the pathogenesis of the organisms producing them [11,12]. Listeriolysin O has been extensively studied, and this CDC has been shown to be an important virulence factor, essential for the cellulosome escape and intracellular multiplication of Listeria monocytogenes [13]. In Streptococcus pyogenes, the spn gene, which encodes an effector protein, is located upstream from the gene encoding Streptolysin (Slo). Cytolysin-mediated translocation involving these two proteins has been described in this bacterium [14]. In this process, Slo acts as a gate when anchored in the target-cell membrane. SPN is thus translocated into the cytoplasm of the target cell, increasing cytotoxicity [14,15]. The study of genes present in the same operons as CDC-encoding genes may therefore increase our understanding of virulence mechanisms in these bacterial pathogens.
CDC have been identified in bacteria of the B. cereus group. These proteins are named cereolysin O (Clo) in B. cereus, thuringiolysin O (Tlo) in B.thuringiensis and anthrolysin O (Alo) in B. anthracis [16-18]. We show here that three paralogous copies of an unannotated gene encoding a 26-amino acid peptide are present in the B. cereus ATCC 14579 genome [19]. One of these paralogues was co-transcribed with the gene encoding cereolysin O, and all three paralogues were expressed in a PlcR-dependent manner.
Results and discussion
Identification of a small gene, co-transcribed with clo
Small peptides often remain unannotated at the time of bacterial sequencing projects [20,21]. However, many such peptides have been shown to play a major role in bacterial physiology. Analysis of the clo chromosomal region of B. cereus ATCC 14579 revealed the presence of a 78 bp ORF between a putative PlcR box and the clo gene (Fig. 1a). This ORF, starting with an ATG codon, was predicted to encode a 26-amino acid peptide and was called pep1. It was preceded by a typical ribosome binding site at an appropriate distance.
Figure 1.
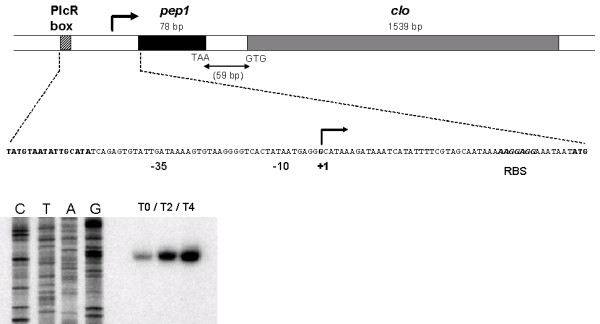
Map of the B. cereus ATCC 14579 pep-clo locus. (A) The PlcR recognition site (bold underlined), -10 and -35 boxes (underlined) and the putative RBS binding site (bold italic) are indicated. The transcription initiation site (+1) is indicated by the arrow. (B) Total RNA (20 μg) extracted from B. cereus at the onset of stationary phase (T0), two hours (T2) and four hours (T4) after T0 was subjected to primer extension analysis, using an oligonucleotide binding 65 nucleotides downstream from the Clo start codon. The same oligonucleotide was used to prime dideoxy sequencing reactions from the corresponding region obtained by PCR amplification (lanes C, T, A, G).
Primer extension was carried out in order to map the transcription start site of the clo gene, using B. cereus total RNA extracted after various culture times. The 5'-end of the mRNA corresponding to clo was located downstream from the PlcR box, and upstream from the pep1 gene, indicating that a bicistronic transcript consisting of pep1-clo had been produced (Fig. 1b). This result suggests that pep1 and clo were co-transcribed from a single transcription start point in the conditions tested. The -10 and -35 regions of this promoter are highly similar to the -35 region (TTGACA) and -10 region (TATAAT) of vegetative promoters recognised by the σA RNA polymerase of B. subtilis (Fig. 1a). Similar experiments were performed with RNA extracted from the B. cereus Δ plcR strain. No signal was detected at T0, T2 and T4 in such conditions (data not shown), indicating that expression of the pep1-clo operon was PlcR-dependent. This result is consistent with the lack of detection of the Clo protein in the extracellular fraction of the B. cereus ΔplcR strain [4].
Identification of pep paralogues and orthologues in the B. cereus group
The deduced amino-acid sequence of the peptide encoded by pep1 (Pep1) was used to screen the complete genome of B. cereus ATCC 14579 by TBLASTN. This search led to the identification of another two paralogues elsewhere on the chromosome, not located close to any particular gene. These paralogues were called pep2 and pep3. The NCBI NR database was also screened by TBLASTN. This analysis showed that ORFs presenting strong sequence similarity with pep1 were identified in all the members of the B. cereus group (Fig. 2). In most of the completed genomic sequences, pep1 orthologues were found in multiple copies, up to three copies, depending on the strain. In all of the genomes in which pep1 orthologues were identified, one copy was located upstream from a CDC-encoding gene (clo, alo or tlo). Recently, the 5'-end of the alo transcript was mapped [22]. Despite a slightly diverging sequence between alo and clo promoter regions, the 5'-end of the alo transcript was positioned downstream from the PlcR box and upstream from the pep1 orthologue, revealing that in B. anthracis, a pep1-alo bicistronic transcript was detected, as in B. cereus (pep1-clo). Thus, the structural organisation of the operon constituted of pep1 and a CDC-encoding gene seems to be conserved between species of the B. cereus group.
Figure 2.
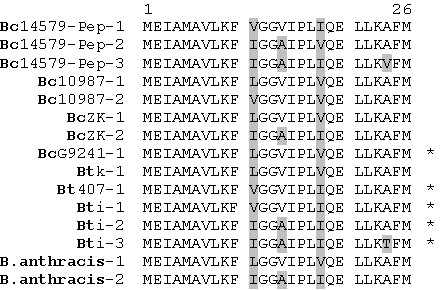
Alignment of Pep sequences identified in members of the B. cereus group. Diverging amino acids are shown in grey boxes. Alignments were performed with the Multalin version 5.4.1 program [40]. The numbers indicate copy number (1, 2 or 3) in the available genome sequences from the B. cereus group. Each number 1 corresponds to a Pep orthologue encoded by an ORF positioned upstream from a cholesterol-dependent cytolysin-encoding gene. Bc14579: B. cereus ATCC14579; Bc10987: B. cereus ATCC10987; BcZK: B. cereus EL33; BcG9241: B. cereus G9241; Btk: B. thuringiensis serovar konkukian strain 97-27; Bt407: B.thuringiensis strain 407Cry-; Bti: B. thuringiensis serovar israelensis ATCC 35646. For B. anthracis, the finished and unfinished genome sequences of the various strains gave the same Pep sequences, which are indicated only once. The B. anthracis strains tested were: strain Ames Ancestor, strain Ames, strain Sterne, strain Kruger B, strain A1055, strain CNEVA-9066, strain Western North America USA6153, strain Vollum, and strain Australia 94. In the B. anthracis strain A2012 unfinished genome sequence, only orthologue number 1 was identified. *: Pep orthologue identified in an unfinished genome sequence.
No sequence displaying significant similarity to Pep1 was identified in bacteria outside the B. cereus group, or in other sequences in the databases, indicating that Pep1 orthologues are probably restricted to the B. cereus group. However, in the genome of the atypical B. cereus strain NVH 391–98, no pep1 orthologue could be identified. In this strain, the genome has a reduced size (4 Mb) compared to the other B. cereus group members [23], and no CDC encoding gene is present. This finding is consistent with the fact that this strain is genetically distant from other B. cereus group members [24].
PlcR-dependent expression of pep1, pep2 and pep3
In silico analysis revealed the presence of a PlcR recognition site (TATGNAN4TNCATA) about 100 nucleotides upstream from the three pep genes in B. cereus ATCC 14579. Alignment of the upstream region of the pep1, pep2 and pep3 genes identified in B.cereus ATCC 14579 showed that the three promoter regions were very similar to the -35 and -10 regions recognised by the σA RNA polymerase of B. subtilis (Fig.3). A PlcR recognition site was also found upstream from all the pep orthologues identified in the other bacteria of the B. cereus group (data not shown).
Figure 3.

Alignment of spp1, spp2 and spp3 promoter regions identified in the B.cereus ATCC 14579 genome. Diverging nucleotides are shown in grey boxes. The PlcR recognition site (bold underlined), -10 and -35 boxes (underlined) and the putative RBS-binding site (bold italic) are indicated. The transcription initiation site of spp1 is shown in bold.
We investigated whether the expression of the pep genes in B. cereus ATCC 14579 depended on PlcR, by inserting about 450 bp, including each of the 5'-pep regions, upstream from the lacZ reporter gene of pHT304-18Z (Table 1). B. cereus strains carrying the three different recombinant plasmids were cultured in LB medium and β-galactosidase activity was measured at various stages, from the exponential growth phase to the late stationary phase (Fig. 4). The kinetics of β-galactosidase production were similar for all three strains, with pep-directed lacZ transcription activated at the end of exponential growth. However, transcription from the pep1 promoter appeared to begin earlier, whereas that from the pep3 promoter was activated later. These slight variations in the time course of expression may reflect differences in promoter efficiency, which might result from differences in the affinity between PlcR and its target sequences. Our results indicate that all three copies of pep are expressed in B. cereus ATCC 14579. The transcriptional activity of the three pep promoters was drastically decreased in the B. cereus ATCC 14579 Δ plcR mutant (Fig. 4). Thus, the expression of the three pep genes is PlcR-regulated. However, weak PlcR-independent expression was detected for pep2'-Z (below 500 Miller Units), pep1'-Z and pep3'-Z (below 100 Miller Units) (Fig. 4). This expression was significantly higher than that observed with the negative control pHT304-18Z without promoter (values < 10 Miller Units, data not shown). In B. anthracis, the PlcR regulator is not functional because the plcR gene is truncated [3]. A weak alo expression was detected by RT-PCR in B. anthracis cells grown in LB medium [25]. alo expression was also detected in B. anthracis cultured in rich media or grown in infected mice [18,22]. Thus, the weak expression of pep1-clo detected in B. cereus ATCC 14579 Δ plcR, may be similar to the alo expression observed in B. anthracis, which does not produce an active PlcR molecule. These peptides were designated Spp, for small PlcR-regulated peptide.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
| Strains | ||
| B. cereus ATCC14579 | laboratory collection | |
| B. cereus Δ plcR | ATCC14579 plcR::Km | [5] |
| B. subtilis 168 | laboratory collection | |
| B. cereus F4430/73 | laboratory collection | |
| B. thuringiensis 407 Cry- | laboratory collection | |
| E. coli ET12567 | (F- dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara14 pacY1 xyl-5 leuB6 thi-1) | laboratory collection |
| Proteus mirabilis | N. Boemare | |
| Pseudomonas aeruginosa | N. Boemare | |
| Salmonella spp. | N. Boemare | |
| Enterococcus faecalis | P. Serror | |
| Listeria innocua | laboratory collection | |
| Streptococcus agalactiae | P. Serror | |
| Staphylococcus aureus | P. Serror | |
| Plasmids | ||
| pHT304-18'Z | ApR and EmR cloning vehicle; lacZ reporter gene | [38] |
| pHT-Ppep1'Z | 433 bp region upstream from clo start codon inserted between PstI and BamHI sites of pHT304-18'Z | this work |
| pHT-Ppep2'Z | 448 bp region upstream from pep2 start codon inserted between HindIII and BamHI sites of pHT304-18'Z | this work |
| pHT-Ppep3'Z | 480 bp region upstream from pep3 start codon inserted between HindIII and BamHI sites of pHT304-18'Z | this work |
Km, kanamycin; Ap, ampicillin; Em, erythromycin.
Figure 4.
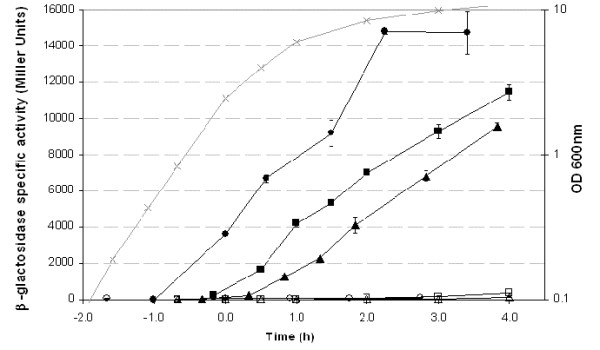
Expression of pep genes in B. cereus ATCC 14579 and B. cereus ATCC 14579 Δ plcR. Expression of Ppep1'-Z (circle), Ppep2'-Z (square), and Ppep3'-Z (triangle) in wild-type (closed symbols) and B. cereus Δ plcR (open symbols) strains. Grey symbols, OD600 of cultures of bacteria. Cells were grown at 37°C in LB medium. Time zero indicates entry into stationary phase. Standard deviations of triplicate measurements are shown for β-galactosidase activity.
Putative role of the Spp peptides
Bacillus species are known to produce and export an abundance of small peptides. Several of these peptides are involved in signalling or have antimicrobial activity [21]. Analysis of the deduced amino-acid sequence (26 aa) of spp1 (pep1) with the SignalP 3.0 server showed there to be no predicted signal peptide. However, a double-glycine motif was found at positions 12 to 13 in all the Spp orthologues (Fig. 2). This double-glycine motif is a characteristic of some secreted peptides, such as competence-stimulating peptides in streptococci and bacteriocins in lactic acid bacteria [26]. The leader region of such peptides is cleaved after the double-glycine motif by an ABC transporter [27]. The presence of the double-glycine motif suggested that Spp is exported. By analogy to the described functions of double-glycine peptides in other Gram-positive bacteria, and given that competence has never been described in B. cereus, we hypothesized that Spp has a bacteriocin-like function.
For analysis of the physiological role of Spp, the entire deduced amino-acid sequence of spp1 (26 aa), and the 13 aa C-terminal region of this peptide (starting after the two glycines) were synthesised chemically, giving Pep26 and Pep13, respectively. These two molecules were tested against various target bacterial cells, to determine whether Spp1 had bacteriocin-like functions. No growth inhibition was observed with the negative control (diluted DMSO) for any bacterial cell (not shown), whereas Pep13 displayed antibacterial activity at high concentrations (7.26 mM) on Bacillus target cells: B. subtilis, B. thuringiensis, B. cereus F4430, and B.cereus ATCC 14579 (Fig. 5). The antibacterial activity of Pep13 was detectable at dilutions down to 1.85 mM. Pep13 (at 7.26 mM) also displayed antibacterial activity against other Gram-positive target bacteria: Enterococcus faecalis, Streptococcus agalactiae and Listeria innocua, but not against Staphylococcus aureus (data not shown). We also assayed activity against Gram-negative indicator bacteria: Pep13 (at 7.26 mM) displayed weak antibacterial activity against Salmonella spp., but not against Escherichia coli K12, Proteus mirabilis or Pseudomonas aeruginosa (data not shown). Antibacterial activity of Pep26 (at 2.45 mM) resulted in only a small growth inhibition zone in assays with Bacillus indicator cells (data not shown), and no effect was observed against other indicator bacteria. The C-terminal region of Spp1 (synthetic Pep13) had stronger antibacterial activity than the entire Spp1 molecule (synthetic Pep26). This suggests that processing by cleavage downstream from the double-glycine motif may be necessary for peptide activation.
Figure 5.
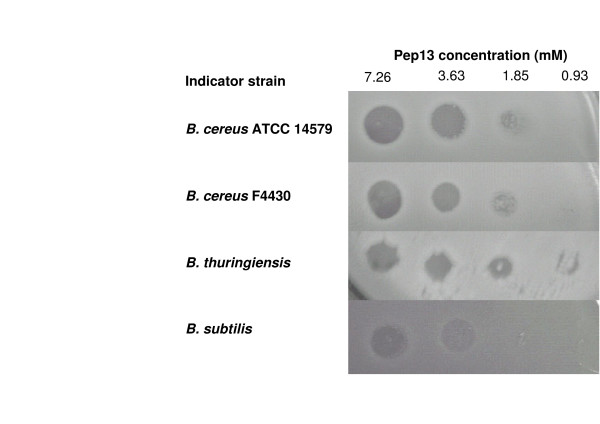
Agar-spot tests showing antibacterial activity of synthetic Pep13. Indicator strains were grown in LB to an OD600= 0.6 and diluted to OD= 0.2 before spreading on LB agar. We spotted 15 μl of Pep13, at dilutions of 7.26 mM to 0.93 mM, on indicator strains. Antibacterial activity was assessed after overnight incubation at 37°C.
Among the indicator strains tested, B. cereus strains which are Spp producers, were the most affected by the Pep13 antibacterial activity. Thus, other maturation process such as posttranslational modifications, are probably required to protect the bacterial cells against their own peptide.
When B. cereus vegetative cells were incubated 1 hour in a phosphate buffer supplemented with Pep13 (to a final concentration of 0.7 mM), the number of CFU decreased from 1.5 (+/-0.1) × 107/ml to 3.3 (+/- 0.7) × 106/ml (experiments were repeated twice). In the same conditions, the number of B. subtilis CFU decreased from 2.1 × 107/ml to 2.3 × 105/ml. This indicates that Pep13 was bactericidal rather than bacteriostatic against these target cells.
However, although spp is expressed, there is no evidence that Spp is actually synthesized and secreted. Furthermore, given the high concentrations of Pep13 required in our assays, we cannot rule out that the antibacterial activity detected is caused by the high Pep13 hydrophobicity rather than by a specific antibacterial activity.
B. cereus has been isolated from soil, and from the gut of insects and nematodes [28]. Like many other bacteria isolated from such ecological niches in which there is strong competition between numerous species of micro-organisms for colonisation, B.cereus has been shown to produce antimicrobial peptides [29,30]. Recently, an antibacterial substance with a molecular mass of 3.4 kDa, active only against Gram-positive bacteria, was described in B. cereus ATCC 14579 [31]. This antibacterial activity is probably not caused by Spp, because its antibacterial spectrum is different and the predicted molecular mass of Spp1 is lower: 2.9 kDa (26 aa), and 1.5 kDa for the C-terminal fragment of Spp1 (13 aa). However, we cannot rule out the possibility that Spp1 undergoes post-translational modifications, accounting for differences in molecular mass and antibacterial spectrum.
Two small peptides with double-glycine leader sequences produced by competent cells of S. pneumoniae were recently shown to be involved in the lysis of non-competent S.pneumoniae cells, leading to the release of pneumolysin, a non-secreted CDC. This work revealed the existence of co-operation between bacteriocins and a CDC [32]. In B. cereus, Clo, which is found in the extracellular fraction [4], is most probably exported by the SEC machinery because it has a signal peptide. Thus, the link between Spp and Clo is probably different from that described in S. pneumoniae.
In S. pyogenes, a co-operative effect between a CDC (Slo) and a protein (Spn) encoded by a gene from the same operon has been observed. This co-operative effect increases toxicity to target cells [14]. We showed that spp1 (pep1) and clo are co-transcribed in B. cereus ATCC 14579. This operon structure was found to be conserved among bacteria belonging to the B. cereus group. These findings suggest that Clo and Spp might co-operate to play a role similar to that of Slo and Spn in S.pyogenes, in specific ecological niches or growth conditions that remain to be determined.
Conclusion
This work has led to the identification of spp genes present in all members of the B. cereus group. We showed that the three spp genes of B. cereus ATCC14579 were expressed in a PlcR-dependent manner. In all the B. cereus group strains, a spp gene is coexpressed with the CDC genes encoding cereolysin, thuringiolysin or anthrolysin. The biological signification of this co-expression and the proposed Spp antibacterial role will have to be clarified.
Methods
Strains and growth conditions
The strains used in this study are listed in Table 1. E. coli, and B. cereus cells were routinely grown in Luria broth (LB), at 37°C with vigorous shaking. The antibiotic concentrations used for bacterial selection were: ampicillin, 100 μg.ml-1; erythromycin, 10 μg.ml-1 and kanamycin, 150 μg.ml-1. Bacteria with the Lac+ phenotype were identified on LB agar containing 40 μg.ml-1X-Gal.
Database comparison and sequence analysis
TBLASTN alignments were performed with the deduced amino-acid sequence of the protein encoded by pep1 from B. cereus ATCC 14579 to screen the NR database [33]. The putative signal peptide in the polypeptide sequence was identified with the SignalP 3.0 server [34].
DNA manipulation
Plasmid DNA was purified from E. coli using QIAprep spin columns (Qiagen). Chromosomal DNA was extracted from B. cereus cells as previously described [35]. Restriction enzymes and T4 DNA ligase were used as recommended by the manufacturer (New England Biolabs). Oligonucleotide primers were synthesised by Proligo-Genset (Paris, France). PCR was performed in a GeneAmp PCR system 2400 thermocycler (Perkin-Elmer), using the high-fidelity Pfx DNA polymerase (Invitrogen). Amplified DNA fragments were purified with the QIAquick PCR Purification Kit (Qiagen), digested and separated on 0.7% agarose gels. Digested DNA fragments were extracted from agarose gels by centrifugation in a filter device (Ultrafree DA, Millipore). All constructs were verified by DNA sequencing (GenomeExpress, France). Electroporation was used to transform E. coli and B. cereus, as previously described [36,37].
Construction of pep'-lacZ transcriptional fusions
We constructed pep'-lacZ transcriptional fusions by inserting a PCR-amplified DNA fragment harbouring the putative pep1, pep2 or pep3 promoter regions, digested at the endonuclease sites introduced in the primers (Table 2), between the corresponding sites of pHT304-18'Z [38]. The recombinant plasmids (Table 1) were introduced into B. cereus ATCC 14579 wild-type and Δ plcR mutant strains by electroporation.
Table 2.
Primers used
| Primer name | 5'-3' sequence* | Restriction sites |
| Ppep1-L | GATACTGCAGCCTTATGGGCCAATAGCAGT | PstI |
| Ppep1-R | CGTCGGATCCTGATTGATAAATGATTGCTAACTAA | BamHI |
| Ppep2-L | CGCAAGCTTTCTAAACAAGGAATCCTACAAAG | HindIII |
| Ppep2-R | CGCGGATCCCTCCTCTTTTTCGTATTAAGATG | BamHI |
| Ppep3-L | CGCAAGCTTGGAAATAGTGGGTCTAGAACAT | HindIII |
| Ppep3-R | CGCGGATCCCCTCTTTTGTTAATACTGGGA | BamHI |
| Extsnclo | CTAACTAATAAACATGCAAGGAAC |
* Restriction enzyme sites are underlined.
β-Galactosidase assay
β-Galactosidase specific activities from cells of B. cereus strains harbouring plasmids with lacZ transcriptional fusions were measured as previously described [35], and were expressed in units of β-galactosidase per milligram of protein (Miller units). The Bradford method (BioRad protein assay) was used for total protein quantification.
RNA extraction and primer extension
Total RNA was extracted from B. cereus ATCC 14579 wild-type and ΔplcR cells grown in LB at 37°C, at the onset of stationary phase (T0), two hours (T2) and four hours (T4) after T0, as previously described [39]. The clo transcription start site was identified by primer extension with the ExtsnClo oligonucleotide (Table 2), as previously described [39]. DNA sequencing was performed by the dideoxy chain termination method, with the same primer and the corresponding PCR product used as the template, with the T7 sequenase PCR product sequencing kit (USB Corporation).
Antibacterial activity
The entire deduced amino-acid sequence of the pep1 ORF (26 aa: MEIAMAVLKFVGGVIPLIQELLKAFM), and the 13 aa C-terminal region of this peptide were synthesised chemically by Millegen (Toulouse, France). These molecules were called Pep26 and Pep13, respectively. Due to their strong hydrophobicity, these molecules were dissolved in DMSO, as recommended by the manufacturer. The resulting stock solution was then diluted with H2O to 7 mg.ml-1 (2.45 mM) in 65% (v/v) DMSO/H2O for Pep26, and to 11 mg.ml-1 (7.26 mM) in 25% (v/v) DMSO/H2O for Pep13. These solutions were further diluted in H2O and assayed on target bacterial cells. Indicator strains were grown in LB at 37°C with vigorous shaking, until an OD600nm of 0.6 was reached. They were then diluted in fresh LB to give an OD of 0.2 and 5 ml were spread on LB-agar plates. The plates were incubated for 10 min and excess liquid was then removed. Plates were allowed to dry at room temperature for 10 min under laminar air flow. Then, 15 μl of Pep26, Pep13, or DMSO (diluted to a final concentration of 65% as negative control) were applied to the plates inoculated with indicator strains. Plates were incubated overnight at 37°C before checking for a putative zone of growth inhibition.
In order to determine whether Pep13 was bactericidal or bacteriostatic, indicator strains were cultured as described above until an OD of 0.7 was reached. They were diluted 10 fold in a 0.1 M potassium phosphate buffer (pH 7) and 200 μl were incubated with 20 μl of a 7.2 mM Pep13 solution for 1 hour at 37°C. Then, the mixture was serially diluted to determine the number of CFU on LB agar medium.
Abbreviations
CDC, cholesterol-dependent cytolysin.
Authors' contributions
JB performed the experiments. JB and DL performed the data analysis, and wrote the manuscript. DL supervised the project. Both authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank Sinda Fedhila for assistance with unpublished experiments. We also thank N. Boemare, C. Laroui, C. Nielsen-Leroux, P. Serror for providing some of the strains used in this study and C. Braun-Breton for providing human blood. The initial stage of the work was supported by a grant "AQS: Caractérisation de la virulence de Bacillus cereus" from the Ministère de la Recherche and the Ministère de l'Agriculture et de la Pêche. This work was supported by INRA (Institut National de la Recherche Agronomique).
Contributor Information
Julien Brillard, Email: brillard@avignon.inra.fr.
Didier Lereclus, Email: lereclus@jouy.inra.fr.
References
- Daffonchio D, Cherif A, Borin S. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the " Bacillus cereus group". Appl Environ Microbiol. 2000;66:5460–5468. doi: 10.1128/AEM.66.12.5460-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis –one species on the basis of genetic evidence. Appl Environ Microbiol. 2000;66:2627–2630. doi: 10.1128/AEM.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H, Gominet M, Okstad OA, Kolsto AB, Lereclus D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol. 1999;32:1043–1053. doi: 10.1046/j.1365-2958.1999.01419.x. [DOI] [PubMed] [Google Scholar]
- Gohar M, Okstad OA, Gilois N, Sanchis V, Kolsto AB, Lereclus D. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics. 2002;2:784–791. doi: 10.1002/1615-9861(200206)2:6<784::AID-PROT784>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Salamitou S, Ramisse F, Brehelin M, Bourguet D, Gilois N, Gominet M, Hernandez E, Lereclus D. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology. 2000;146:2825–2832. doi: 10.1099/00221287-146-11-2825. [DOI] [PubMed] [Google Scholar]
- Callegan MC, Kane ST, Cochran DC, Gilmore MS, Gominet M, Lereclus D. Relationship of plcR-regulated factors to Bacillus endophthalmitis virulence. Infect Immun. 2003;71:3116–3124. doi: 10.1128/IAI.71.6.3116-3124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D, Agaisse H, Gominet M, Salamitou S, Sanchis V. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J Bacteriol. 1996;178:2749–2756. doi: 10.1128/jb.178.10.2749-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol Microbiol. 2001;40:963–975. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. Embo J. 2002;21:4550–4559. doi: 10.1093/emboj/cdf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington SJ, Jost BH, Songer JG. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol Lett. 2000;182:197–205. doi: 10.1016/S0378-1097(99)00536-4. [DOI] [PubMed] [Google Scholar]
- Palmer M. The family of thiol-activated, cholesterol-binding cytolysins. Toxicon. 2001;39:1681–1689. doi: 10.1016/S0041-0101(01)00155-6. [DOI] [PubMed] [Google Scholar]
- Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/S0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol. 2002;44:257–269. doi: 10.1046/j.1365-2958.2002.02876.x. [DOI] [PubMed] [Google Scholar]
- Alouf JE. Cholesterol-binding cytolytic protein toxins. Int J Med Microbiol. 2000;290:351–356. doi: 10.1016/S1438-4221(00)80039-9. [DOI] [PubMed] [Google Scholar]
- Alouf JE, Billington SJ, Jost BH. Repertoire and general features of the family of cholesterol-dependent cytolysins. In: Alouf JE, Popoff MR, editor. The comprehensive sourcebook of bacterial protein toxins. Burlington, USA: Elsevier; 2006. pp. 643–658. [Google Scholar]
- Shannon JG, Ross CL, Koehler TM, Rest RF. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect Immun. 2003;71:3183–3189. doi: 10.1128/IAI.71.6.3183-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D'Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- Dirix G, Monsieurs P, Marchal K, Vanderleyden J, Michiels J. Screening genomes of Gram-positive bacteria for double-glycine-motif-containing peptides. Microbiology. 2004;150:1121–1126. doi: 10.1099/mic.0.27040-0. [DOI] [PubMed] [Google Scholar]
- Zuber P. A peptide profile of the Bacillus subtilis genome. Peptides. 2001;22:1555–1577. doi: 10.1016/S0196-9781(01)00492-2. [DOI] [PubMed] [Google Scholar]
- Ross CL, Koehler TM. plcR papR-independent expression of anthrolysin O by Bacillus anthracis. J Bacteriol. 2006;188:7823–7829. doi: 10.1128/JB.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus A, Goltsman E, Auger S, Galleron N, Segurens B, Dossat C, Land ML, Broussolle V, Brillard J, Guinebretiere MH, Sanchis V, Nguen-The C, Lereclus D, Richardson P, Wincker P, Weissenbach J, Ehrlich SD, Sorokin A. Extending the Bacillus cereus group genomics to putative food-borne pathogens of different toxicity. Chem Biol Interact. 2007 doi: 10.1016/j.cbi.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sorokin A, Candelon B, Guilloux K, Galleron N, Wackerow-Kouzova N, Ehrlich SD, Bourguet D, Sanchis V. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl Environ Microbiol. 2006;72:1569–1578. doi: 10.1128/AEM.72.2.1569-1578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Mock M, Robichon D, Landier A, Lereclus D, Fouet A. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol Microbiol. 2001;42:1189–1198. doi: 10.1046/j.1365-2958.2001.02692.x. [DOI] [PubMed] [Google Scholar]
- Dirix G, Monsieurs P, Dombrecht B, Daniels R, Marchal K, Vanderleyden J, Michiels J. Peptide signal molecules and bacteriocins in Gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides. 2004;25:1425–1440. doi: 10.1016/j.peptides.2003.10.028. [DOI] [PubMed] [Google Scholar]
- Havarstein LS, Holo H, Nes IF. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- Jensen GB, Hansen BM, Eilenberg J, Mahillon J. The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol. 2003;5:631–640. doi: 10.1046/j.1462-2920.2003.00461.x. [DOI] [PubMed] [Google Scholar]
- Bizani D, Brandelli A. Characterization of a bacteriocin produced by a newly isolated Bacillus sp. Strain 8 A. J Appl Microbiol. 2002;93:512–519. doi: 10.1046/j.1365-2672.2002.01720.x. [DOI] [PubMed] [Google Scholar]
- Oscariz JC, Lasa I, Pisabarro AG. Detection and characterization of cerein 7, a new bacteriocin produced by Bacillus cereus with a broad spectrum of activity. FEMS Microbiol Lett. 1999;178:337–341. doi: 10.1111/j.1574-6968.1999.tb08696.x. [DOI] [PubMed] [Google Scholar]
- Risoen PA, Ronning P, Hegna IK, Kolsto AB. Characterization of a broad range antimicrobial substance from Bacillus cereus. J Appl Microbiol. 2004;96:648–655. doi: 10.1046/j.1365-2672.2003.02139.x. [DOI] [PubMed] [Google Scholar]
- Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci USA. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SignalP 3.0 http://www.cbs.dtu.dk/
- Bouillaut L, Ramarao N, Buisson C, Gilois N, Gohar M, Lereclus D, Nielsen-LeRoux C. FlhA Influences Bacillus thuringiensis PlcR-Regulated Gene Transcription, Protein Production, and Virulence. Appl Environ Microbiol. 2005;71:8903–8910. doi: 10.1128/AEM.71.12.8903-8910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D, Arantes O, Chaufaux J, Lecadet M. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;51:211–217. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- Agaisse H, Lereclus D. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J Bacteriol. 1994;176:4734–4741. doi: 10.1128/jb.176.15.4734-4741.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H, Lereclus D. STAB-SD: a Shine-Dalgarno sequence in the 5' untranslated region is a determinant of mRNA stability. Mol Microbiol. 1996;20:633–643. doi: 10.1046/j.1365-2958.1996.5401046.x. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]


