Abstract
Background
A number of studies have shown that peptidases and in particular cysteine peptidases constitute major pathogenicity factors in Entamoeba histolytica. Recent studies have suggested that a considerable number of genes coding for proteolytic enzymes are present within the E. histolytica genome and questions remain about the mode of expression of the various molecules.
Results
By homology search within the recently published amoeba genome, we identified a total of 86 E. histolytica genes coding for putative peptidases, including 46 recently described peptidase genes. In total these comprise (i) 50 cysteine peptidases of different families but most of which belong to the C1 papain superfamily, (ii) 22 different metallo peptidases from at least 11 different families, (iii) 10 serine peptidases belonging to 3 different families, and (iv) 4 aspartic peptidases of only one family. Using an oligonucleotide microarray, peptidase gene expression patterns of 7 different E. histolytica isolates as well as of heat stressed cells were analysed. A total of 21 out of 79 amoeba peptidase genes analysed were found to be significantly expressed under standard axenic culture conditions whereas the remaining are not expressed or at very low levels only. In heat-stressed cells the expression of 2 and 3 peptidase genes, respectively, were either decreased or increased. Only minor differences were observed between the various isolates investigated, despite the fact that these isolates were originated from asymptomatic individuals or from patients with various forms of amoebic diseases.
Conclusion
Entamoeba histolytica possesses a large number of genes coding for proteolytic enzymes. Under standard culture conditions or upon heat-stress only a relatively small number of these genes is significantly expressed and only very few variations become apparent between various clinical E. histolytica isolates, calling into question the importance of these enzymes in E. histolytica pathogenicity. Further studies are required to define the precise role of most of the proteolytic enzyme for amoeba cell biology but in particular for E. histolytica virulence.
Background
The faecal-oral spread protozoan parasite Entamoeba histolytica is an important human pathogen. Normally, this parasite resides and multiplies in the large bowel and can persist there for months and years causing only an asymptomatic luminal gut infection. However, occasionally E. histolytica penetrates the intestinal mucosa, which leads to ulcerative colitis or it disseminates to other organs, most commonly to the liver, where it induces abscess formation. Cysteine peptidases are considered to play a major role for the pathogenicity of E. histolytica as suggested by a large number of in vitro and in vivo studies [1-9]. Most convincing are results from infections of laboratory animals indicating that E. histolytica trophozoites that have reduced cysteine peptidase activity are greatly impaired in their ability to induce amoebic liver abscesses [8,9]. In addition, overexpression of cysteine peptidases led to an increase in cytopathic activity, measured by in vitro monolayer disruption, as well as to a significant increase in amoebic liver abscess formation in laboratory animals in comparison to matching controls [10]. Furthermore, the discovery that amoeba cysteine peptidases possess interleukin-1β converting enzyme activity suggests a novel mechanism of these enzymes in amoebic virulence [11].
Homology searches based on the conservation of active site regions revealed that the E. histolytica genome contains a multitude of at least 50 genes coding for cysteine peptidases (reviewed by Clark et al [12]). Of these, the majority is structurally related to the C1 papain superfamily, whereas a few others are more similar to family C2 (calpain-like cysteine proteinases), C19 (ubiquitinyl hydrolase), C48 (Ulp1 peptidase), C54 (autophagin), and C65 (otubain), respectively [12].
Phylogenetic analyses of the 37 C1-family members revealed that they represent 3 distinct clades (A, B, C), each consisting of 13, 11 and 13 members, respectively [12]. EhCP-A and EhCP-B family members are organised as classical pre-pro enzymes with an overall cathepsin L-like structure. They differ in length of the pro regions as well as of the catalytic domains and have specific sequence motifs within the N-terminal regions of the mature enzymes. In addition, most members of the EhCP-B contain hydrophobic stretches near or at the C-terminus [12,13]. The primary structure prediction of the 13 EhCP-C members indicated a hydrophobic region located 11 to 28 amino acid residues apart from the N-terminus, which is predicted to form a signal anchor. As there is no example of a structural related cysteine peptidase corresponding to the EhCP-C subfamily, any function of this group of molecules remains to be determined.
In addition, two genes encoding for putative cysteine peptidases of the family C2 (calpain-like proteases) were identified within the genome (EhCALP1 and EhCALP2). These molecules are involved in several cellular processes including signal transduction pathways, remodelling of the cytoskeleton and membranes and apoptosis [14].
Another 4 genes were identified coding for enzymes with homology to the peptidase family C54 also termed autophagins (EhAUTO1-4). The process of autophagy has initially been described in other eukaryotic cells as a rescue mechanism that is induced upon starvation or oxidative stress. It is a process by which cells digest parts of their own cytosolic material. This allows the recycling of molecules under conditions of nutritional limitation and remodelling of intracellular structure for cell differentiation [15-17].
Four other genes putatively encoding cysteine peptidases in E. histolytica show homology to members of the C19 and C65 families. These two groups of enzymes are known to be involved in ubiquitin degradation. In addition 3 genes with homology to Ulp1 peptidase (C48 family) were found. Ulp1 is a member of a family of peptidases that control the function of SUMO a small ubiquitin like modifier protein [18].
Only preliminary data are available for other peptidase family members in E. histolytica. So far, a collagenase [19], a high molecular weight proteinase [20], a serine-metallo proteinase [21], a tripeptidyl peptidase I [22] and a serine protease [23] have been reported.
In this study we have analysed the genome of E. histolytica for the presence of additional peptidases belonging to the aspartate, serine and metallo peptidase families. Furthermore, the expression profile of the amoeba genes for the various proteolytic enzymes was assessed in 7 different E. histolytica isolates as well as under heat stress conditions using an oligonucleotide-based microarray and quantitative real time PCR.
Results
Peptidase genes in E. histolytica
Homology search within the E. histolytica genome revealed a total of 86 genes coding for putative peptidases. These comprise 50 cysteine peptidases of various families, all of which belonging to clan CA. In addition, 4 aspartic, 10 serine and 22 metallo peptidase genes were identified (Figure 1, Table 1). Structural details of the various E. histolytica cysteine peptidases have been described recently [12].
Figure 1.
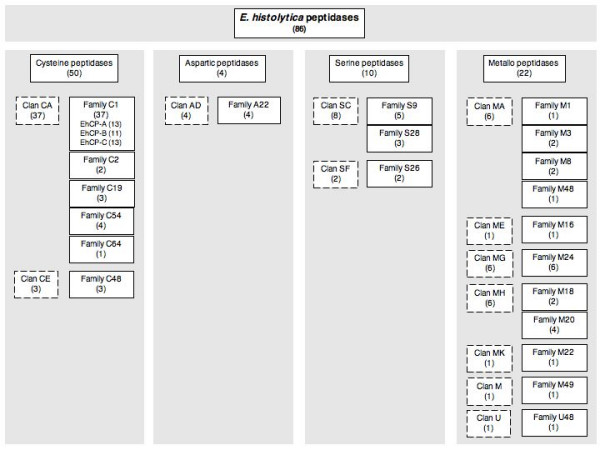
Summary of the peptidase gene families identified within the Entamoeba histolytica genome. The identified enzymes were grouped into the corresponding peptidase clans and families according to the MEROPS nomenclature.
Table 1.
Peptidases of Entamoeba histolytica
| Protein name | Clan, family, subfamily | Accession No. | Protein length | Active site residues | Remarks | Name, NCBI | |
| Cysteine peptidases | |||||||
| 1 | EhCP-A1 | CA, C1, A |
XP_650156 XM_645064 |
315 | Q112C118H259N279 | SP: 13, Pro: 80, TM: - | cysteine protease 1 |
| 2 | EhCP-A2 | CA, C1, A |
XP_650642 XM_645550 |
315 | Q112C118H259N279 | SP: 13, Pro; 80, TM: - | cysteine proteinase 2 |
| 3 | EhCP-A3 | CA, C1, A |
XP_653254 XM_648162 |
308 | Q111C115H251N271 | SP: 13, Pro: 79, TM: - | cysteine proteinase acp1 |
| 4 | EhCP-A4 | CA, C1, A |
XP_656602 XM_651510 |
311 | Q112C118H253N273 | SP: 20, Pro: 73, TM: - | cysteine proteinase |
| 5 | EhCP-A5 | CA, C1, A |
XP_650937 XM_645845 |
318 | Q113C119H261N271 | SP: 20, Pro: 72, TM: - | cysteine proteinase |
| 6 | EhCP-A6 | CA, C1, A |
XP_657364 XM_652272 |
320 | Q115C122H261N281 | SP: 17, Pro: 79, TM: - | cysteine proteinase |
| 7 | EhCP-A7 | CA, C1, A |
XP_648996 XM_643904 |
315 | Q112C118H259N279 | SP: 13, Pro: 80, TM: - | cysteine protease 8 |
| 8 | EhCP-A8 | CA, C1, A |
XP_657446 XM_652354 |
317 | Q116C122H260N280 | SP: 15, Pro: 82, TM: - | cysteine protease 9 |
| 9 | EhCP-A9 | CA, C1, A |
XP_655675 XM_650583 |
297 | Q126C132H269N290 | SP: 17, Pro: 90, TM: - | cysteine protease 10 |
| 10 | EhCP-A10 | CA, C1, A |
XP_651147 XM_646598 |
420 | Q185C191H336N357 | SP: 18, Pro: 148, TM: - | cysteine protease 17 |
| 11 | EhCP-A11 | CA, C1, A |
XP_651690 XM_646598 |
324 | Q118C124H-N287 | SP: 17, Pro: 79, TM: - | cysteine protease 19 |
| 12 | EhCP-A12 | CA, C1, A |
XP_653823 XM_648731 |
317 | not identified, N281 | SP: 14, Pro: 83, TM: - | cysteine proteinase |
| 13 | EhCP-A13 | CA, C1, A | not annotated | 250 (IS) | Q125C130 | SP: 18, Pro: 108 | |
| 14 | EhCP-B1 | CA, C1, A |
XP_651581 XM_646489 |
426 | Q105C101H308N328 | SP: 15, Pro: 106, hydroph. C-term. | cysteine proteinase 7 |
| 15 | EhCP-B2 | CA, C1, A | AAO03568 | 431 | Q145C151H308S328 | SP: 15, Pro: 106, GPI | cysteine protease 11 |
| 16 | EhCP-B3 | CA, C1, A |
XP_656747 XM_651655 |
474 | Q155C161H304N324 | SP: 16, Pro: 107, TM: 444–466 | cysteine protease 12-related |
| 17 | EhCP-B4 | CA, C1, A |
XP_648501 XM_643409 |
379 | Q153C159H302N322 | SP: 16, Pro: 105, TM: 355–377 or GPI | cysteine protease 13 |
| 18 | EhCP-B5 | CA, C1, A |
XP_652671 XM_647579 |
434 | Q151C157H311N326 | SP: 12, Pro: 108, GPI | cysteine protease 14-related |
| 19 | EhCP-B6 | CA, C1, A |
XP_652465 XM_647373 |
300 | Q84C90H232N252 | SP: 14, Pro: 55, hydroph. C-term. | cysteine protease 15 |
| 20 | EhCP-B7 | CA, C1, A |
XP_650400 XM_645308 |
650 | Q-C171H312N332 | SP: 18, Pro: 144, hydroph. C-term. | cysteine protease 16 |
| 21 | EhCP-B8 | CA, C1, A |
XP_651049 XM_645957 |
473 | Q110C156H329N249 | SP: 15, Pro: 105, GPI | cysteine protease 18 |
| 22 | EhCP-B9 | CA, C1, A |
XP_652993 XM_647901 |
446 | Q161C167H328N348 | SP: 19, Pro: 112, hydroph. C-term | cysteine protease |
| 23 | EhCP-B10 | CA, C1, A |
XP_648306 XM_643214 |
372 (IS) | Q67C76H244N264 | hydroph. C-term. | cysteine protease |
| 24 | EhCP-B11 | CA, C1, A |
XP_648013 XM_642921 |
133 (IS) | cysteine protease 11-related | ||
| 25 | EhCP-C1 | CA, C1, A |
XP_654453 XM_649361 |
586 | Q70C76H-N345 | SA: 12–34 | hypothetical protein |
| 26 | EhCP-C2 | CA, C1, A |
XP_656632 XM_651540 |
567 | Q87C93H306N326 | SA: 27–49 | hypothetical protein |
| 27 | EhCP-C3 | CA, C1, A |
XP_655128 XM_650036 |
572 | Q94C100H322N337 | SA: 17–39 | hypothetical protein |
| 28 | EhCP-C4 | CA, C1, A | XP_655800 XM_650708 | 502 | Q32C38H246N271 | SP: 15 | hypothetical protein |
| 29 | EhCP-C5 | CA, C1, A | XP_654800 XM_649708 | 557 | Q90C96H302N327 | SA: 20–42 | hypothetical protein |
| 30 | EhCP-C6 | CA, C1, A | XP_651553 XM_646461 | 557 | Q93C99H293N- | SA: 14–36 | hypothetical protein |
| 31 | EhCP-C7 | CA, C1, A | XP_657273 XM_652181 | 595 | Q89C95H297N322 | SA: 19–41 | hypothetical protein |
| 32 | EhCP-C8 | CA, C1, A | XP_655479 XM_652181 | 627 | Q91C96H317N366 | SA: 29–51 | hypothetical protein |
| 33 | EhCP-C9 | CA, C1, A | XP_655011 XM_649919 | 518 | not identified | SA: 12–34 | hypothetical protein |
| 34 | EhCP-C10 | CA, C1, A | XP_654829 XM_649737 | 530 | Q87C93H299N324 | SA: 15–37 | hypothetical protein |
| 35 | EhCP-C11 | CA, C1, A | XP_648083 XM_642991 | 526 | not identified | SA: 20–42 | hypothetical protein |
| 36 | EhCP-C12 | CA, C1, A | XP_650829 XM_645737 | 473 | not identified | SA: 26–48, TM: 449–471 | hypothetical protein |
| 37 | EhCP-C13 | CA, C1, A | XP_656556 XM_651464 | 564 | Q89C95H266N290 | SA: 21–43 | hypothetical protein |
| 38 | EhCALP1 | CA, C2 | XP_649922 XM_644830 | 591 | not identified | SP: -, TM: - | calpain-like cysteine protease |
| 39 | EhCALP2 | CA, C2 | XP_657312 XM_652220 | 473 | Q51C57H206N227 | SP: -, TM: - | calpain family cysteine protease |
| 40 | EhUBHY | CA, C19 | XP_657356 XM_652264 | 444 | not identified | SP: -, TM: - | peptidase |
| 41 | EhUBP | CA, C19 | XP_654028 XM_648936 | 352 | N30C35H330D348 | SP: -, TM: - | ubiquitin-specific protease |
| 42 | EhUCH | CA, C19 | XP_655880 XM_650788 | 386 | N37C42H345D360 | SP: -, TM: - | ubiquitin carboxyl-terminal hydrolase |
| 43 | EhUlp1-1 | CA, C48 | XP_650529 XM_645437 | 197 | H95D112Q146C152 | SP: -, TM: - | Ulp1 protease |
| 44 | EhUlp1-2 | CA, C48 | XP_651052 XM_645960 | 538 | H399D435Q482C488 | SP: -, TM: - | Ulp1 protease |
| 45 | EhUlp1-3 | CA, C48 | XP_657158 XM_652066 | 285 | H174D191Q234C240 | SP: -, TM: - | Ulp1 protease |
| 46 | EhAUTO1 | CA, C54 | XP_651386 XM_646294 | 325 | Y71C100D251H253 | SP: -, TM: - | peptidase |
| 47 | EhAUTO2 | CA, C54 | XP_653798 XM_648706 | 364 | Y93C103D278H280 | SP: -, TM: - | peptidase |
| 48 | EhAUTO3 | CA, C54 | XP_652043 XM_646951 | 364 | Y92C103D279H281 | SP: -, TM: - | hypothetical protein |
| 49 | EhAUTO4 | CA, C54 | XP_656724 XM_651632 | 348 | Y92C112D265H267 | SP: -, TM: - | hypothetical protein |
| 50 | EhOTU | CA, C65 | XP_654013 XM_648921 | 259 | D105C108H212 | SP: -, TM: - | OTU-like cysteine protease |
| Aspartic peptidase | |||||||
| 51 | EhAsP22-1 | AD, A22, A | XP_654079 XM_648987 | 340 | D178D223 | SP:24 or TM: 7–26 + 7 × TM | Intramembrane protease |
| 52 | EhAsP22-2 | AD, A22, A | XP_652820 XM_647728 | 316 | D157, D209 | SP: 19 or TM: 2–19 + 8 × TM | Signal peptide peptidase |
| 53 | EhAsP22-3 | AD, A22, A | XP_657563 XM_652471 | 320 | D157D205 | SA:7–29 + 8 xTM | Signal peptide peptidase |
| 54 | EhAsP22-4 | AD, A22, A | XP_653696 XM_648604 | 396 | D245D318 | SP: -, 7 × TM | Presenilin 1-related peptidase |
| Serine peptidase | |||||||
| 55 | EhSP9-1 | SC, S9, C | XP_655265 XM_650173 | 653 | S514D595H627 | SP: 15, TM: - | dipeptidyl-peptidase |
| 56 | EhSP9-2 | SC, S9, C | XP_655222 XM_650130 | 665 | S526D607H639 | SP: 15, TM: - | dipeptidyl-peptidase |
| 57 | EhSP9-3 | SC, S9, C | XP_656380 XM_651288 | 656 | S516D599H632 | SP: -, TM: - | prolyl oligopeptidase |
| 58 | EhSP9-4 | SC, S9, C | XP_649111 XM_644019 XP_648413 XM_643321 XP_655473 XM_650381 | 669 | not identified | SP: 16, TM: - | dipeptidyl-peptidase |
| 59 | EhSP9-5 | SC, S9 | XP_655676 XM_650584 | 102 (IS) | SP: -, TM: - | prolyl oligopeptidase family | |
| 60 | EhSP26-1 | SF, S26, B | XP_653142 XM_648050 | 189 | S65H106 | SP: -, TM: 34–56, 163–185 | signal peptidase (signalase) |
| 61 | EhSP26-2 | SF, S26 | XP_651791 XM_646699 | 121 | not identified | SP: -, TM: - | microsomal signal peptidase |
| 62 | EhSP28-1 | SC, S28 | XP_656762 XM_651670 | 457 | S165D395H421 | SP: 15, TM: - | serine peptidase |
| 63 | EhSP28-2 | SC, S28 | XP_648991 XM_643899 | 480 | S165D418H444 | SP: 15, TM: - | serine peptidase |
| 64 | EhSP28-3 | SC, S28 | XP_652089 XM_646997 | 466 | S155D404H431 | SP: -, TM: - | serine peptidase |
| Metallo peptidases | |||||||
| 65 | EhMP1-1 | MA, M1 | XP_652558 XM_647466 | 827 | H295E296H299E318Y481 | SP: -, TM: - | aminopeptidase |
| 66 | EhMP3-1 | MA, M3 | XP_649877 XM_644785 | 675 | H463E464H467E493 | SP: -, TM: - | oligopeptidase A |
| 67 | EhMP3-2 | MA, M3 | XP_649600 XM_644508 | 710 | H498E499H502E527 | SP: -, TM: - | oligopeptidase A |
| 68 | EhMP8-1 | MA, M8 | XP_655394 XM_650302 | 643 | H206E207H210H270M281 | SP: 18, TM: 605–627 | leishmaniolysin-related peptidase |
| 69 | EhMP8-2 | MA, M8 | XP_652632 XM_647540 | 662 | H207E208H211H267M278 | SP: -, TM: 598–620 | leishmaniolysin-related peptidase |
| 70 | EhMP48-1 | MA, M48, A | XP_648770 XM_643678 | 416 | H274E275H278E353 | SA: 4–21 + 6 × TM | CAAX prenyl protease |
| 71 | EhMP16-1 | ME, M16, C | XP_654849 XM_649757 | 970 | H59E62H63E137E158 | SP: -, TM: - | Zn-dependent peptidase, eupitrilysin |
| 72 | EhMP24-1 | MG, M24, A | XP_651539 XM_646447 | 409 | H165D186D197H266E299E394 | SP: -, TM: - | methionine aminopeptidase |
| 73 | EhMP24-2 | MG, M24, B | XP_657085 XM_651993 | 471 | H244D265D276H354H358H365E401E441 | SP: -, TM: - | Xaa-Pro dipeptidase |
| 74 | EhMP24-3 | MG, M24, B | XP_654211 XM_649119 | 563 | H364D384D396H461H465H470E491E505 | SP: -, TM: - | aminopeptidase |
| 75 | EhMP24-4 | MG, M24, B | XP_650646 XM_645554 | 559 | H346D366D378H443H447H452E473E487 | SP: -, TM: - | aminopeptidase |
| 76 | EhMP24-5 | MG, M24, B | XP_649980 XM_644888 | 589 | H369D389D401H466H475E496E510 | SP: -, TM: - | aminopeptidase |
| 77 | EhMP24-6 | MG, M24, B | XP_653331 XM_648239 XM_649891 XM_644799 | 371 | not identified | SP: -, TM: - | peptidase |
| 78 | EhMP18-1 | MH, M18 | XP_656618 XM_651526 | 435 | H79D81D222E258E259D307H402 | SP: -, TM: - | aminopeptidase |
| 79 | EhMP18-2 | MH, M18 | XP_650466 XM_645374 | 431 | H81D82D228E262E263D311H401 | SP: -, TM: - | aspartyl aminopeptidase |
| 80 | EhMP20-1 | MH, M20, B | XP_656428 XM_651336 | 379 | H75D133E163E164D186H353 | SP: -, TM: - | peptidase T |
| 81 | EhMP20-2 | MH, M20, B | XP_650152 XM_645060 | 401 | H79D139E172E173D195H376 | SP: -, TM: - | peptidase T |
| 82 | EhMP20-3 | MH, M20, C | XP_656545 XM_651453 XP_655596 XM_650504 XP_652163 XM_647071 | 516 | H95D97D134E165E166D192H490 | SP: -, TM: - | aminoacyl-histidine dipeptidase |
| 83 | EhMP20-4 | MH, M20, C | XP_655616 XM_650524 | 505 | H95D97D-E154E155D181H477 | SP: -, TM: - | aminoacyl-histidine dipeptidase |
| 84 | EhMP22-1 | MK, M22 | XP_652292 XM_647200 | 335 | unknown | SP: -, TM: - | glycoprotein endopeptidase |
| 85 | EhMP49-1 | M, M49 | XP_654273 XM_649181 | 645 | H412E413H417E467 | SP: -, TM: - | dipeptidyl-peptidase III |
| 86 | EhU48-1 | U, U48 | XP_656466 XM_651374 | 216 | H274E275H278E353 | SA: 4–26 + 6 × TM | CAAX prenyl protease |
Primary structure prediction of the other 3 groups of proteolytic enzymes are as follows:
Aspartic peptidases
The 4 aspartic peptidases (EhAsP22-1 to EhAsP22-4) share 30 to 40% sequence identity and are homologous to intramembrane-cleaving peptidases (clan AD, family A22). All of them have the specific active site residues TyrAsp and GlyLeuGlyAsp and contain 7 or 8 transmembrane domains but only EhAsP22-1 and EhAsP22-2 have recognizable signal peptides, whereas EhAsP22-3 contains a predicted signal anchor motif. EhAsp22-1, -2 and -3 have significant homology to signal peptide peptidases of various organisms including Trypanosoma cruzi and Arabidopsis thaliana and in addition, EhAsP22-1 and EhAsP22-3 contain the signal peptide peptidase-specific motif GlnProAlaLeuLeuTyr [24,25]. The primary structure of EhAsp22-4 revealed highest identity (35–40%) to putative presenilins of various organisms including Dictyostelium discoideum, Arabidopsis thaliana and Homo sapiens, but a signal peptide or signal anchor is absent.
Serine peptidases
Of the 10 E. histolytica genes coding for putative serine peptidases, 5 are predicted to belong to clan SC, family S9 (Figure 1, Table 1), with the active site residues Ser, Asp, His. According to the amino acid residues adjacent to the active site Ser (GGSYGG), EhSP9-1, -2, and -3 can be grouped into subfamily C. The sequences of EhSP9-1 and EhSP9-2 are identical except for a 12 amino acid insertion present in EhSP9-2. In contrast, EhSP9-3, -4 or -5 share only 20% sequence identity with EhSP9-1 or EhSP9-2. The active site residues of EhSP9-4 are not conserved and for EhSP9-5 only a partial sequence of 102 amino acid residues is available. Thus, a reliable assignment of these two enzymes to a specific S9 subfamily is not possible. Signal peptides were identified only for EhSP9-1, EhSP9-3 and EhSP9-4, respectively.
Another 3 enzymes were classified into clan SC but represent most likely members of family S28 of serine peptidases (EhSP28-1, -2, -3), which are also known as lysosomal Pro-Xaa carboxypeptidases. All 3 molecules have a predicted signal peptide and are of similar size comprising betwen 457 and 480 amino acid residues. EhSP28-1 (EhSp1) and EhSP28-2 (EhSp2) have been previously characterized [23]. Both are highly similar as they share 89% sequence identity, but only 35% to EhSP28-3.
Two other serine peptidases (EhSP26-1 and EhSP26-2) have homology to members of the signal peptidase family S26B (clan SF) containing the catalytic dyad Ser and His. EhSP26-1 has a calculated molecular mass of approximately 20 kDa and contains 2 hydrophobic regions located near the N- and C-terminus, respectively. Sequence similarity to other members of this family is approximately 45%. In contrast, EhSP26-2 shares only 20% sequence identity with members of the S26 family. Moreover, it does not contain predicted transmembrane regions and the active site is not conserved.
Metallo peptidases
A considerable number of 22 E. histolytica genes are predicted to encode putative metallo peptidases. These are relatively diverse and can be attributed to 7 different clans and 11 different families (Figure 1, Table 1). Six of the enzymes group into clan MA, with the characteristic zinc binding-motif consisting of two histidine residues encompassing the sequence HEXXH. One member is assigned to family M1 (EhMP1-1) and two others to family M3 (EhMP3-1, EhMP3-2). The latter are known as Glu-zincins with the third Zn-binding site being a glutamate residue.
Another two clan MA members (EhMP8-1, EhMP8-2) are homologous to metzincins, which are characterized by a C-terminal His residue being a third zinc-binding site. The two enzymes share 34% sequence identity and both contain a predicted C-terminal transmembrane region but only EhMP8-1 has a signal sequence.
A further clan MA member belongs to family M48 (EhMP48-1) and contains a predicted signal anchor and 6 additional transmembrane domains. The structure of EhMP48-1 is homologous to ste24, an endopeptidase from yeast. Like the yeast enzyme, the amoeba molecule contains the conserved HEXXH zinc-binding motif, located between the fourth and the fifth transmembrane domain.
Another putative metallo peptidase was assigned to clan ME, family M16C containing the characteristic zinc-binding motif HXXEH. Members of this family are falcilysin from Plasmodium falciparum, eupitrilysin from Homo sapiens and CYM1 peptidase from Saccharomyces cerevisiae.
A group of 6 enzymes (EhMP24-1 to EhMP24-6) was predicted to constitute metallo peptidases of clan MG, family 24, which usually represent cytosolic exopeptidases that require co-catalytic ions such as cobalt or manganese. Another 6 peptidases were identified, with homology to metallo peptidases of clan MH. Of these, 2 constitute most likely aspartyl aminopeptidases belonging to family M18 (EhMP18-1, EhMP18-2). They share 40% identity and approximately 35% with members of this family from other organisms. The other 4 amoeba enzymes of clan MH were attributed to family 20 (EhMP20-1 to EhMP20-4). In general, enzymes of this family hydrolyse the late products of protein degradation to complete the conversion of proteins into free amino acids.
The deduced amino acid sequence of a further amoeba gene revealed homology to clan MK, family 22 of metallo peptidases. The only enzyme belonging to this family known so far is the O-sialoglycoprotein endopeptidase from Pasteurella haemolytica. At present, the nature of the active site residues is unknown [26]. Like the amoeba homologue, the bacterial peptidase does not possess a signal peptide.
In addition, one E. histolytica enzyme was identified belonging to family M49, clan M. The mammalian homologues are cytosolic dipeptidyl peptidases, which sequentially release N-terminal dipeptides [27]. Moreover, an enzyme designated EhU48-1 were annotated, which is similar to EhMP48-1. Like EhMP48-1, it contains a signal anchor sequence and six transmembrane domains. Nevertheless, homology search grouped this peptidase into the U48 family. However, the specificities of the two families are overlapping but not identical [28,29].
Peptidase gene expression of various E. histolytica isolate under standard axenic culture conditions
To allow detailed expression analyses of the various E. histolytica peptidase genes, a small microarray was designed. This array contains 86 specific oligonucleotides representing 4 different E. histolytica houskeeping genes, 3 peptidase-inhibitor genes as well as 79 of the 86 identified peptidase genes. Genes coding for the serine peptidase EhSP9-4 or for the cysteine peptidases EhCP-A7, EhUBP, EhUCH, EhUlp-1, EhUlp-2 and EhUlp-3, respectively, were not included because it was either not possible to design a specific oligonucleotide or they were identified after the array was already spotted. In a first attempt, labelled cDNA from the widely used laboratory strain HM-1:IMSS was hybridized to the array (Figure 2). The results from multiple experiments using RNA preparations from cells grown under standard axenic culture conditions were highly reproducible and indicated that only 3 peptidase genes were expressed at high levels (mean spot intensity >8000), all of them encoding cysteine proteinases (EhCP-A1, EhCP-A2, EhCP-A5). A set of 17 peptidase genes revealed intermediate expression levels (mean spot intensity 800 to 3000). This group comprised the genes for the cysteine peptidases EhCP-A6, -A10, -A11, -B2, -C4 and EhCALP1, the aspartic peptidase EhAsP22-1, the serine peptidase EhSP9-2 and the metallo peptidases EhMP1-1, 16-1, 18-1, 20-3, 20-4, 24-1, 24-2, 24-6 and 48-1, respectively. All other peptidase genes were expressed at levels below the detection limit of Northern blots (mean spot intensity <700). The reliability of the results obtained by array hybridization was confirmed by qRT-PCR using a set of 22 pairs of primers amplyfing cDNAs of the 3 highly expressed genes as well as a representative number of the intermediate or low expressed peptidase genes (Table 2).
Figure 2.
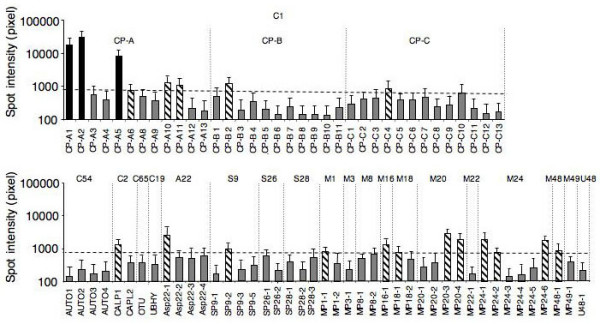
Expression of E. histolytica peptidase genes of the E. histolytica isolate HM-1:IMSS as determined by microarrays. Error bars represent the standard error of the mean of nine hybridizations (biological replicates).
Table 2.
Confirmation of microarray results via real time PCR.
| Gene accession | Gene name | Microarray data | Real time PCR data | |
| Signal intensity (pixel) | relative expression | relative expression | ||
| High expression | ||||
| actin | 33512 | 1 | 1 | |
| XM_645064 | ehcp-a1 | 17691 | 0.528 | 0.853 |
| XM_645550 | ehcp-a2 | 32474 | 0.969 | 0.853 |
| XM_645845 | ehcp-a5 | 8628 | 0.257 | 0.368 |
| Intermediate | ||||
| XM_648987 | ehasp22-1 | 2156 | 0.064 | 0.020 |
| XM_652272 | ehcp-a6 | 802 | 0.024 | 0.026 |
| XM_650130 | ehsp 9-2 | 974 | 0.029 | 0.035 |
|
XM_651453 XM_650504 XM_647071 |
ehmp20-3 | 2838 | 0.085 | 0.360 |
| XM_651374 | ehmp48-1 | 895 | 0.027 | 0.020 |
| Low | ||||
| XM_648162 | ehcp-a3 | 549 | 0.016 | 0.000 |
| XM_651510 | ehcp-a4 | 414 | 0.012 | 0.013 |
| XM_652354 | ehcp-a8 | 510 | 0.015 | 0.010 |
| XM_647728 | ehasp 22-2 | 458 | 0.014 | 0.000 |
| XM_652471 | ehasp 22-3 | 450 | 0.013 | 0.010 |
| XM_648604 | ehasp 22-4 | 516 | 0.015 | 0.000 |
| XM_651670 | ehsp28-1 | 380 | 0.011 | 0.010 |
| XM_646997 | ehsp 28-4 | 525 | 0.016 | 0.020 |
| XM_650302 | ehmp8-1 | 542 | 0.016 | 0.015 |
| XM_647540 | ehmp8-2 | 686 | 0.020 | 0.010 |
| XM_649181 | ehmp49-1 | 406 | 0.012 | 0.023 |
| XM_647901 | ehcp-b9 | 144 | 0.004 | 0.000 |
| XM_651336 | ehmp20-1 | 282 | 0.008 | 0.000 |
| XM_643899 | ehsp 28-2 | 230 | 0.007 | 0.009 |
Gene expression profile of selected peptidases in E. histolytica isolate HM-1:IMSS.
For microarray analysis nine independent experiments were performed. The signal intensity is indicated in pixel. For real time PCR analysis all primer sets were run in duplicates. Two biological replicates were investigated. The expression of actin as normalizer was set to 1. The relative expression of the peptidases were related to actin.
In order to determine the extend of inter-strain variation in the expression of peptidase genes, HM-1:IMSS was compared with 6 different E. histolytica isolates all of them cultivated under axenic conditions. These isolates originated from different parts of the world and were obtained from patients with different forms of amoebic disease or in at least one case from an asymptomatic E. histolytica carrier. Pairwise comparison of the various isolates with HM-1:IMSS revealed only minor differences in the expression of the various peptidase genes (Table 3). Three isolates including the one from an asymptomatic carrier showed no differences and two isolates differed only in the expression of one gene. In isolate HK-9, expression of the of the gene for cysteine peptidase EhCP-A5 was decreased by 2.3 fold and in isolate DRP expression of the the gene for the metallo peptidase EhM48-1 was increased by 5.2 fold. The only exception was isolate EGG, which revealed differences in expression for 4 peptidase genes. This isolate was obtained from a patient who simultaneously developed amoebic colitis and liver abscess. Compared to HM-1:IMSS isolate EEG showed decreased expression of the genes for the cysteine peptidase EhCP-A1 as well as for the metallo peptidase EhMP20-3 by about 2 fold, and an increase in the expression of the genes for serine peptidase EhSP9-2 and for the metallo peptidase EhMP20-1 by about 2.8 and 8.6 fold, respectively.
Table 3.
Genes differentially expressed in various E. histolytica isolates in comparison to the E. histolytica isolate HM-1:IMSS.
| Gene accession | Gene name | Change of signal intensity HM-1:IMSS/strain of interest (pixel) | Fold change | p-value |
| Expression of HM-1:IMSS/NIH:200 | ||||
| No | ||||
| Expression of HM-1:IMSS/HK-9 | ||||
| XM_645845 | EhCP-A5 | 5.860 → 2.500 | 2.3 × decrease | 0.0001 |
| Expression of HM-1:IMSS/EGG | ||||
| XM_651336 | EhMP20-1 | 570 → 4.900 | 8.6 × increase | 0.0004 |
| XM_650130 | EhSP9-2 | 1.700 → 4.700 | 2.8 × increase | < 0.0001 |
| XM_645064 | EhCP-A1 | 7.300 → 3.700 | 2.0 × decrease | 0.0006 |
| XM_651453 | EhMP20-3 | 1.600 → 700 | 2.3 × decrease | 0.0001 |
| Expression of HM-1:IMSS/DRP | ||||
| XM_643678 | EhM48-1 | 850 → 4.400 | 5.2 × increase | < 0.0001 |
| Expression of HM-1:IMSS/452 | ||||
| No | ||||
| Expression of HM-1:IMSS/32 | ||||
| No | ||||
The gene accession number, gene name, fold change and p-value are shown. All transcripts are statistically significant, having a p-value < 0.001.
Peptidase gene expression in response to heat stress
Previous studies have suggested that the level of expression of a number of cysteine peptidase genes is sensitive to heat shock [30,31]. To further characterize the influence of heat stress on the expression pattern of the various E. histolytica peptidase genes, amoeba were cultured at 42°C for 4 hours and compared with amoebae cultivated under standard culture conditions at 36°C. The results indicated that only 5 of the 79 peptidase genes investigated were differentially expressed upon heat shock. The amount of RNA for the highly expressed genes ehcp-a1 and ehcp-a2 was found to be decreased by about 6 and 4 fold, respectively, whereas the expression of ehcp-a5, ehcp-a6 or ehmp8-2 was increased by approximately 2 fold (Table 4). Similar results were obtained by qRT-PCR. However, there were no significant differences in expression for the remaining 74 peptidase genes.
Table 4.
Genes differentially expressed in E. histolytica HM-1:IMSS isolate under heat shock.
| Gene accession | Gene name | Change of signal intensity 36°C → 42°C (pixel) | Fold change | p-value |
| Expression of HM-1:IMSS 36°C/HM-1:IMSS 42°C | ||||
| XM_645845 | EhCP-A5 | 14.000 → 32.000 | 2.3 × increase | 0.0001 |
| XM_652272 | EhCP-A6 | 1.050 → 2.200 | 2.1 × increase | 0.0001 |
| XM_647540 | EhMP8-2 | 680 → 1.500 | 2.2 × increase | < 0.0001 |
| XM_645064 | EhCP-A1 | 25.000 → 4.000 | 6.3 × decrease | < 0.0001 |
| XM_645550 | EhCP-A2 | 45.000 → 11.000 | 4.1 × decrease | < 0.0001 |
Only those genes are listed that show a differential expression pattern during heat shock. The gene accession number, gene name, signal intensity, fold change and p-value are shown. All transcripts are statistically significant, having a p-value < 0.001.
Discussion
In an attempt to annotate all E. histolytica peptidase genes, a total of 86 putative or known proteolytic enzymes were identified within the E. histolytica genome. Such a great number of peptidase genes is not unusual for protozoans. So far, 110 annotated peptidases were found for Plasmodium falciparum and 70 for Giardia lamblia. Entamoeba, Plasmodium and Giardia contain aspartic peptidases of the A22 family. In addition, P. falciparum contains genes belonging to the A1 family known as plasmepsins. Of the various cysteine peptidases, the autophagin-like as well as the OTU-like enzymes are only present in E. histolytica. On the other hand, several cysteine peptidase families found in P. falciparum, such as C2, C12, C13, C14, C44 and C56 have no counterpart in Entamoeba. Regarding the serine and metallo peptidases no striking differences between the families of Entamoeba, Plasmodium and Giardia became obvious, except 10 additional peptidase families, that are peculiar for P. falciparum. The leishmanolysin-like peptidases of the M8 family are specific for E. histolytica and absent in P. falciparum and G. lamblia, respectively.
Since only a fraction of the 86 putative amoeba peptidases have been biochemically and functionally characterized so far, the function and localization of most of the molecules can only be predicted from the deduced primary structure.
All four aspartic peptidases identified within the Entamoeba histolytica genome may belong to intramembrane-cleaving proteases, which usually perform downstream functions such as cell signalling, regulation and intercellular communications [32]. So far, three families of peptidases are known to promote intramembrane cleavage. These are metallo peptidases represented by the human site-2 protease [33], serine peptidases represented by Drosophila melanogaster rhomboid-1 [34], and aspartic proteases including human presenilins [35], as well as signal peptide peptidases [24]. Within the Entamoeba genome, homologous to only the presenilins and signal peptide peptidases have been found.
One of the amoebic aspartic peptidases (EhAsp22-4) shows highest identity to presenilins. So far, the physiological function of presenilins is not fully understood. Presenilin is one of the subunits that form a multiprotein complex called gamma-secretase [36]. Homologues are found in various organisms of different origin such as Caenorhabditis elegans, Drosophila melanogaster, and even plants [37,38]. It has been shown that mutations within this protein are associated with Alzheimer's disease [36]. However, homologues to other subunits of this complex, such as nicastrin have not been identified within the Entamoeba genome. EhAsp22-4 contains seven putative transmembrane domains. Interestingly, the active site residues were found within one predicted outside loop of the protein. This is different to the other known presenilins [39].
The 11 identified serine peptidases can be grouped into 3 families. The two amoeba serine peptidases characterized so far, belong to family S28, previously designated EhSP1 and EhSP2 and now renamed EhSP28-1 and EhSP28-2 [23]. Biochemical analysis revealed that these peptidases prefer the substrate Suc-AAF-AMC. This enzymatic feature is identical to that of the E. histolytica tripeptidyl serine peptidase purified by Flockenhaus and colleagues [22]. Unfortunately, no sequence of the purified tripeptidyl peptidase is available. In addition to these two described serine peptidases, one more serine peptidase gene belonging to family S28 has been identified. Microarray analysis indicated that EhSP28-1, EhSP28-2 and EhSP28-3 are not expressed or expressed at a very low level, which is in contrast to the results of Barrios-Ceballos and colleagues [23]. They postulated that the identified peptidase activity corresponds to EhSP28-2 and that this protein is associated with the trophozoite membrane. Using bioinformatic tools, no hydrophobic stretches or transmembrane domains could be deduced within EhSP28-2. Due to these controversial results, the amoeba serine peptidases require further investigation.
The function of the two S26 family serine peptidases identified within E. histolytica is unknown. Peptidases of this family are usually membrane proteins and their function is the processing of newly synthesised secretory proteins. They remove the hydrophobic, N-terminal signal peptides as the proteins are translocated across membranes [40].
Four genes belong to the S9 family (homologous to dipeptidyl-peptidases). One peptidase (SP9-2) shows the highest expression of all serine peptidases analysed in this study. However, the function of these enzymes in E. histolytica remains to be determined.
It is likely that some of the E. histolytica serine peptidases might play a role during the encystation process is it was shown for E. invadens, the in vitro model organism for en- and excystation [41]. Unfortunately, the serine peptidases involved in the E. invadens encystations processes have not been identified so far.
A total of 22 genes were identified encoding metallo peptidases, which are predicted to belong to 11 different families. The members of two of the identified metallo peptidase families contain transmembrane domains. These are the leishmanolysin-like peptidases and the CAAX prenyl peptidases. EhMP8-1 and EhMP8-2 are homologous to leishmanolysin found in kinetoplastids. Leishmanolysin occurs mainly as a heavily-glycosylated protein that is attached to the outer membrane of Leishmania promastigates by a glycosylphosphatidylinositol anchor. It has been demonstrated that leishmanolysin plays a role in resistance of promastigotes to complement-mediated lysis and in receptor-mediated uptake of the parasite by phagocytic host cells [42]. There are other eukaryotes, including Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens that have homologues of this protein. Nevertheless, highest degree of sequence similarity to the classical leishmanolysin is found for the enzymes of E. histolytica and Dictyostelium discoideum. However, the proteins of these two organisms have not been characterised so far.
Interestingly, under standard axenic culture conditions only a relatively small number of peptidase genes is significantly expressed. The results are in agreement with a recent study by Ehrenkaufer et al., in which the expression pattern of 38 of the 50 different cysteine peptidase genes were analysed in the standard laboratory E. histolytica isolate HM-1:IMSS [43]. However, in contrast to the results presented here, Ehrenkaufer et al., found differences in the expression of a considerable number of peptidase genes when recent clinical isolates were compared with strain HM-1:IMSS. The discrepancy between the two studies is most likely due to differences in the culture media used. In the study presented here, all E. histolytica isolates were grown under axenic conditions in a monophasic medium, whereas Ehrenkaufer et al. cultured their recent clinical isolates xenically using a diphasic medium and compared the results with HM-1:IMSS grown under axenic condition in a monophasic medium. As proteolytic enzymes are considered to be involved in nutrition uptake and digestion, differences in the composition of the culture medium and in particular the presence of microorganisms should considerably influence expression of petidase genes in Entamoeba.
However, questions remain about possible functions of all the different peptidases present in E. histolytica. At least some of them may be involved in encystation- or exystation processes, as described for E. invadens or for a cathepsin C like peptidase of G. lamblia, which is involved in processing of cyst-wall specific proteins [44]. Aggressive and invading Entamoeba trophozoites should be endowed with adequate mechanisms that ensure their protection against host defence strategies. In this study, the trophozoites were exposed to a temporary heat stress, which partly mimics the situation during tissue invasion. Heat stressed amoebae revealed downregulation of the genes for EhCP-A1 and EhCP-A2 and elevated expression of the genes for EhCP-A5, EhCP-A6 and EhMP8-2, respectively, which is in accordance with a recent report by Weber and colleagues [31]. As EhCP-A6 and EhMP8-2 are expressed at very low levels during in vitro cultivation, these enzymes are obviously not essential for parasite growth at least at standard culture conditions. It has been postulated that the upregulation of the gene for EhCP-A6 during heat stress is due to its potential role in the degradation of damaged proteins [31]. Recently, in few other studies, regulation of peptidase expression in response to various conditions has been described. In HM-1:IMSS clone L6, which is deficient in virulence, phagocytosis as well as cysteine peptidase activity, expression of the genes for EhCP-A1, EhCP-A2 and EhCP-A5 was significantly decreased [45]. In contrast, during intestinal colonisation expression of the genes for EhCP-A1, EhCP-A4 and EhCP-A6 was found to be increased [46]. This further highlights the importance of peptidases for E. histolytica pathogenicity.
Conclusion
Under standard culture conditions only a relatively small number of at least 86 identified peptidase genes is expressed and only very few variations become apparent between various clinical E. histolytica isolates. Nevertheless, here and in few other studies, it was shown that the peptidase expression can be regulated in response to various conditions. Therefore, further studies are necessary to understand the role of all or at least most of the peptidases in the biochemistry and especially for the virulence of E. histolytica.
Methods
E. histolytica isolates and parasite culture
Seven E. histolytica isolates were used in this study. Strain HM-1:IMSS was isolated in 1967 from a patient with amoebic dysentery, strain NIH:200 was isolated in 1949 from a patient with colitis, strain HK-9 was isolated from a patient with amoebic dysentery (year unknown), strain DRP was isolated in 1985 from a patient with an amoeboma, strain EGG was isolated in 1988 from a patient with colitis and amoebic liver abscess, strain 452 was isolated in 1983 from an asymptomatic carrier. Origin of strain 32 is unknown. HM-1:IMSS, NIH:200 and HK-9 are standard laboratory strains obtained from the American Type Culture Collection. They were original isolated in Mexico, India and Korea respectively. All other strains were isolated in Brasil and kindly provided by Prof. E. F. Silva, University of Minas Gerais, Belo Horizonte, Brasil. Different genotypes of the E. histolytica strains were confirmed by PCR-based genotyping based on variation in the numbers of short tandem repeats that are linked to E. histolytica tRNAs [47]. Trophozoites of the various isolates were cultured axenically in TYI-S-33 medium supplemented with 10% adult bovine serum [48]. Cells were harvested by chilling on ice and subsequent centrifugation at 430 × g at 4°C for 5 min. The resulting pellet was washed twice with phosphate-buffered saline (6.7 mM NaHPO4, 3.3 mM NaH2PO4, 140 mM NaCl, pH7.2). For heat shock experiments incubation temperature of cultures was shifted from 36°C to 42°C for 4 hours.
Identification of peptidase homologous of E. histolytica
Conserved domains of cysteine-, serine-, metallo-, or aspartic-peptidases were used for homology search [49] against the E. histolytica genome as provided by The Sanger Centre and The Institute of Genomic Research [50-52]. With the help of MEROPS [53] the identified enzymes were grouped into the corresponding peptidase clans and families.
Microarray design
For microarray experiments a 60-base oligonucleotide array was designed containing probes for 79 of the 86 identified putative peptidase genes. The various oligonucleotides contain similar GC-contents of 35.5% and an average Tm of 71.6°C, with a standard deviation of 1.17 (range 66–74°C). The oligonucleotides were designed and synthesized by Eurogentec. Each oligonucleotide was printed in quadruplicate on glass slides (Advalytix Epoxy AD100) in a concentration of 50 μM. The spotting procedure was done in cooperation with the University of Marburg (Genomic Solutions OmniGrid). The oligonucleotide sequences are listed [see Additional file 1].
RNA isolation, microarray hybridization, sample labelling, and visualization
Total amoeba RNA was isolated using TRIZOL reagent (InVitrogen) according to standard protocols. For microarray analysis 5 μg of total RNA was used. Two biological replicates including dye swap experiments were performed. The reverse transcription of RNA into cDNA was performed according to the Atlas Superscript Fluorescent Labeling Kit (TaKaRa) followed by indirect labelling.
The cDNA labelling was performed with the Cy3- and Cy5-monoreactive dyes (Amersham). In a typical oligoarray the control cDNA was labelled with Cy3, while the experimental second cDNA was labelled with Cy5 (and vice versa for dye swap experiment). Prehybridization and hybridization was performed using standard protocols.
Microarray data analysis
Each array was scanned at 550 nm (Cy3) and at 650 nm (Cy5) at a resolution of 5 μm. Calculation and output of the data was done using ScanArray software, version 3.0 (PerkinElmer). For calculation, the mean signal intensity (pixel) minus local background (pixel) of each spot was used. Flagged spots were eliminated. Two methods for normalizing the data were applied. i) normalization among the totality of genes and ii) normalization among housekeeping genes. For the first normalization method, the signal intensities of each spot of the experiment as well as of each spot of the control were totalised. The sum of the experiment over the sum of the control gives the normalization factor. In order to normalise the control, the signal intensity of each spot of the control was divided by this normalization factor. For housekeeping normalization, the calculation was performed in the same manner as described above but on the basis of the housekeeping gene spot intensities of the control. Spots with a signal intensity (pixel) = 300 were excluded from data analysing. Genes with a ratio of more than 2 and less than 0.5 were considered as differentially expressed.
Quantitative RT-PCR
In order to validate the results obtained by microarray analyses, quantitative RT-PCR (qRT-PCR) was performed by random sampling. Sense and antisense primers were designed to amplify approximately 100 base pairs [see Additional file 2]. These primers were designed independently from the oligonucleotides used on the microarray. Thus, they represent different regions of the same gene. cDNA synthesis was carried out with SuperScriptIII Reverse Transcriptase (InVitrogen). In a final volume of 20 μl, 1 μg of RNase-free DNase-treated total RNA was mixed with 5 × First-Strand buffer, 500 μM dNTPs, 500 nM OdT-T71 (5'-GAG AGA GGA TCC AAG TAC TAA TAC GAC TCA CTA TAG GGA GAT24), 2 mM DTT, 40 U RnaseOut (Invitrogen) and SuperScriptIII (200 U/μl). Incubation was performed for 1 h at 42°C. Quantitative amplification was performed in a Rotor-Gene (Corbett) using RealMasterMix (Eppendorf) SYBR Green kit. 1 μl of the synthesized cDNA was mixed with 2,5× RealMasterMix/20 × SYBR, 5 pmol/μl of the respective sense-primer and antisense-primer to a final volume of 20 μl. Amplification conditions were as follows: 35 cycles at 95°C for 15 s, 58°C for 20 s and 68°C for 20 s and an adjacent melting step (42°C–95°C). Two biological replicates were analysed in triplicate. Relative quantification was carried out with the use of the delta delta ct method provided by the Rotor-Gene software [54] and E. histolytica actin gene RNA as normalizer.
Authors' contributions
IB and ET conceived the study. IB coordinated the study and performed the data analysis together with MT. IB and ET drafted the manuscript. MT, LB, MAG, MF, and HI carried out the laboratory component. MT helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
List of oligonucleotides used for the microarray design. The table shows a list of all oligonucleotides present of the microarray used.
List of oligonucleotides used for RT-PCR. The table shows a list of all oligonucleotides used for RT-PCR.
Acknowledgments
Acknowledgements
This work was supported by the German Science Foundation (DFG) (BR1744/7-1), the German Accademic Exchange Service (DAAD) (D/04/40412) and the Evangelische Studienstiftung, Villigst. It includes part of the doctoral thesis of MT.
Contributor Information
Manuela Tillack, Email: tillack@bni-hamburg.de.
Laura Biller, Email: biller@bni-hamburg.de.
Henriette Irmer, Email: irmer@bni-hamburg.de.
Michelle Freitas, Email: michellerfreitas@hotmail.com.
Maria A Gomes, Email: magomes@icb.ufmg.br.
Egbert Tannich, Email: tannich@bni-hamburg.de.
Iris Bruchhaus, Email: bruchhaus@bni-hamburg.de.
References
- Gadasi H, Kessler E. Correlation of virulence and collagenolytic activity in Entamoeba histolytica. Infect Immun. 1983;39:528–531. doi: 10.1128/iai.39.2.528-531.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushbaugh WB, Hofbauer AF, Pittman FE. Entamoeba histolytica: purification of cathepsin B. Exp Parasitol. 1985;59:328–336. doi: 10.1016/0014-4894(85)90088-8. [DOI] [PubMed] [Google Scholar]
- Luaces AL, Barrett AJ. Affinity purification and biochemical characterization of histolysin, the major cysteine proteinase of Entamoeba histolytica. Biochem J. 1988;250:903–909. doi: 10.1042/bj2500903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SL, Keene WE, McKerrow JH. Thiol proteinase expression and pathogenicity of Entamoeba histolytica. J Clin Microbiol. 1989;27:2772–2777. doi: 10.1128/jcm.27.12.2772-2777.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte W, Scholze H. Action of the major protease from Entamoeba histolytica on proteins of the extracellular matrix. J Protozool. 1989;36:538–543. doi: 10.1111/j.1550-7408.1989.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Keene WE, Hidalgo ME, Orozco E, McKerrow JH. Entamoeba histolytica: correlation of the cytopathic effect of virulent trophozoites with secretion of a cysteine proteinase. Exp Parasitol. 1990;71:199–206. doi: 10.1016/0014-4894(90)90022-5. [DOI] [PubMed] [Google Scholar]
- Li E, Yang WG, Zhang T, Stanley SL., Jr Interaction of laminin with Entamoeba histolytica cysteine proteinases and its effect on amebic pathogenesis. Infect Immun. 1995;63:4150–4153. doi: 10.1128/iai.63.10.4150-4153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SL, Jr, Zhang T, Rubin D, Li E. Role of the Entamoeba histolytica cysteine proteinase in amebic liver abscess formation in severe combined immunodeficient mice. Infect Immun. 1995;63:1587–1590. doi: 10.1128/iai.63.4.1587-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankri S, Stolarsky T, Bracha R, Padilla-Vaca F, Mirelman D. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect Immun. 1999;67:421–422. doi: 10.1128/iai.67.1.421-422.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillack M, Nowak N, Lotter H, Bracha R, Mirelman D, Tannich E, Bruchhaus I. Increased expression of the major cysteine proteinases by stable episomal transfection underlines the important role of EhCP5 for the pathogenicity of Entamoeba histolytica. Mol Biochem Parasitol. 2006;149:58–64. doi: 10.1016/j.molbiopara.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang L, Seydel KB, Li E, Ankri S, Mirelman D, Stanley SL., Jr Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ICE) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol Microbiol. 2000;37:542–548. doi: 10.1046/j.1365-2958.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- Clark GC, Alsmark UCM, Hofer M, Saito-Makano Y, Aloi V, Marion S, Weber C, Mukherjee C, Bruchhaus I, Tannich E, Leippe M, Sicheritz-Ponten T, Foster PG, Samuelson J, Noel CJ, Hirt RP, Embley TM, Gilchrist CA, Mann BJ, Singh U, Ackers JP, Bhattacharya S, Bhattacharya A, Lohia A, Guillen N, Duchene M, Nozaki T, Hall N. Stucture and content of the Entamoeba histolytica genome. Advances in Parasitology. 2007. [DOI] [PubMed]
- Bruchhaus I, Loftus BJ, Hall N, Tannich E. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot Cell. 2003;2:501–509. doi: 10.1128/EC.2.3.501-509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Bhaskar V, Fernandez J, Courey AJ. Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J Biol Chem. 2004;279:43805–43814. doi: 10.1074/jbc.M404942200. [DOI] [PubMed] [Google Scholar]
- Munoz ML, Calderon J, Rojkind M. The collagenase of Entamoeba histolytica. J Exp Med. 1982;155:42–51. doi: 10.1084/jem.155.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze H, Frey S, Cejka Z, Bakker-Grunwald T. Evidence for the existence of both proteasomes and a novel high molecular weight peptidase in Entamoeba histolytica. J Biol Chem. 1996;271:6212–6216. doi: 10.1074/jbc.271.11.6212. [DOI] [PubMed] [Google Scholar]
- Padilla-Vaca F, Martinez-Gallardo N, Blanco-Labra A, Shmuely H, Mirelman D. Novel thermo-stable serine-metallo proteinase of Entamoeba histolytica. Arch Med Res. 2000;31:S221–223. doi: 10.1016/S0188-4409(00)00232-0. [DOI] [PubMed] [Google Scholar]
- Flockenhaus B, Pietersma M, Bakker-Grunwald T, Scholze H. The occurrence of a tripeptidyl peptidase I in trophozoites of Entamoeba histolytica. Arch Med Res. 2000;31:S67–68. doi: 10.1016/S0188-4409(00)00138-7. [DOI] [PubMed] [Google Scholar]
- Barrios-Ceballos MP, Martinez-Gallardo NA, Anaya-Velazquez F, Mirelman D, Padilla-Vaca F. A novel protease from Entamoeba histolytica homologous to members of the family S28 of serine proteases. Exp Parasitol. 2005;110:270–275. doi: 10.1016/j.exppara.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Hutton M, Nyborg A, Baker M, Jansen K, Golde TE. Identification of a novel family of presenilin homologues. Hum Mol Genet. 2002;11:1037–1044. doi: 10.1093/hmg/11.9.1037. [DOI] [PubMed] [Google Scholar]
- Jiang P, Mellors A. O-Sialoglycoprotein endopeptidase. In: Barrett AJ, Rawlings ND, Woessner JF, editor. Handbook of Proteolytic Enzymes. 2. Elsevier, London; 2004. pp. 977–980. [Google Scholar]
- Chen JM, Barrett AJ. Dipeptidyl-peptidase III. In: Barrett AJ, Rawlings ND, Woessner JF, editor. Handbook of Proteolytic Enzymes. 2. Elsevier, London; 2004. pp. 809–812. [Google Scholar]
- Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry. 2000;39:4096–4104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- Trueblood CE, Boyartchuk VL, Picologlou EA, Rozema D, Poulter CD, Rine J. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol Cell Biol. 2000;20:4381–92. doi: 10.1128/MCB.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane RC, Shah PH, Singh U. Transcriptional profiling of Entamoeba histolytica trophozoites. Int J Parasitol. 2005;35:533–542. doi: 10.1016/j.ijpara.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, Gouyette C, Mirelman D, Coppee JY, Guillen N. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2006;5:871–875. doi: 10.1128/EC.5.5.871-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann E, Lemberg MK, Weihofen A, Dev KK, Dengler U, Rovelli G, Martoglio B. Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J Biol Chem. 2004;279:50790–5078. doi: 10.1074/jbc.M407898200. [DOI] [PubMed] [Google Scholar]
- Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/S1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/S0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Zhang YW, Xu H, Thinakaran G. Pathological and physiological functions of presenilins. Mol Neurodegener. 2006;1:4. doi: 10.1186/1750-1326-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smialowska A, Baumeister R. Presenilin function in Caenorhabditis elegans. Neurodegener Dis. 2006;3:227–232. doi: 10.1159/000095260. [DOI] [PubMed] [Google Scholar]
- Mahoney MB, Parks AL, Ruddy DA, Tiong SY, Esengil H, Phan AC, Philandrinos P, Winter CG, Chatterjee R, Huppert K, Fisher WW, L'Archeveque L, Mapa FA, Woo W, Ellis MC, Curtis D. Presenilin-based genetic screens in Drosophila melanogaster identify novel notch pathway modifiers. Genetics. 2006;172:2309–2324. doi: 10.1534/genetics.104.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Nyborg AC, Iwata N, Diehl TS, Saido TC, Golde TE, Wolfe MS. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- Carlos JL, Paetzel M, Brubaker G, Karla A, Ashwell CM, Lively MO, Cao G, Bullinger P, Dalbey RE. The role of the membrane-spanning domain of type I signal peptidases in substrate cleavage site selection. J Biol Chem. 2000;275:38813–3822. doi: 10.1074/jbc.M007093200. [DOI] [PubMed] [Google Scholar]
- Riahi Y, Ankri S. Involvement of serine proteinases during encystation of Entamoeba invadens. Arch Med Res. 2000;31:S187–189. doi: 10.1016/S0188-4409(00)00220-4. [DOI] [PubMed] [Google Scholar]
- Yao C, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) of Leishmania sp. Biosynthesis, regulation of expression, and function. Mol Biochem Parasitol. 2003;132:1–16. doi: 10.1016/S0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechnaisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007 doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- Touz MC, Nores MJ, Slavin I, Carmona C, Conrad JT, Mowatt MR, Nash TE, Coronel CE, Lujan HD. The activity of a developmentally regulated cysteine proteinase is required for cyst wall formation in the primitive eukaryote Giardia lamblia. J Biol Chem. 2002;277:8474–8481. doi: 10.1074/jbc.M110250200. [DOI] [PubMed] [Google Scholar]
- Hirata KK, Que X, Melendez-Lopez SG, Debnath A, Myers S, Herdman DS, Orozco E, Bhattacharya A, McKerrow JH, Reed SL. A phagocytosis mutant of Entamoeba histolytica is less virulent due to deficient proteinase expression and release. Exp Parasitol. 2006;115:192–199. doi: 10.1016/j.exppara.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, Hamano S, Ehrenkaufer G, Okada M, Singh U, Nozaki T, Mann BJ, Petri WA., Jr Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol Biochem Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Ali IK, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol. 2005;43:5843–5847. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LS, Harlow DR, Cunnick CC. A new medium for axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-X. [DOI] [PubMed] [Google Scholar]
- Blast http://www.ncbi.nlm.nih.gov/BLAST/
- Welcome Trust Sanger Institute, Entamoeba Whole Genome Shotgun http://www.sanger.ac.uk/Projects/E_histolytica/
- Entamoeba histolytica GeneDB http://www.genedb.org/genedb/ehistolytica
- TIGR Database, Entamoeba histolytica Genome Project http://www.tigr.org/tdb/e2k1/eha1/
- MEROPS, the peptide database http://merops.sanger.ac.uk/
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of oligonucleotides used for the microarray design. The table shows a list of all oligonucleotides present of the microarray used.
List of oligonucleotides used for RT-PCR. The table shows a list of all oligonucleotides used for RT-PCR.


