Abstract
Cucurbita moschata, a cucurbit species responsive to inductive short-day (SD) photoperiods, and Zucchini yellow mosaic virus (ZYMV) were used to test whether long-distance movement of FLOWERING LOCUS T (FT) mRNA or FT is required for floral induction. Ectopic expression of FT by ZYMV was highly effective in mediating floral induction of long-day (LD)–treated plants. Moreover, the infection zone of ZYMV was far removed from floral meristems, suggesting that FT transcripts do not function as the florigenic signal in this system. Heterografting demonstrated efficient transmission of a florigenic signal from flowering Cucurbita maxima stocks to LD-grown C. moschata scions. Real-time RT-PCR performed on phloem sap collected from C. maxima stocks detected no FT transcripts, whereas mass spectrometry of phloem sap proteins revealed the presence of Cm-FTL1 and Cm-FTL2. Importantly, studies on LD- and SD-treated C. moschata plants established that Cmo-FTL1 and Cmo-FTL2 are regulated by photoperiod at the level of movement into the phloem and not by transcription. Finally, mass spectrometry of florally induced heterografted C. moschata scions revealed that C. maxima FT, but not FT mRNA, crossed the graft union in the phloem translocation stream. Collectively, these studies are consistent with FT functioning as a component of the florigenic signaling system in the cucurbits.
INTRODUCTION
Leaf-to-apex communication initiates flowering in response to environmental cues such as photoperiod. Grafting, phloem-girdling, timed removal of leaves, and fractional and photoperiodic induction experiments have been used to show that photoperiod is perceived in the leaves and a phloem-mobile floral stimulus (florigen), or inhibitor, is transported through the phloem translocation stream to the shoot apical meristem (Chailakhyan, 1936; Zeevaart, 1962, 1976; Lang, 1965, 1977; Bernier, 1988; Colasanti and Sundaresan, 2000; Bernier and Périlleux, 2005).
The molecular genetics of photoperiodic floral induction are best characterized in Arabidopsis thaliana (Mouradov et al., 2002; Bastow and Dean, 2003; Searle and Coupland, 2004; Corbesier and Coupland, 2005). Key elements involved in integrating the input signal(s) from the photoperiodic pathway are the zinc-finger protein CONSTANS (CO) (Putterill et al., 1995) and the RAF kinase inhibitor–like protein FLOWERING LOCUS T (FT) (Kardailsky et al., 1999; Kobayashi et al., 1999). Promoter β-glucuronidase analyses have revealed vascular- rather than meristem-specific expression patterns for these genes, findings consistent with long-distance signaling roles for both CO and FT in floral induction (Takada and Goto, 2003; An et al., 2004).
A signaling role for CO was confirmed using tissue-specific promoters in which it was demonstrated that its expression, within companion cells located in source leaves, promoted flowering in the co mutant background (An et al., 2004). Grafting studies confirmed that CO controls the production of a phloem-borne substance (presumably florigen) essential for floral induction. Assignment of CO, per se, as florigen was discounted as companion cell-specific expression of a CO:green fluorescent protein (GFP) construct did not reveal a capacity for CO-GFP to traffic beyond the cells in which it was transcribed (An et al., 2004; Ayre and Turgeon, 2004). However, the CO-GFP fusion protein was still able to rescue flowering in the co mutant background.
A role for FT and/or FT mRNA as the phloem-mobile florigenic signaling agent(s) was tested using a range of tissue-specific promoters. In contrast with CO, FT promoted flowering not only when it was expressed in source companion cells, but also in numerous other tissues, including those of the meristem, a tissue in which it is not normally detected by in situ hybridization (An et al., 2004). Given that FT expression is controlled by CO (Samach et al., 2000) and is required to promote flowering (Kardailsky et al., 1999; Kobayashi et al., 1999), these results strongly implicated FT as a component of the florigenic signaling system.
As the phloem sap has been demonstrated to contain a range of mRNA species and RNA binding proteins (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Yoo et al., 2004; Gomez et al., 2005), some of which appear to influence events taking place in the meristem (Kim et al., 2001; Haywood et al., 2005), the possibility exists that FT mRNA could act as the signaling molecule. Lifschitz et al. (2006) conducted a series of heterografting studies to test this possibility. Although expression of SINGLE FLOWER TRUSS (SFT), a tomato (Solanum lycopersicum) FT homolog, in a clade orthologous to the FT/TWIN SISTER OF FT (TSF) clade, in the tomato stock could rescue flowering in the mutant scion, analysis of RNA extracted from scion tissues failed to detect SFT mRNA in grafts in which floral induction had occurred. These results did not support a role for the long-distance trafficking of FT mRNA in floral induction.
Two recent studies conducted on Arabidopsis and rice (Oryza sativa) examined whether FT protein and the rice ortholog Hd3a participate in long-distance signaling to induce flowering (Corbesier et al., 2007; Tamaki et al., 2007). Here, transgenic plant lines expressing FT-GFP and Hd3a-GFP in vascular tissues were used to test whether FT and Hd3a move in the phloem to the shoot apical meristem (SAM) to induce flowering. Evidence consistent with such long-distance movement was presented, but neither study provided definitive proof against a role for FT mRNA.
In this study, we used a combination of Cucurbita moschata, a cucurbit (squash) species responsive to inductive short-day (SD) photoperiods, and a potyvirus vector, Zucchini yellow mosaic virus (ZYMV), to test whether long-distance movement of FT mRNA and/or FT is required for floral induction. The choice of a potyvirus was important, as these plant viruses are polycistronic, encoding for a polyprotein and, hence, do not produce subgenomic RNA species. Ectopic expression of FT by ZYMV was highly effective in mediating floral induction of long-day (LD)–treated C. moschata plants. Analysis of such induced plants revealed that the infection zone of ZYMV was not coincident with the floral meristems, indicating that FT transcripts are unlikely to function as the florigenic signal in this system. Heterografting studies demonstrated the efficient transmission of a florigenic signal from flowering pumpkin (Cucurbita maxima) stocks to LD-grown C. moschata scions. Real-time RT-PCR performed on phloem sap collected from these flowering C. maxima stocks failed to detect the presence of FT transcripts, whereas mass spectrometry analysis revealed the presence of FT protein in the translocation stream. Importantly, parallel mass spectrometry analysis of the phloem sap from florally induced C. moschata plants and scions also detected the presence of FT proteins. These studies are consistent with FT functioning as the florigenic signal in photoperiodic induction of flowering.
RESULTS
Screening Cucurbits for Photoperiodically Controlled Flowering
Ideally, to test for the presence of FT mRNA and/or FT protein in the phloem translocation stream, a photoperiodically inducible model plant system should be employed in which it is possible to gain direct access to the phloem sap. Given that the cucurbits have been widely used to investigate the phloem translocation stream (Golecki et al., 1998, 1999; Lough and Lucas, 2006), we performed a photoperiodic screen of 97 cucurbit accessions, obtained from Plant Genetic Resources Conservation Unit (www.ars-grin.gov/ars/SoAtlantic/Griffin/pgrcu). One accession, C. moschata PI441726, representing an undomesticated squash species, remained vegetative under LD conditions but flowered when grown under SD growth conditions. Seed collected from C. moschata PI441726 was rescreened and, as shown in Figure 1A, floral induction was confirmed under SD conditions; plants grown under LD conditions, by use of daylength extension, remained vegetative and failed to develop floral buds (Figure 1B). Seed stock obtained from this second screening was used for subsequent viral expression and C. maxima (stock):C. moschata PI441726 (scion) heterografting studies.
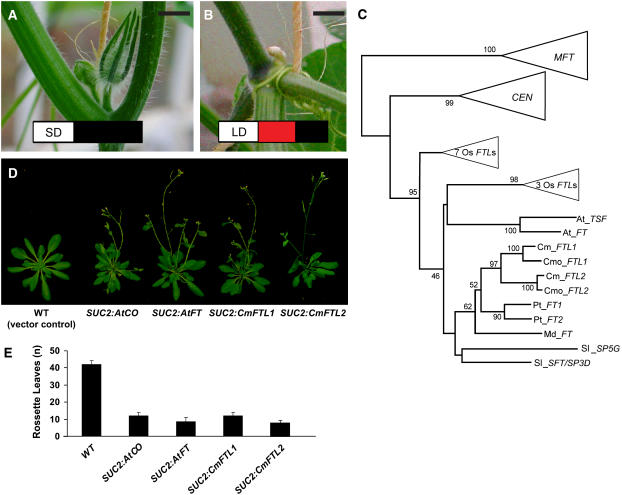
Figure 1.
Analysis of Cucurbit FT-Like Orthologs.
(A) and (B) Flowering in C. moschata PI441726 was induced when plants were grown under SD (A) but repressed under LD (B) conditions. White box represents daylight, black box represents darkness, and red box represents daylength extension interrupting the dark period. Bars = 1 cm.
(C) Phylogenetic analysis of the relationship between Cmo-FTL1/FTL2, Cm-FTL1/ FTL2, and the FT-TFL family in angiosperms. The tree was constructed based on a nucleic acid alignment of ∼500 protein coding nucleotides using the neighbor-joining method implemented in MEGA 3.1. Numbers at each branch point are the bootstrap values for percentages of 1000 replicate trees. The portions of the tree corresponding to clusters that include Arabidopsis MOTHER OF FT (MFT), Antirrhinum majus CENTRORADIALIS (CEN), and two clusters of rice genes are compressed and can be viewed in Supplemental Figure 1 online (see Supplemental Figures 2 and 3 online for additional details). The CEN cluster includes Arabidopsis FT homologs TFL1, BFT, and ATC. Gene accession numbers are in Supplemental Table 1 online. Abbreviations for species are as follows: Arabidopsis thaliana (At), Cucurbita maxima (Cm), Cucurbita moschata (Cmo), Solanum lycopersicum (Sl), Malus × domestica (Md), Oryza sativa (Os), and Populus trichocarpa (Pt).
(D) The SUC2 promoter was employed to drive companion cell–specific expression of CO, FT, Cm-FTL1, and Cm-FTL2 in transgenic Arabidopsis plants grown under noninductive SD conditions.
(E) Timing of floral initiation in transgenic Arabidopsis plants was determined by counting the number of rosette leaves formed at the time of bolting (mean ± se, n = 20 plants per treatment).
Cucurbit FT Homologs Activate Flowering in Arabidopsis
The basic tenet of the long-distance florigenic signal is that in some form it will be common to all plant species (Chailakhyan, 1937; Lifschitz et al., 2006). This would suggest that cucurbit FT homologs would function in Arabidopsis and vice versa. To test this principle, we first employed C. moschata PI441726 (hereafter referred to as simply C. moschata) and C. maxima vascular strip cDNA libraries to isolate related cDNA clones that were being expressed in flowering plants. Two full-length clones were obtained for each species and were named C. moschata FLOWERING LOCUS T-LIKE1 (Cmo-FTL1) and Cmo-FTL2, and C. maxima FLOWERING LOCUS T-LIKE1 (Cm-FTL1) and Cm-FTL2. The Arabidopsis FT, Cm-FTL1, Cm-FTL2, Cmo-FTL1, and Cmo-FTL2 conceptual amino acid sequences (see Supplemental Figure 1 online) all exhibited an extremely high level of conservation.
A molecular phylogenetic analysis of the FT family of proteins was performed, and the results demonstrate that Cmo-FTL1/Cm-FTL1 and Cmo-FTL2/Cm-FTL2 form clades that are orthologous to a clade formed by At-FT and At-TSF (Figure 1C) (i.e., there is no one-to-one orthologous relationship between the C. moschata and C. maxima genes and either Arabidopsis gene). Rather, there exists a two-for-two relationship in which the cucurbit FTL1/FTL2 clade is orthologous to the Arabidopsis FT/TSF clade. In other words, we infer that there was an FT homolog in the most recent common ancestor of cucurbits and Arabidopsis inherited by each lineage and then was duplicated independently in the lineages leading to both the cucurbits and Arabidopsis. Thus, subsequent to duplication, each gene pair had the potential for subfunctionalization and/or neofunctionalization; if either occurred in either lineage, an exact functionally equivalent relationship would not exist between either cucurbit gene and FT (or TSF). FT and TSF are known to be partially redundant, but although they differ functionally, their differences may be limited to transcriptional patterns in response to other flowering regulators, and there may be no difference in function of their mRNA or protein products (Michaels et al., 2005; Yamaguchi et al., 2005).
A functional analysis was next performed, using the companion cell-specific Arabidopsis SUC2 promoter (Truernit et al., 1996), to determine whether Cm-FTL1 and Cm-FTL2 were able to induce early flowering in Arabidopsis. In contrast with the vector control, transgenic Arabidopsis plants expressing At-SUC2pro:Cm-FTL1 or At-SUC2pro:Cm-FTL2 exhibited early flowering phenotypes (Figure 1D). The timing of floral induction was determined by counting the number of rosette leaves at the time of bolting. These studies established that the Cm-FTL1 and Cm-FTL2 gene products accelerated floral initiation in a manner equivalent to that observed in Arabidopsis lines expressing CO or FT under the SUC2 promoter (Figure 1E).
Viral Vector-Mediated Induction of Flowering in LD-Grown C. moschata
A cucurbit-infecting potyvirus, ZYMV, was used as a vector (Hsu et al., 2004) to ectopically express At-FT in C. moschata plants (Figures 2A and 2B). ZYMV was employed because it encodes for a polyprotein (Figure 2A) and does not produce subgenomic RNAs; thus, only FT protein would be free to move ahead of the viral infection front. In addition, the ZYMV infectious clone used was earlier mutated to produce an attenuated strain that caused only very mild symptoms (Hsu et al., 2004). Plants of C. moschata were grown under noninductive LD conditions using a 10-h photoperiod and a 14-h night period that was interrupted by 15-min light breaks every hour. Microprojectile bombardment was used to initiate infection with either the ZYMV (empty vector), or ZYMV clones carrying GFP (ZYMV-GFP; control) or At-FT (ZYMV-At-FT), and plants were then observed for the vegetative-to-floral transition. Infection by ZYMV-At-FT led to floral induction, whereas ZYMV-GFP or ZYMV (empty vector) treated plants remained vegetative under these LD noninductive conditions (Figures 2C to 2E).
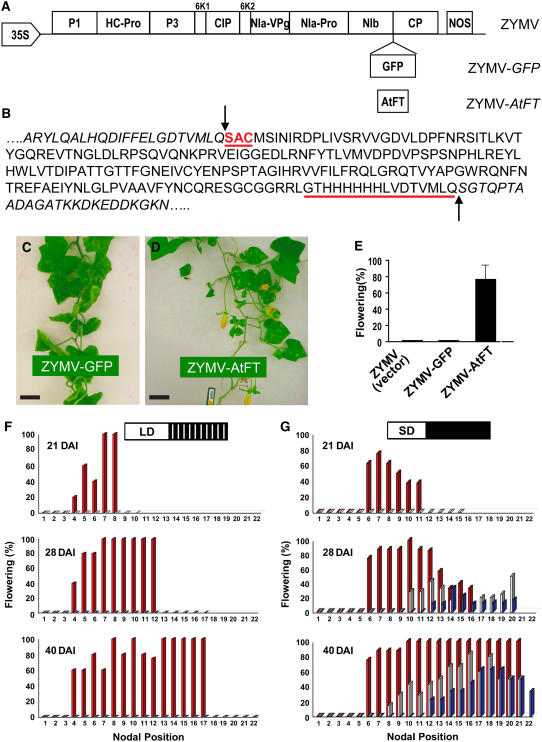
Figure 2.
ZYMV-Mediated Expression of At-FT Induces Flowering in C. moschata PI441726 Grown under LD Noninductive Conditions.
(A) Genome arrangement of the ZYMV vector indicating the sites of insertion of GFP and At-FT.
(B) ZYMV-At-FT conceptual peptide sequence. Residues fused to the N and C terminus of At-FT are underlined, and arrows indicate the positions of autocatalytic NIa-mediated cleavage.
(C) ZYMV-mediated expression of GFP did not induce flowering in LD-grown C. moschata plants. Bar = 10 cm.
(D) ZYMV-mediated expression of At-FT–induced flowering in LD-grown C. moschata plants. Bar = 10 cm.
(E) Flowering rate was scored as the percentage of plants in each treatment having floral buds/flowers at 40 DAI (mean ± se, n = 30 plants per treatment).
(F) C. moschata plants, maintained under noninductive LD conditions, were inoculated with ZYMV-At-FT (red bars), ZYMV-GFP (gray bars), or mock treatment (blue bars). Bud formation was first observed at 21 DAI on ZYMV-At-FT plants and occurred from the 4th and all subsequent nodes. Mock-inoculated and ZYMV-GFP plants did not undergo floral initiation under these noninductive LD conditions.
(G) C. moschata plants, maintained under inductive SD conditions, were inoculated with ZYMV-At-FT (red bars), ZYMV-GFP (gray bars), or mock treatment (blue bars). Note that mock-inoculated and ZYMV-GFP plants were induced to flower ∼1 week later and four to six nodes higher on the plant than ZYMV-At-FT plants.
Data in (F) and (G) are expressed as percentage of plants (n = 5 to 9) forming stage 2 floral buds as a function of nodal position and DAI. White box represents daylight, black box represents darkness, and white lines represent hourly night breaks of 15 min with incandescent (white) light.
Time-course experiments for floral induction, mediated by ZYMV-At-FT or an inductive SD photoperiod, were next performed. As illustrated in Figures 2F and 2G, by 21 d after inoculation (DAI), early bud formation was detected on ZYMV-At-FT–inoculated C. moschata plants grown under either LD or SD conditions. Upon floral induction, bud formation was observed at all subsequent nodes. Importantly, under LD conditions, plants inoculated with ZYMV (vector-only) or ZYMV-GFP did not form buds at 28 or 40 DAI (Figure 2F). However, under SD conditions, these control plants underwent the vegetative-to-floral transition 1 week and four to six nodes later than the ZYMV-At-FT–inoculated C. moschata plants (Figure 2G). Finally, the percentage of plants exhibiting floral initiation, at each node, was always highest in the ZYMV-At-FT–inoculated plants.
ZYMV Infection Domains Do Not Include Meristems
The observed ZYMV-At-FT–mediated floral induction may have resulted from cell autonomous activation in the SAM and/or lateral meristems. To test for this possibility, a series of experiments was next performed to establish the physical relationship between the ZYMV infection front and the SAM/lateral meristems on the C. moschata plants. The infection pathway was first defined in ZYMV-GFP–infected plants grown under LD conditions. Analysis of GFP fluorescence in freehand sections, taken 30 DAI, indicated that the virus was present in all tissues except for the last four nodes located at the shoot apex. A fluorescent signal was not detected in the C. moschata apical or lateral meristems.
Immunolocalization studies were next performed using polyclonal antibodies directed against the ZYMV coat protein (CP). As shown in Figures 3A and 3B, CP accumulation, associated with ZYMV-GFP and ZYMV-At-FT infection, could be detected in young developing sink leaves located at the fifth or sixth node back from the apex. Note that uninfected control plants were devoid of CP signal (Figure 3C). An examination of longitudinal sections, derived from the apices of ZYMV-GFP (Figure 3D) and ZYMV-At-FT–infected plants (70 sections examined from 10 apices per treatment; Figure 3E), confirmed that the virus had not entered into these meristematic tissues. Uninfected control plants were similarly devoid of CP staining (Figure 3F). Analysis of lateral meristems of ZYMV-At-FT and ZYMV-GFP (Figure 3G) infected C. moschata plants identified the presence of virus in developing leaves, but signal was again absent from the apical tissues. Collectively, these studies are consistent with the hypothesis that ZYMV-At-FT infection induces the floral transition in the C. moschata meristem by means of non-cell-autonomous signaling through the phloem. As the FT sequence is present only in the ZYMV infectious RNA, our inability to detect virus in the apex and lateral meristems suggested that FT mRNA, per se, may not be directly involved in floral induction. However, based on these experiments alone, we cannot exclude the possibility that a very low level of ZYMV went undetected and that this was the causal agent for floral induction.
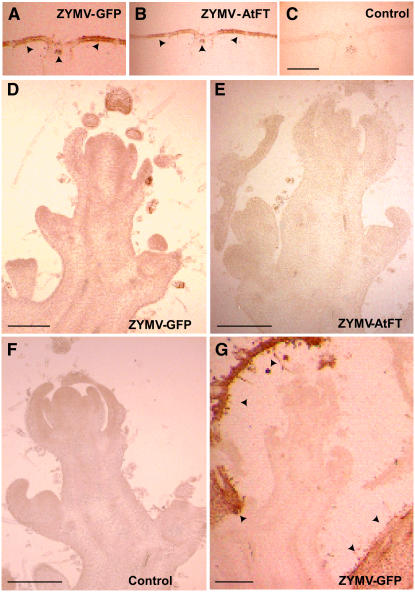
Figure 3.
ZYMV Infection Domains Do Not Include Meristems.
Virus accumulation and localization were determined by immunohistochemical staining of sections using antisera specific for the ZYMV coat protein. Tissues were collected 35 to 40 DAI, and images are representative of those observed from six plants (>30 sections per plant) per treatment. Bars = 500 μm.
(A) to (C) Transverse sections of lamina from young expanding leaves located at the ninth node of plants having 12 to 14 visible nodes. Plants were inoculated with ZYMV-GFP (A), ZYMV-At-FT (B), or mock inoculated (control) (C). Note the presence of CP (dark brown stain; see arrowheads) in plants inoculated with ZYMV-GFP or ZYMV-At-FT.
(D) to (F) Longitudinal sections from apical meristems of the main shoot of ZYMV-GFP (D), ZYMV-At-FT (E), and mock-inoculated (F) plants.
(G) Longitudinal section of an apical meristem from a lateral shoot of a ZYMV-GFP–infected plant. Note the presence of CP in the developing leaves (arrowheads).
Heterograft Experiments Confirm Phloem Delivery of Florigenic Signal
The nature of florigen as a phloem-mobile signal was earlier established using grafting experiments performed with florally induced stocks and noninduced scions (Chailakhyan, 1937; Zeevaart, 1976; Lang, 1977). As C. maxima (a day-neutral species) flowers under LD conditions, whereas C. moschata remains vegetative under this same growth regime, heterograft experiments were next preformed using these two plant species. As illustrated in Table 1, in the control experiment in which C. moschata scions were grafted onto C. moschata stocks, the scions remained vegetative when grown under LD noninductive conditions. By contrast, C. moschata scions grafted onto flowering C. maxima stocks were all induced to produce floral buds. Hence, the phloem sap of flowering C. maxima carries a compatible florigenic signal that mediates the vegetative-to-floral transition in C. moschata.
Table 1.
Graft Transmission of Florigenic Signal from C. maxima Induces Flowering in C. moschata PI441726 Scions
| Stock:Sciona | C. moschata PI441726:C. moschata PI441726 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nodeb | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Floral Budsc | 0 (6) | 0 (6) | 0 (6) | 1 (6) | 1 (6) | 1 (6) | 1 (6) | 0 (6) | 0 (6) | 0 (6) |
| Stock:Scion
|
C. maxima:C. moschata PI441726
|
|||||||||
| Node | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Floral Buds | 1 (6) | 2 (6) | 2 (6) | 3 (6) | 6 (6) | 6 (6) | 6 (6) | 6 (6) | 5 (6) | 5 (6) |
All grafted plants (six per treatment) were grown under noninducing LD conditions.
Nodes 1 and 2 were present at time of grafting; nodes 3 to 10 developed after grafting.
Floral buds/flowers were scored as a function of nodal position using the same criteria used for Figures 2F and 2G.
FT mRNA Was Not Detected in Phloem Sap of Florally Induced C. maxima Plants
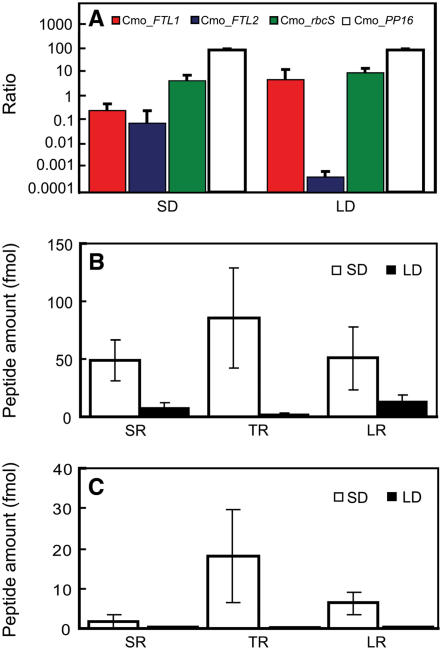
To further investigate the role of FT mRNA and/or FT protein as a component of the florigenic long-distance signaling system, we next examined the relationship between expression of the C. maxima FT/TSF orthologs and their capacity to enter the phloem translocation stream. For these experiments, LD-grown nonflowering (4-week-old) and flowering (6- to 12-week-old) C. maxima plants were used to obtain mRNA from stem vascular tissue. Real-time RT-PCR analysis was employed, and in these experiments, Cm-FTL1 and Cm-FTL2 mRNA levels were normalized to Cm-PP16, as this mRNA species was earlier shown to be expressed in companion cells prior to its entry into the sieve elements for long-distance translocation (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Haywood et al., 2005). Furthermore, the Cm-PP16 mRNA levels were found to be equivalent over the range of growth conditions employed. As shown in Figure 4A, the levels of Cm-FTL1 and Cm-FTL2 mRNA were low in 4-week-old pumpkin plants. Under the growth conditions employed for our studies, C. maxima plants produced their first floral buds ∼5 weeks after germination. Consistent with this observation, the levels of Cm-FTL1 and Cm-FTL2 mRNA increased in stem vascular tissue of 6- to 8-week-old plants. Based on these findings, all subsequent analyses were performed on 6-week-old LD-grown pumpkin plants.
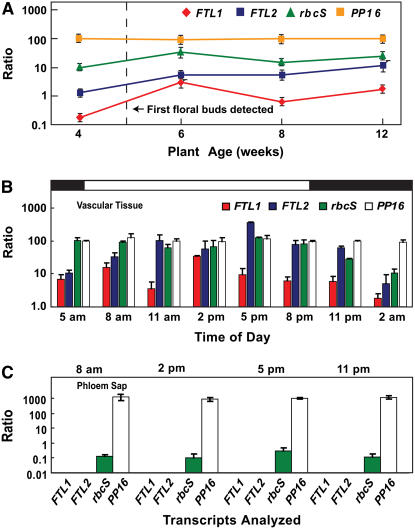
Figure 4.
Cm-FTL1 and Cm-FTL2 Transcripts Are Detectable in Stem Vascular Tissue but Not in Phloem Sap of Flowering C. maxima Plants.
(A) Real-time RT-PCR analysis of Cm-FTL1, Cm-FTL2, Cm-rbcS, and Cm-PP16 transcript levels in vascular tissue excised from mature stems of 4- to 12-week-old pumpkin plants. Note that floral buds were first detected on 5-week-old plants. Transcript abundance is expressed as a ratio with Cm-PP16, which was set arbitrarily at 100. RNA was extracted from vascular tissue collected from 2 to 3 pm. Data represent means ± se for three independent replicate experiments.
(B) Time course of Cm-FTL1, Cm-FTL2, Cm-rbcS, and Cm-PP16 expression in vascular tissue excised from mature stems of 6-week-old pumpkin plants, determined by real-time RT-PCR analysis. Transcript abundance is expressed as a ratio with Cm-PP16, which was set arbitrarily at 100. Data represent means ± se for three independent replicate experiments.
(C) Real-time RT-PCR analysis of Cm-FTL1, Cm-FTL2, Cm-rbcS, and Cm-PP16 levels in phloem sap collected from mature stems of 6-week-old C. maxima plants. Transcript abundance is expressed as a ratio with Cm-PP16, which was set arbitrarily at 1000. Note that Cm-FTL1 and Cm-FTL2 transcripts were not detected in phloem sap at a level 10 times lower than represented by Cm-rbcS mRNA contamination. Data represent means ± se for three independent replicate experiments.
A time course for Cm-FTL1 and Cm-FTL2 expression was next established to ascertain the appropriate period to collect phloem sap for RNA analysis. Vascular tissues were collected over a 24-h period and real-time RT-PCR performed. In contrast with LD-grown Arabidopsis, where the levels of FT mRNA undergo a considerable increase upon the onset of darkness (Suarez-Lopez et al., 2001; Searle and Coupland, 2004), neither Cm-FTL1 nor Cm-FTL2 transcript levels exhibited marked peaks at the end of the photoperiod (Figure 4B). Based on these findings, we selected four time points for the collection of phloem sap to be used for RNA (and protein) analysis.
Our RNA analysis of the phloem sap confirmed that, as in previous studies (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Haywood et al., 2005), Cm-PP16 mRNA was readily detected (Figure 4C). However, by contrast, Cm-FTL1 and Cm-FTL2 transcripts were undetectable in the phloem sap collected from flowering pumpkin plants. Note that Cm-rbcS transcripts were detected at a level 10,000 times below that for Cm-PP16. These results suggested that, in pumpkin, Cm-FTL1 and Cm-FTL2 mRNA may not be components of the phloem long-distance signaling system involved in floral induction.
Cm-FTL1 and Cm-FTL2 Are Present in Phloem Sap of Flowering Pumpkin
To test for the presence of Cm-FTL1 and Cm-FTL2 in the vascular tissue and phloem sap of flowering pumpkin plants, proteins were extracted and then separated by fast protein liquid chromatography (FPLC) methods. Conceptual translations for Cm-FTL1 and Cm-FTL2 predicted positively charged proteins. Consequently, proteins were dialyzed, clarified by centrifugation, and loaded onto a HiTrap SP cation-exchange column, connected to an Amersham Biosciences FPLC system; proteins were eluted with a linear NaCl gradient. Figures 5A and 5C illustrate the profiles of the FPLC-fractionated vascular tissue and phloem sap proteins, respectively.
Figure 5.
Pumpkin FPLC-Fractionated Proteins Interrogated for Cm-FTL1 and Cm-FTL2 by MS.
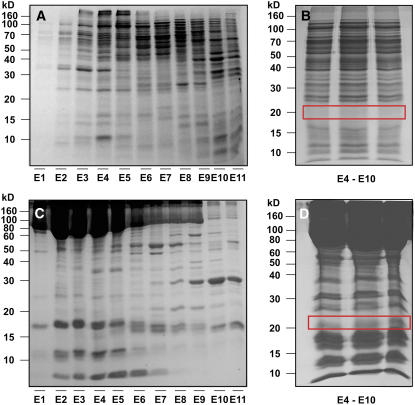
(A) and (C) Proteins extracted from C. maxima stem vascular tissue (A) and phloem sap (C) were separated using cation-exchange FPLC, and aliquots of each elution fraction (E1 to E11) were separated by SDS-PAGE. The 20-kD region associated with each elution fraction was excised, from individually run gels, for in-gel tryptic digest, followed by tandem MS analysis.
(B) and (D) Fractions E4 to E10 from vascular tissue (B) or phloem sap (D) were pulled and then resolved on SDS-PAGE. Rectangular boxes indicate the regions that were excised for in-gel tryptic digest, followed by tandem MS analysis for analysis of Cm-FTL1 and Cm-FTL2.
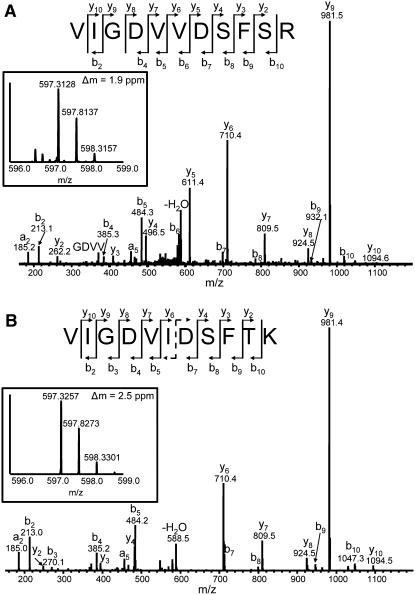
A combination of liquid chromatography–tandem mass spectrometry (LC-MS/MS) was used to interrogate the FPLC-fractionated proteins for the presence of Cm-FTL1 and/or Cm-FTL2. To this end, vascular tissue and phloem sap fractions were separated on SDS-PAGE gels, the 20-kD regions excised, and the proteins then trypsin digested, in-gel, for LC-MS/MS analysis. Our experiments established that, for both vascular tissue and phloem sap, Cm-FTL1 and Cm-FTL2 were located predominantly in fractions E9 and E5, respectively (Figure 5). Figure 6 presents tandem MS spectra of doubly protonated VIGDVVDSFSR and VIGDVIDSFTK, representative tryptic peptides for Cm-FTL1 and Cm-FTL2, respectively. These two peptides have very similar amino acid sequences and happened to have the same nominal mass (mass-to-charge ratio [m/z] of 597 for doubly protonated ions). However, their theoretical accurate masses (m/ztheor 597.3116 and m/ztheor 597.3242) are 21 ppm apart and could be readily distinguished with a high-accuracy mass spectrometer (LTQ-FT), as shown in the inset mass spectra (m/zexp 597.3128 and m/zexp 597.3257, respectively). The two tandem MS spectra have many common fragment ions, y6∼y10 and b2∼b5 ions, due to their close sequence homology. However, some fragment ions, such as y5 of VIGDVVDSFSR at m/z 611.4, clearly distinguish the two peptides in the tandem MS spectra. Additional mass spectrometry data are presented in Supplemental Figure 5 online.
Figure 6.
MS Characterization of Cm-FTL1– and Cm-FTL2–Derived Peptides.
(A) Tandem MS spectrum of doubly protonated VIGDVVDSFSR (m/z 597.3116) from the Cm-FTL1 protein. Almost all y and b series fragments were observed. Specific fragment peaks (e.g., y2∼y5 and b6∼b10) clearly differentiate this spectrum from that obtained for VIGDVLDSFTK presented in (B). Note that the y5 fragment at m/z 611.4 is very intense and explicable only on the basis of this peptide fragment. Inset mass spectrum shows parent ion scan obtained with the LTQ-FT. Here, the mass difference (Δm) between the measured and theoretical mass (m/z 597.3116) was only 1.9 ppm, which is within the 5-ppm experimental error range typical for the LTQ-FT. Note that the mass difference with the theoretical mass of VIGDVLDSFTK (m/z 597.3242) was 19 ppm, well beyond the instrument error.
(B) Tandem MS spectra of doubly protonated VIGDVLDSFTK (m/z 597.3242) from the Cm-FTL2 protein. All the b and y series fragment ions were clearly observed, except for b6 (m/z 597.4) and y5 (m/z 597.3), which have almost the same mass as the parent ion and are unstable in the tandem MS condition, and b1 and y1 ions, which must have been discriminated out due to low mass cutoff by the ion trap mass spectrometer. Inset mass spectrum shows parent scan obtained with LTQ-FT. Δm with the theoretical mass was only 2.5 ppm, which is within the typical experimental error range of 5 ppm of this instrument, confirming the sequence is correct, while the Δm with the theoretical mass of VIGDVVDSFSR (m/z 597.3116) was 25 ppm, well beyond the instrument error.
FTL1/FTL2 Entry in the C. moschata Phloem Translocation Stream Is under Photoperiodic Not Transcriptional Control
Parallel studies were next performed on 6-week-old C. moschata plants grown under either LD noninductive or SD inductive conditions. As illustrated in Figure 7A, real-time RT-PCR performed on mRNA extracted from stem vascular tissue established that Cmo-FTL1 and Cmo-FTL2 were present in both LD- and SD-grown plants. Interestingly, the Cmo-FTL1 mRNA level was found to be as high, or higher, in LD as in SD plants. In situ hybridization experiments confirmed that the Cmo-FTL1 and Cmo-FTL2 transcripts were confined to the phloem in mature petioles and stems. Supplemental Figure 6 online illustrates in situ experiments performed on sections taken from the same region of the plant as used for the mRNA analyses presented in Figure 7A.
Figure 7.
Comparative Analysis of Cmo-FTL1/FTL2 Transcript and Protein Levels in Vascular Tissue and Phloem Sap Collected from Plants Grown under Inductive SD and Noninductive LD Conditions.
(A) Real-time RT-PCR analysis of Cmo-FTL1, Cmo-FTL2, Cmo-rbcS, and Cmo-PP16 transcript levels in vascular tissue excised from mature stems of C. moschata plants subjected to SD and LD conditions. Tissue samples were collected at 3 pm. Transcript abundance is expressed as a ratio with Cmo-PP16, which was set arbitrarily at 100. Values represent means ± se from five independent replicate experiments.
(B) Levels of Cmo-FTL1 and Cmo-FTL2 in excised vascular tissue extracted from mature stems of C. moschata grown under SD and LD conditions. Protein quantification was achieved using the Protein-AQUA method (Kirkpatrick et al., 2005). Values represent means ± se from three independent replicate experiments.
(C) Levels of Cmo-FTL1 and Cmo-FTL2 in phloem sap collected from mature stems of C. moschata grown under SD and LD conditions. Protein quantification was achieved using the Protein-AQUA method (Kirkpatrick et al., 2005). Values represent means ± se from three independent replicate experiments.
Protein was also extracted from stem vascular tissue and phloem sap collected from the same C. moschata plants used for the RNA analysis. Although the Cmo-FTL1 mRNA levels were nearly equivalent in stems of LD and SD plants (Figure 7A), our tandem MS experiments performed on protein samples extracted from these same tissues revealed that, based on the levels of the representative tryptic peptide, VIGDVIDSFSR, Cmo-FTL1 was some fivefold higher in SD- compared with LD-grown plants (Figure 7B). Quantification of the levels of the C. moschata VIGDVIDSFSR, VIGDVIDSFTR (representative tryptic peptide for Cmo-FTL2), and VEIGGTDLR (common tryptic peptide for both Cmo-FTL1 and Cmo-FTL2) peptides was achieved using isotopically labeled synthetic peptides as internal standards (Kirkpatrick et al., 2005). As might be expected, based on the observed levels of Cmo-FTL2 mRNA, only an extremely low level of the VIGDVIDSFTR peptide was detected in stem vascular tissue sampled from LD-grown plants. However, under SD conditions, the level of this VIGDVIDSFTR peptide, and thus Cmo-FTL2, increased some 40-fold (Figure 7B).
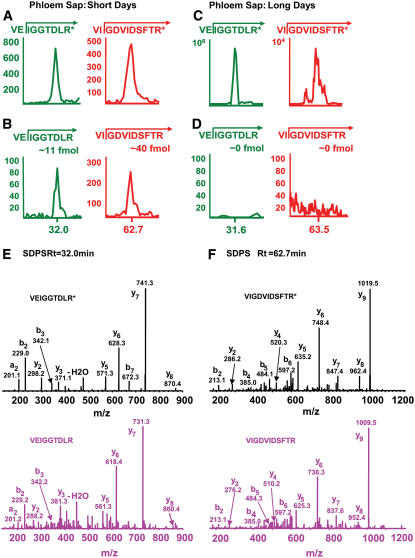
Analysis of the phloem sap collected from SD- and LD-grown C. moschata revealed that Cmo-FTL1 and Cmo-FTL2 could be detected only in SD plants. Again, for these analyses, isotopically labeled synthetic peptides, VIGDVIDSFSR*, VIGDVIDSFTR*, and VEIGGTDLR*, were employed as internal standards. Three replicate experiments were conducted and representative MS data are presented in Figure 8, while Figure 7C contains the summation for all the data collected. In all cases, signal associated with the endogenous VIGDVIDSFSR peptide from the phloem sap was extremely low, and for this reason, individual scans were not included in Figure 8. Single reaction monitoring experiments performed on phloem sap collected from SD-grown plants clearly identified VIGDVIDSFTR and VEIGGTDLR peptides (cf. Figures 8A and 8B). By contrast, no VIGDVIDSFSR, VIGDVIDSFTR, or VEIGGTDLR peptides could be detected in phloem sap collected from LD-grown plants (cf. Figures 8C and 8D). Confirmation of the identities for the endogenous VIGDVIDSFTR and VEIGGTDLR peptides was obtained by tandem MS analysis at retention times of 32 and 62.7 min, respectively (Figures 8E and 8F). Note that the VIGDVIDSFTR and VEIGGTDLR peptides have exactly the same MS/MS spectra as VIGDVIDSFTR* and VEIGGTDLR* except that they have a 10-D shift for y series fragments. Taken together, these experiments establish that the entry of Cmo-FTL1 and Cmo-FTL2 into the phloem translocation stream is controlled by daylength and correlates with flowering. Finally, based on our quantitative analysis of the phloem sap collected from SD-grown flowering C. moschata, the level of Cmo-FTL2 is ∼10-fold higher than that of Cmo-FTL1 (Figure 7C).
Figure 8.
Photoperiodic Control of Cmo-FTL1/FTL2 Entry into the Phloem Translocation Stream.
Phloem sap was collected from mature stems of C. moschata grown under inductive SD or noninductive LD conditions. Proteins contained within the 18- to ∼22-kD region of SDS-PAGE gels were in-gel digested and total peptides extracted for analysis by MS. Peptide aliquots (7 μg) for each treatment were subjected to LC-single reaction monitoring (SRM). Arrows indicate the reaction monitored for each trace ion chromatogram. All LC-SRM traces are from the same data set. The x and y axes in LC-SRM traces are retention times, in minutes, and ion counts, respectively.
(A) LC-SRM traces of isotopically labeled synthetic peptides (AQUA peptides; 100 fmol; R* represents Arg[13C615N4]) added to the 7-μg aliquot of peptides derived from phloem sap collected from SD-grown plants. The AQUA peptides served as internal standards for measuring the levels of VIGDVIDSFTR (TR) and VEIGGTDLR (LR) peptides derived from Cmo-FTL2.
(B) LC-SRM traces of Cmo-FTL2 native peptides, VIGDVIDSFTR and VEIGGTDLR, derived from phloem sap collected from SD-grown plants. Peptide quantification was made by comparing peak abundances at the same retention times between the traces presented in (A) and (B).
(C) LC-SRM traces of AQUA peptides (100 fmol) added to the 7-μg aliquot of peptides derived from phloem sap collected from LD-grown plants.
(D) LC-SRM traces of Cmo-FTL2 native peptides derived from phloem sap collected from LD-grown plants. Note the absence of peaks at the same retention times illustrated for the traces in (C).
(E) Tandem MS spectra of AQUA (top) and native (bottom) peptides of VEIGGTDLR from SD-grown plant phloem sap (SDPS) at the retention time of 32 min. Native peptides have the same tandem MS spectra as the AQUA peptides, except for a 10-D shift for y series fragments.
(F) Tandem MS spectra of AQUA (top) and native (bottom) peptides of VIGDVIDSFTR from SDPS at the retention time of 62.7 min.
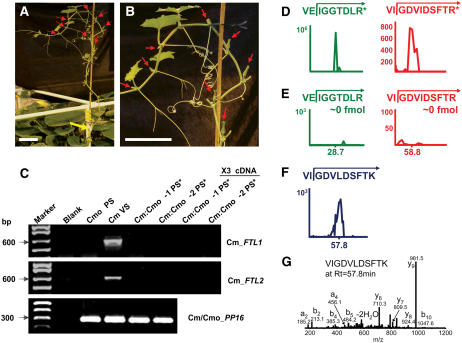
Detection of Pumpkin FT in Phloem Sap of Florally Induced C. moschata Scions
To further investigate the relationship between FT and floral induction, we next performed experiments, under LD conditions, in which C. moschata scions were grafted onto flowering pumpkin stocks. Upon induction to flowering (Figures 9A and 9B), each scion was harvested to collect phloem sap and both mRNA and protein extracted. RT-PCR experiments were performed using primers specific for Cm-FTL1 and Cm-FTL2, and as shown in Figure 9C, we obtained no evidence for import of these transcripts across the graft union.
Figure 9.
Analysis of Phloem Sap from Florally Induced C. moschata Scions Grafted onto Pumpkin Failed to Detect Cm-FTL1 or Cm-FTL2 Transcripts, Whereas Cm-FTL Peptides Were Present and Correlated with Floral Induction.
(A) and (B) Photographs of a C. moschata scion that was grafted onto a flowering C. maxima plant. Note the presence of the floral buds (red arrows); yellow arrow in (A) indicates the graft union. All grafted plants were grown under noninductive LD conditions. Scion leaves were excised as they approached maturity to ensure delivery of photosynthate from the pumpkin stock. Bars = 10 cm.
(C) RT-PCR analysis failed to detect C. maxima FT mRNA in phloem sap collected from florally induced C. moschata scions. Gene-specific primers were employed to amplify Cm-FTL1, Cm-FTL2, and Cm/Cmo-PP16. Expected product sizes are indicated on the left. cDNAs prepared from poly(A)+ RNA extracted from C. maxima vascular tissue (Cm VS; positive control), phloem sap cDNAs prepared from poly(A)+ RNA extracted from C. moschata grown under LD conditions (Cmo PS; negative control), and two individual florally induced C. moschata scions grafted on C. maxima were used as templates. Amplification of Cm/Cmo-PP16 transcripts served as an internal control (bottom panel). PCR conditions were as follows: 2 min at 94°C (one cycle), 30 s at 94°C; 30 s at 60°C and 50 s at 72°C (40 cycles). Specific tissues analyzed are indicted by an asterisk.
(D) to (G) Presence of Cm-FTL peptides in the phloem sap of C. moschata scions, grafted onto C. maxima stocks, correlate with floral induction. Phloem sap was collected from flowering C. moschata scions grown under noninductive LD conditions. Proteins contained within the 18- to ∼22-kD region of SDS-PAGE gels were in-gel digested and total peptides extracted for analysis by mass spectrometry. Peptide aliquots (7 μg) for each treatment were subjected to LC-SRM. Arrows indicate the reaction monitored for each trace ion chromatogram. All LC-SRM traces are from the same data set. The x and y axes in LC-SRM traces are retention times, in minutes, and ion counts, respectively.
(D) LC-SRM traces of isotopically labeled synthetic peptides (AQUA peptides; 100 fmol, R* represents Arg[13C615N4]) added to the 7-μg aliquot of phloem-derived peptides. The AQUA peptides served as internal standards for measuring the levels of VIGDVIDSFTR (TR) and VEIGGTDLR (LR) peptides present in the scion phloem sap derived from Cmo-FTL2.
(E) LC-SRM traces of Cmo-FTL2 native peptides, VIGDVIDSFTR and VEIGGTDLR, derived from phloem sap collected from florally induced C. moschata scions. Note the absence of peaks at the expected retention times.
(F) LC-SRM trace of Cm-FTL2 native peptide derived from phloem sap collected from florally induced C. moschata scions. Note the presence of the VIGDVLDSFTK peptide indicating the import of Cm-FTL2 across the graft union.
(G) Tandem MS spectrum at the retention time corresponding to VIGDVLDSFTK confirms the presence of the C. maxima FT peptide.
Single reaction monitoring experiments were next performed on the phloem proteins obtained from each individual C. moschata scion. As illustrated in Figures 9D and 9E, no VIGDVIDSFTR or VEIGGTDLR peptides derived from Cmo-FTL2 could be detected. Additionally, no VIGDVIDSFSR peptide derived from Cmo-FTL1 could be detected (data not shown). This result was expected, based on our previous finding that these proteins do not enter the phloem translocation stream when C. moschata is maintained under LD conditions. However, as shown in Figure 9F, our single reaction monitoring data detected a strong signal for the VIGDVIDSFTK peptide derived from Cm-FTL2. Tandem MS analysis unambiguously identified this VIGDVIDSFTK peptide as being derived from Cm-FTL2 present within the phloem proteins collected from these florally induced C. moschata scions (Figure 9G). The VIGDVIDSFTK peptide was clearly identified in five out of the six scion phloem protein preparations analyzed.
DISCUSSION
The role of the phloem in coordination of developmental events, at the whole-plant level, has long been known (Oparka and Turgeon, 1999; Oparka and Santa Cruz, 2000; Lucas and Lee, 2004; Lough and Lucas, 2006). However, molecular information on the nature of the signaling agents that move through the translocation stream has remained elusive. An excellent example is florigen, the hormonal signal that transmits photoperiodic information from mature leaves to the SAM to regulate flowering time. Seven decades after Chailakhyan (1936) coined the term florigen, molecular genetic studies have now begun to identify the key players in this pathway. Studies on Arabidopsis earlier implicated FT mRNA as florigen (Huang et al., 2005), but this work was recently retracted (Böhlenius et al., 2007). Furthermore, experiments with tomato and tobacco (Nicotiana tabacum) provided results that failed to support a role for FT mRNA in phloem long-distance signaling (Lifschitz et al., 2006). Here, we provide direct evidence that FT, but not FT mRNA, is present in the phloem sap of flowering cucurbits and correlates with floral induction.
The Gene Pair Cm-FTL1 and Cm-FTL2 Is Orthologous to the Arabidopsis Pair FT and TSF
We isolated two FT homologs from C. maxima and C. moschata, which we named FTL1 and FTL2. Molecular phylogenetic analysis shows that these gene pairs are orthologous to the Arabidopsis gene pair FT and TSF. Each gene duplication occurred independently (i.e., after divergence of the lineages leading to Arabidopsis, C. maxima, and C. moschata), and so neither cucurbit gene is strictly orthologous to FT or TSF. Similarly, our analysis shows that the clade with two poplar FT homologs, known as FT1 and FT2, is most likely orthologous to the Arabidopsis FT/TSF clade as well as the C. maxima and C. moschata FTL1/FTL2 clades (i.e., there is a two-for-two relationship between the poplar gene pair and the Arabidopsis gene pair and between the poplar gene pair and the cucurbit gene pairs). Importantly, there is no one-for-one orthologous relationship between either Arabidopsis gene or either cucurbit gene and any other plant FT homolog (including tomato SFT) in our analysis. Interestingly, rice has 10 FT homologs that group strongly with these dicot FT homologs (Figure 1C), suggesting the possibility of greater diversification of FT genes and possibly FT function in rice than in Arabidopsis, cucurbits, or poplar.
Parallel experiments in which we ectopically expressed Cm-FTL1, Cm-FTL2, FT, or CO in Arabidopsis companion cells yielded early flowering phenotypes (Figures 1D and 1E). A similar situation was recently reported in which expression of poplar FT2 in Arabidopsis resulted in early flowering phenotypes (Böhlenius et al., 2006; Hsu et al., 2006).
Cucurbits Function as a Model System for the Dissection of Florigenic Signaling
In this study, a novel cucurbit system was developed to investigate the molecular nature of florigen. By screening a large population of cucurbit accessions, we identified C. moschata PI441726 that flowers only under inductive SD conditions (Figures 1A and 2G). Here, we should note that this SD attribute allowed us to perform photoperiodic induction experiments without significantly altering the carbon budget of plants grown under either SD or LD conditions. This is an important feature, as the level of photosynthate has also been implicated as a component of the florigenic signaling pathway (Bernier, 1988; Bernier and Périlleux, 2005). An additional feature of this cucurbit system is the ease with which grafting experiments can be performed (Golecki et al., 1998, 1999; Ruiz-Medrano et al., 1999). Finally, as a host for the ZYMV vector, C. moschata allowed us to test the influence of heterologous/foreign genes on floral induction.
As viral infection can, by itself, alter the host flowering time, it was important to establish whether the ZYMV vector, or ZYMV-GFP, would alter the flowering response in C. moschata. We obtained clear evidence that neither ZYMV (vector) nor ZYMV-GFP had any effect on flowering in C. moschata plants being grown under noninductive LD conditions: all infected plants remained vegetative (Figures 2C, 2E, and 2F). By contrast, equivalent plants inoculated with ZYMV-At-FT were all induced to flower (Figures 2D to 2F). These results establish that, when ectopically expressed, At-FT has a similar effect in C. moschata as it does in Arabidopsis. Parallel studies performed on plants grown under inductive SD conditions (Figure 2G) confirmed that ZYMV-At-FT–inoculated plants flowered earlier than ZYMV-GFP and mock-inoculated plants. Interestingly, ZYMV-GFP inoculated plants were found to begin flowering two to four nodes earlier than mock-inoculated plants. This suggests that under SD conditions, ZYMV-GFP infection may have altered the time and/or level of Cmo-FTL1/Cmo-FTL2 expression. In any event, our findings are consistent with the notion that the long-distance florigenic signal, either FT mRNA or FT, is common to all flowering plants (Chailakhyan, 1937; Lifschitz et al., 2006).
ZYMV-mediated At-FT protein expression is based on autocatalytic processing of a high molecular weight viral polyprotein precursor (Carrington et al., 1993). The processed At-FT protein product has N-terminal S-A-C and C-terminal G-T-H-H-H-H-H-H-L-V-D-T-V-M-L-Q amino acid residues fused to the At-FT (Figure 2B). As this processed At-FT product was highly effective at inducing flowering, it would appear that At-FT is tolerant of N- and C-terminal extensions. This result will be of importance for future studies on the trafficking of FT within the meristem.
Our ZYMV immunolocalization studies (Figure 3) confirmed that potyvirus infection domains do not include meristematic tissues (Matthews, 1991; Jones et al., 1998). Here, we should stress that, in our studies, the CP served as an effective reporter for the presence of the viral RNA within the tissues of interest. Thus, the absence of ZYMV from the plant apex (Figures 3D and 3E) proves that the influence of At-FT on floral induction must be non-cell-autonomous in nature. The likelihood that At-FT RNA sequences, embedded within the ∼9-kb viral vector, contribute to floral induction appears remote. A more likely scenario is that At-FT was the non-cell-autonomous (mobile) signal or it acted to propagate a signal that then mediated floral induction in the C. moschata meristem(s).
Cucurbit Heterografts Confirm Phloem Transfer of Florigenic Stimulus
Grafting experiments are a simple, but powerful, method for establishing the transmission of a floral stimulus through the phloem. In this regard, our heterografting experiments performed with C. moschata and C. maxima demonstrated the delivery of a long-distance florigenic signal that induced flowering in C. moschata plants being grown under noninductive LD conditions (Table 1). The C. moschata homografted plants grown under exactly the same LD conditions served as a control for this study. Here, five out of the six plants remained vegetative, with one plant forming aborted floral buds at nodes 4 through 7. It is important to note that removal of the mature leaves, from the C. moschata scions, resulted in highly efficient floral induction at nodes 5 to 10; nodes 1 and 2 existed at the time of grafting, while 3 to 10 developed after grafting. Our results indicated that the phloem translocation stream in C. maxima stock plants should contain Cm-FTL1/Cm-FTL2 mRNA and/or Cm-FTL1/Cm-FTL2, if this is indeed a component of the florigenic signaling system.
FT mRNA Could Not Be Detected in Phloem Sap of Flowering C. maxima
The diurnal expression patterns for both Cm-FTL1 and Cm-FTL2 indicated that transcript levels were highest during the daylight hours and at their lowest in the early morning (Figure 4B). As there was no pronounced peak in expression, phloem sap was collected at four discrete times to assay for the presence of Cm-FTL1/Cm-FTL2 mRNA. Our real-time RT-PCR experiments, performed on phloem sap–extracted RNA, indicated that neither Cm-FTL1 nor Cm-FTL2 transcripts could be detected (Figure 4C). Hence, if these transcripts were to function in florigenic signaling, they must be present in the phloem sap at extremely low levels. Such a scenario could explain why Lifschitz et al. (2006) were unable to detect the tomato SFT transcripts in scions whose phenotype(s) was rescued by a graft-transmissible signal(s). However, a more likely scenario is that, for the cucurbits, tomato, and tobacco, the florigenic signal is not RNA based.
FTL1 and FTL2 in the Phloem Sap of C. maxima Is Consistent with a Role in Long-Distance Signaling
Analytical quantities of phloem sap can be routinely collected from the stems of flowering C. maxima, which made this plant an ideal system to explore whether FT is present in the phloem translocation stream. As the cucurbit phloem sap is a highly complex mixture of proteins (Figure 5; Golecki et al., 1998, 1999; Yoo et al., 2004), a combination of FPLC methods and MS was used to identify tryptic peptides of Cm-FTL1 and Cm-FTL2. The spectral data obtained from these studies provided direct evidence for the presence of both Cm-FTL1 and Cm-FTL2 in phloem sap collected from the long-distance translocation stream of flowering C. maxima (Figure 6; see Supplemental Figure 5 online). Thus, our results are consistent with the hypothesis that, in the cucurbits, FTL1 and FTL2 can enter the phloem translocation stream.
Photoperiodic Control of FTL1/FTL2 Entry into the C. moschata Phloem Is Consistent with a Role in Floral Induction
Our comparative analysis of Cmo-FTL1 and Cmo-FTL2 transcript and protein levels, under inductive SD and noninductive LD conditions, indicated the operation of a complex control mechanism acting on FTL1 and FTL2 synthesis and trafficking into the phloem translocation stream (Figures 7 and 8). Although high Cmo-FTL1 mRNA levels were detected in stem vascular tissue of LD-grown plants, the level of protein in this same tissue was low compared with the SD situation (Figures 7A and 7B). For Cmo-FTL2, transcript levels were low under LD conditions and increased ∼100-fold in plants treated with SD inductive conditions; a near parallel increase in FTL2 levels was observed under SD conditions. These findings support the notion that Cmo-FTL2 may be more important in terms of contributing to the long-distance floral stimulus. Our discovery that Cmo-FTL1 and Cmo-FTL2 are present only in the phloem sap of SD-induced plants is consistent with a role in florigenic signaling in this species. Here, it is interesting to note that Brassica napus FT homologs have also been detected in exudate obtained from inflorescence stems (Giavalisco et al., 2006).
Pumpkin FTL2 Passes the Graft Union to Induced Flowering in C. moschata Scions
The value of using cucurbits to examine the molecular nature of the florigenic signal is exemplified by the results obtained using the C. maxima:C. moschata grafting system. By maintaining heterografted plants under noninductive LD conditions, we could observe pumpkin-mediated floral induction in the C. moschata scions (Figures 9A and 9B) and examine, by MS, the contents of the scion phloem sap for the presence of pumpkin FTL1 and FTL2 (Figures 9D to 9G). The fact that we could detect a clear VIGDVLDSFTK peptide signal from Cm-FTL2, in the scion phloem sap, implicates it as a signaling component that gives rise to floral induction of C. moschata scions maintained under noninductive LD conditions. The absence of Cm-FTL1 and Cm-FTL2 mRNA in the phloem sap of these scions adds further evidence against a role for the mRNA in this particular long-distance signaling process.
Generality of FT Acting as a Long-Distance Signal?
Our results obtained with C. maxima and C. moschata are consistent with those recently reported for rice and Arabidopsis. In the rice study, Tamaki et al. (2007) used an Hd3a-GFP reporter to compare the accumulation pattern of Hd3a with that of Hd3a mRNA. The Hd3a-GFP was detected in the apex, where endogenous Hd3a transcript levels were extremely low, suggesting movement of the reporter protein from the leaves; however, no direct evidence was obtained for the actual trafficking of Hd3a in the phloem translocation stream. In addition, no tests were performed to ascertain whether the Hd3a:GFP transcripts actually move, long-distance, from the presumptive site of synthesis in leaves to the plant apex. Hence, a role for Hd3a mRNA in floral induction, while seemingly unlikely, cannot be discounted (Tamaki et al., 2007). Lastly, as the expression level of the Hd3a:GFP transgene was ∼10-fold higher than that measured for endogenous Hd3a mRNA, caution may be called for in the interpretation of these data.
In the Arabidopsis study, Corbesier et al. (2007) presented further evidence indicating that FT may function as a phloem-transported component of the florigenic signaling pathway. As the endogenous level of FT expression is known to be low, Corbesier et al. (2007) used the strong SUC2 companion cell–specific promoter to drive the expression of an FT:GFP reporter system in an ft-7 background. Unfortunately, the SUC2 promoter is active not only in the loading zones, but extends along the long-distance transport pathway and has also been shown to be active in various sink tissues (Martens et al., 2006). Consistent with this activity in sink regions, parallel in situ hybridization experiments probing for FT:GFP transcripts and confocal microscopy analyzing FT-GFP fluorescence clearly showed congruence in the two signals at a distance of some 50 to 100 μm from the SAM. Thus, detection of FT-GFP in the apex does not provide compelling evidence for the hypothesis that FT-GFP induction of flowering results from its import from distant tissues.
The SUC2:FT:GFP ft-7 transgenic plant line was also used in heterografting experiments with wild-type Arabidopsis plants to test for the passage of FT-GFP across the graft union. Analysis of wild-type roots grafted onto SUC2:FT:GFP ft-7 transgenic scions indicated that FT-GFP, but not FT:GFP mRNA, move long distance into the rootstock. Y-graft experiments also demonstrated import of FT-GFP into the receiver shoot apex, but the caveat to these experiments is that FT:GFP was overexpressed compared with endogenous FT, and even so, the effect on floral induction was quite weak. It is also unfortunate that the Y-graft system was not used to test for the delivery of FT:GFP transcripts into this scion system. In any event, the conclusion drawn that FT:GFP transcripts are not phloem mobile needs to be considered in light of our previous work indicating the presence of a phloem-based RNA surveillance system to control transcript import into specific sink tissues (Haywood et al., 2005).
Mechanism of Cucurbit FTL1 and FTL2 Trafficking into the Sieve Tube System
Two modes of transport are possible for FTL1 and FTL2 to enter the cucurbit phloem translocation stream (Oparka and Santa Cruz, 2000; Lucas and Lee, 2004). Plasmodesmata connecting functional sieve elements to their neighboring companion cells allow for the exchange diffusion of the 27-kD GFP (Imlau et al., 1999). When expressed under the SUC2 promoter, GFP enters the phloem translocation stream and is delivered to sink tissues where it can pass out through plasmodesmata into the surrounding cells (Imlau et al., 1999; Oparka et al., 1999; Stadler et al., 2005). Thus, as FTL1 and FTL2 are 20 kD, they too would have the potential to diffuse from their site of synthesis in the companion cells into the sieve elements.
The companion cell–sieve element plasmodesmata also mediate the selective trafficking of proteins. In this situation, protein–protein interaction is required and specific mutations in the non-cell-autonomous protein can inhibit cell-to-cell trafficking (Lucas and Lee, 2004). That Cmo-FTL1 and Cmo-FTL2 are present in the stem vascular tissues of plants maintained under LD conditions, but undetectable in the phloem sap collected from these same plants (Figures 7 and 8), implicates an additional mechanism acting to control the trafficking of these two proteins from the companion cells into the sieve tube system. Determination as to the nature of this control process and the actual mode of trafficking employed by FTL1 and FTL2 to enter the phloem will be an important next step in unraveling the mysteries of florigen. Of equal importance will be the elucidation of the mechanism by which FT egress from the terminal phloem to begin the signal cascade that must propagate all the way to the SAM (Abe et al., 2005; Wigge et al., 2005).
Finally, although our results with the cucurbits are consistent with FTL1 and FTL2 acting as a component in the long-distance florigenic signaling pathway, a role for the FT transcripts in other plant systems should not be discounted. Indeed, presently there are excellent examples, such as Cm-PP16 (Ruiz-Medrano et al., 1999; Haywood et al., 2005), where both protein and the encoding mRNA have been demonstrated to move through the phloem translocation stream. It will certainly be of considerable interest if future studies do indeed identify species in which FT transcripts, or both FT and FT mRNA, are present in the phloem sap and move to the plant apex. Identifying the molecular events involved in switching between these two forms of long-distance macromolecule signals would provide considerable insight into the evolution of processes that regulate events at the whole-plant level.
METHODS
Plant Materials
Arabidopsis thaliana (ecotype Columbia) plants were grown hydroponically using Grodan blocks (Grodania) in a pathogen-free controlled environment chamber under LDs (250 μmol m−2 s−1 PAR, 25/20°C day/night temperatures, daylength of 16 h) or SDs (conditions as above, but with a daylength of 10 h). Flowering time was measured by scoring the number of rosette leaves at the time of bolting; at least 20 individual plants were used per treatment. Data are expressed as mean ± se. Cucurbita moschata PI441726 and Cucurbita maxima cv Big Max plants were grown under both a pathogen-free greenhouse environment (natural daylight conditions: 1000 μmol m−2 s−1 PAR, 30/20°C day/night temperatures, daylength of 16 h [LD]) and in a controlled environment (350 μmol m−2 s−1 PAR, 28/20°C day/night temperatures, and either 10 h SD or LD [10 h daylength and a dark period that was interrupted by a 15-min incandescent light treatment each hour]).
Identification of a cucurbit species in which floral induction was photoperiodically controlled was achieved through screening of seeds for 97 accessions provided by the Agricultural Research Service Plant Genetic Resources Conservation Unit (www.ars-grin.gov/ars/SoAtlantic/Griffin/pgrcu). Ten seedlings for each accession were grown in greenhouses under winter conditions, when the daylength was <12 h (i.e., SD conditions), or in equivalent greenhouses in which incandescent lighting was used to provide daylength extension such that the plants were exposed to a 16-h photoperiod (i.e., LD conditions). Ninety-six accessions were found to flower under both SD and LD conditions, whereas plants for one accession, C. moschata PI441726, flowered only under SD conditions. Fruit from these plants were collected and the resultant seed rescreened under both LD and SD conditions to confirm that floral induction in C. moschata PI441726 was indeed under photoperiodic control.
Cloning of Cucurbita maxima, C. moschata, and Arabidopsis Genes
Vascular tissue, excised from mature (6-week-old) C. maxima or C. moschata PI441726 stems, was used in the cloning of the respective FT homologs. High molecular weight RNA was obtained using either an RNAqueous kit (Ambion) or TRIZOL (Invitrogen). Aliquots (1 μg) of this RNA were then used for RT-PCR with SuperScript RT III (Invitrogen) according to the manufacturer's instructions. Samples were treated with DNase I (Invitrogen) for 15 min, at room temperature, to remove any possibility of genomic DNA contamination, diluted 10-fold, and aliquots (1 μL) used for PCR amplification. The same general procedure was used for cloning FT and CO from Arabidopsis leaf tissue. The following gene-specific primers were used: Cm-FTL1, forward 5′-ATATGCATGCATGCCGAGAAATCGTGAC-3′ and reverse 5′-GGTTGATAATATAATGTGAAGTGAAGTGT-3′; Cm-FTL2, forward 5′-TTGGGAGAGTCATCGGCGACGTTA-3′ and reverse 5′-TCTTCTCCTTCCACCAGACCCAC-3′; Cmo-FTL1, forward 5′-AGGTCCATCTCGATTAGGGTTGCTTACAACTCG-3′ and reverse 5′-CGTAGCACACTATCTCTTGTCCAAAGGTTGCCT-3′; Cmo-FTL2, forward 5′-CACGAGGTCCATTTCCATTAGGGCTACTTACAACAAT-3′ and reverse 5′-CATAGCACACGATCTCTTGACCAAAGGTCGCTC-3′; FT, forward 5′-ATATGCATGCATGTCTATAAATATAAGAGAC-3′ and reverse 5′-ATATGGTACCAAGTCTTCTTCCTCCGCA-3′; CO, forward 5′-ATATGCATGCATGTTGAAACAAGAGAGTAAC-3′ and reverse 5′-ATATGGTACCGAATGAAGGAACAATCCCA-3′. PCR conditions were as follows: 2 min at 94°C (one cycle), 10 s at 94°C; 30 s at 58°C and 60 s at 68°C (36 cycles).
FT Phylogenetic Analysis
Public databases, including The Institute for Genomic Research and Joint Genome Initiative, were searched using various Arabidopsis and rice (Oryza sativa) flowering-time genes as queries. Gene names and their nucleotide accessions can be found in Supplemental Table 1 online. Os-FT-L5 was not included in this study because it has been identified as being a pseudogene. Nucleotide sequences were aligned based on a protein sequence alignment generated using Muscle 3.52 (Edgar, 2004). The alignments were edited using GeneDoc (http://www.psc.edu/biomed/genedoc/) to remove nonaligning regions (see Supplemental Figure 4 online). Neighbor-joining trees were constructed using the computer program MEGA3.1 (Kumar et al., 2004). The Kimura two-parameter substitution model was used, and a consensus tree was built from 1000 bootstrap replicates (see Supplemental Figure 2 online). In addition, Bayesian phylogenetic analyses were performed using MrBayes 3.0B4 (Huelsenbeck and Ronquist, 2001). Searches were run using four different Markov chains for 3,000,000 generations using the GTR model with a substitution rate that varies according to the gamma distribution and allows for a proportion of sites to be invariable (GTR + IG). Sampling occurred every 100 trees, and the first 10,000 trees were discarded to build a consensus tree (see Supplemental Figure 3 online).
Plant Transformation
The SUC2 promoter sequence (Truernit et al., 1996) and the CO, FT, Cm-FTL1, and Cm-FTL2 gene sequences were subcloned into the binary vector pART27 (Gleave, 1992). All binary plasmids were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis (Columbia ecotype) plants by the floral dip method (Clough and Bent, 1998). Seeds from primary transformed kanamycin-resistant transgenic control (vector only; three lines), CO (three lines), FT (six lines), Cm-FTL1 (six lines), and Cm-FTL2 (12 lines) plants were used to generate T1 progeny that were then employed for flowering-time assays.
ZYMV Constructs
The ZYMV-based viral vector, p35SZYMVNIbMCS (Lin et al., 2002), was first digested with BglII and KpnI. Next, PCR fragments encoding FT, CO, and eGFP full-length sequences were digested with SphI and KpnI restriction enzymes followed by ligation into p35SZYMVNIbMCS. For these studies, the eGFP template was obtained from a pEGFP plasmid (BD Biosciences) using the following primer pair: forward 5′-ATATGCATGCATGGTGAGCAAGGGCGAGGAGCT-3′; reverse 5′-ATATGGTACCCTTGTACAGCTCGTCCATGCCGA-3′. All viral constructs were verified by sequence analysis.
Floral Induction Mediated by ZYMV Expression of At-FT
Viral vector inoculations were initiated by microprojectile bombardment using a Helios gene gun system (Bio-Rad Laboratories). ZYMV-based constructs were precipitated onto gold particles (DNA loading ratio of 2 μg/mg gold). Briefly, 1-μm gold particles (15 mg) were resuspended in 100 μL of 50 mM spermidine. DNA (25 μg) was then precipitated onto the gold with 100 μL of 1 M CaCl2. This gold:DNA mixture was washed three times with 500 μL of 100% ethanol, and the final pellet was resuspended in 2.5 mL of 25 μg/mL polyvinylpyrrolidone in 100% ethanol for loading into GoldCoat tubing at a ratio of 1 mL of DNA/gold particle suspension per 15 × 0.5-inch cartridge. A helium pressure of 100 p.s.i., delivered at a distance of 3 cm, was used to inoculate ZYMV constructs into C. moschata PI441726 seedlings that had developed two true leaves.
The ZYMV vector was inoculated onto one true leaf and one cotyledon, and plants were then monitored weekly for floral bud initiation at each node. The stage of floral bud induction was recorded according to the following scoring system: 0, no visible bud; 1, bud <1 mm; 2, bud 1 to 5 mm; 3, 5 to 15 mm; 4, bud >15 mm; and 5, open flowers. Flowering rate was defined as the ratio of plants in each group with a stage ≥2.
The stability of the inserted sequence within the ZYMV vector was monitored by RT-PCR. RNA was extracted (TRIZOL; Invitrogen) from apical leaves of systemically infected plants 30 DAI. The RT reaction was performed with 0.5 μg total RNA using Superscript RT III (Invitrogen) with random primers (Invitrogen). The PCR conditions were as follows: 2 min at 94°C (one cycle), 10 s at 94°C; 30 s at 54°C and 90 s at 70°C (32 cycles) using Taq DNA polymerase (Invitrogen) and ZYMV-specific primers flanking the insertion site (forward 5′-AGAGGCTATTTGCGCTGCGATG-3′; reverse 5′-CTTTCACGCGTGGCAGTGACAT-3′). The diagnostic size of the amplified insert was evaluated using ethidium bromide–stained 1% agarose gels. These assays established that the ZYMV-At-FT vector stably expressed At-FT for up to 40 DAI.
Epifluorescence Microscopy
Spatial distribution of GFP in ZYMV-inoculated C. moschata PI441726 plants was determined using epifluorescence microscopy (Leica MZFLIII stereomicroscope; Leica Microsystems). Plants 4 to 30 DAI were used to obtain fresh, unfixed, sections of lamina, petiole, and stem tissues. Images were captured and analyzed using a Leica DC300F camera (excitation filter, 480 nm; emission filter, 510 nm) in conjunction with Adobe Photoshop (Adobe Systems).
Immunolocalization of ZYMV Coat Protein
Tissues from C. moschata PI441726 plants were fixed in FAA (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) overnight at 4°C, dehydrated through an ethanol series and Neo-Clear (Merck), and finally embedded in paraffin. Sections (10 μm) were cut and mounted on electrostatically charged Superfrost Plus slides (British Drug House). Slides were used immediately or stored at −20°C. Sections were deparaffinized in Neo-Clear (2 × 10 min), rehydrated through an ethanol series, and then transferred into distilled water. Endogenous peroxidase activity was blocked using a 10-min treatment in 0.5% H2O2 in methanol. Slides were then pretreated in boiling 1 mM EDTA, pH 7.5, for 5 to 15 min, cooled to room temperature, and then washed in distilled water for 5 min, followed by TBST (0.01% Tween-20 in Tris-buffered saline, pH 7.5) for 5 min. Sections were next incubated with blocking reagent (0.1% BSA in TBST) for at least 10 min, excess blocking reagent was removed, and then they were irrigated with anti-ZYMV antibody solution (Agdia) diluted 1:100 in blocking reagent. After overnight treatment in anti-ZYMV antibody solution, slides were washed in TBST (2 × 5 min) and sections covered with the secondary antibody (anti-rabbit IgG; Sigma-Aldrich) diluted in blocking reagent (1:300). After a 1-h incubation, slides were washed in TBST (2 × 5 min) and placed in a humid chamber, and sections were covered with DAB substrate (Roche) and incubated until desired staining was achieved (usually 5 to 15 min). Slides were rinsed with distilled water and mounted and photographed on a Zeiss Axioskop mounted with a Zeiss Axiocam MRc camera.
Grafting Protocol
Heterografting experiments were performed with C. moschata PI441726 and C. maxima plants essentially as previously described (Ruiz-Medrano et al., 1999; Yoo et al., 2004). Each excised C. moschata PI441726 scion (∼15 cm in length with two nodes) was carefully inserted into an incision made in the main stem of a 5-week-old stock (C. moschata PI441726 or C. maxima) plant. The graft site was then fastened and sealed with Parafilm and the scion covered with a clear plastic bag that was removed 1 week later. As the scions grew, mature leaves were excised to maintain the sink status of this tissue. Homo- and heterografted scions were observed until they had developed a total of 10 visible nodes (i.e., eight new leaves), and the scoring system for flowering was as described above.
Collection of RNA and Proteins from Phloem Sap and Vascular Tissue
Phloem sap collection was performed as previously described (Ruiz-Medrano et al., 1999; Yoo et al., 2004), with modifications. Briefly, C. maxima or C. moschata PI441726 stems were excised with a sterile razor blade and the cut surface blotted, several times, with sterile filter paper (3 MM; Whatman). Phloem sap exuded thereafter was collected using sterile micropipette tips and immediately mixed with 9 volumes of protein sap collection buffer (7 M urea, 2 M thiourea, 4% CHAPS, and 67 mM DTT), or 200-μL aliquots were mixed with 500 μL of TRIZOL reagent (Invitrogen) for phloem sap RNA extraction. All buffers and samples were kept on ice during phloem sap collection.
Vascular tissue was excised from the same region of the plant as used for the collection of phloem sap. Stem sections (10 cm) were excised from the plant and vascular bundles removed using forceps. Vascular tissue was first ground under liquid nitrogen and then divided into two aliquots. The first was mixed with TRIZOL reagent and processed for RNA extraction according to the manufacturer's protocol (Invitrogen). The second aliquot was mixed with phloem sap collection buffer, followed by sonication (eight pulses for 2 min using a Sonic Dismembrator; Fisher Scientific) and centrifugation (10,000g for 30 min); the supernatant was collected and filtered. All buffers and samples were kept on ice during RNA and protein extraction.
Real-Time PCR
Transcript levels of C. maxima and C. moschata FTL1, FTL2, rbcS, and PP16 were determined on a Bio-Rad iCycler IQ using POWER SYBR Green PCR Master Mix (Applied Biosystems) in 25-μL reactions. An aliquot (1 μL) of the RT reaction was used as template. PCR conditions were as follows: 10 min at 95°C (one cycle); 30 s at 95°C, 30 s at 60°C, and 50 s at 72°C (50 cycles). Gene-specific primers used were as above and as follows: Cm-rbcS, forward 5′-AGGCACCAATGGCTTCCTCCATCGTCTC-3′ and reverse 5′-GCACTGGACTTGGCGGATGTTGTCGAAG-3′; Cm-PP16, forward 5′-GTGGTAAAGGACTTCAAGCCCACGACC-3′ and reverse 5′-ATGGGTTTGAAGAAGCCAAGCCACTTA-3′; Cmo-rbcS, forward 5′-ATGGCTTCCTCCATCGTCTCATCC-3′ and reverse 5′-GAGCGGAGAAGGTACGCAATTTCC-3′; Cmo-PP16, forward 5′-ATGGGTTTGAAGAAGCCAAGCCAC-3′ and reverse 5′-AGTTGCTAAAAATGCAGGACCAGATCCATTATGGG-3′. Each RT sample was assayed in triplicate. Individual threshold cycles were located in 15 to 35 cycles, which is in the linear range of PCR amplification for each gene. Data were further analyzed in Excel. Transcript levels were normalized to C. maxima and C. moschata PP16, respectively.
Protein Preparation and LC-MS/MS Analysis
Proteins isolated from C. maxima phloem sap and vascular tissue were fractionated using chromatographic methods. Proteins were first dialyzed against buffer A (20 mM Na2HPO4/NaH2PO4, pH 7.0, 1 mM EDTA, 10% glycerol, and 1% 2-mercaptoethanol) and then clarified by centrifugation (17,000g for 30 min). Samples were then loaded onto a buffer A–equilibrated HiTrap SP column connected to an FPLC system (Amersham Biosciences). After washing the column with 10 volumes of buffer A, proteins were eluted with a linear gradient of 0 to 500 mM NaCl in buffer A supplemented with 1 M NaCl. Proteins isolated from C. moschata PI441726 phloem sap and vascular tissue were dialyzed against buffer B (20 mM Tris, pH 7.5, 1 mM EDTA, and 1% 2-mercaptoethanol). FPLC-fractionated vascular tissue and phloem sap proteins of C. maxima, as well as dialyzed vascular tissue and phloem sap proteins of C. moschata PI441726, were resolved on 12% SDS-PAGE gels. The 20-kD regions on these gels were then excised, and the proteins therein were prepared for MS analysis using standard reduction, alkylation, and tryptic digest procedures (Rosenfeld et al., 1992).
All quantification data for Cmo-FTL1 and Cmo-FTL2 and semiquantification data for Cm-FTL1 and Cm-FTL2 were acquired using a Nano-LC-2D system (Eksigent) coupled with an LTQ ion trap mass spectrometer (Thermo Finnigan) with an in-laboratory fabricated fritless reverse phase microcapillary column (75 μm × 180 mm packed with Magic C18AQ, 3 μm 100 Å; Michrom Bio Resources) and vented column configuration.
Each digested sample was transferred by an autosampler to the online trap column (Zorbax 300SB-C18, 5 μm, 0.3 mm × 5 mm; Agilent) and desalted. Peptides were then eluted from the trap, separated by capillary column by reverse phase gradient, at a flow rate of 300 nL per min, and directly sprayed into the mass spectrometer. Buffer compositions used for reverse phase chromatography were as follows: buffer A, 0.1% formic acid in water; buffer B, 0.1% formic acid in 100% acetonitrile. A 120-min (2 to 40% buffer B for 95 min, 40 to 80% buffer B for 12 min, and 80% buffer B for 13 min) or 168-min gradient (2% buffer B for 3 min, 2 to 10% buffer B for 7 min, 10 to 40% buffer B for 130 min, 40 to 80% buffer B for 15 min, and 80% buffer B for 13 min) was used for the Nano-LC-2D/LTQ system.
A high-resolution mass spectrometer (LTQ-FT; Thermo Finnigan) was used to acquire accurate mass spectra of VIGDVVDSFSR from Cm-FTL1 and VIGDVIDSFTK from Cm-FTL2.
Semiquantification of Cm-FTL1/FTL2 and Quantification of Cmo-FTL1/FTL2
Semiquantitative information of Cm-FTL1 and Cm-FTL2 was obtained by monitoring MS/MS of representative peptides VIGDVVDSFSR (m/z 597.3) and VIGDVIDSFTK (m/z 597.3), respectively, plus peptide VEIGGTDLR (m/z 480.3) that is common to both proteins. Cmo-FTL1 and Cmo-FTL2 were quantified using the Protein-AQUA method in which isotopically labeled synthetic peptides were used (Kirkpatrick et al., 2005). Absolute quantification (AQUA) peptides, VIGDVIDSFSR* (SR*; representative of Cmo-FTL1), VIGDVIDSFTR* (TR*; representative of Cmo-FTL2), and VEIGGTDLR* (LR*; common to Cmo-FTL1 and Cmo-FTL2) were prepared by Sigma-Aldrich Genosys (http://www.sigmaaldrich.com/Brands/Sigma_Genosys/Custom_Peptides.html) and used as internal standards for tandem MS analyses. The R* represents Arg residues containing six 13C and four 15N; the synthetic peptides and their y series fragments have 10-D higher mass than the corresponding native peptides and their fragments. Prior to their use in AQUA experiments, all synthetic peptides were tested to confirm that the preparations were devoid of contaminating mono-isotopic peptides. Concentrations of in-gel–digested C. moschata PI441726 proteins were determined on a NanoDrop ND-1000 (NanoDrop Technologies) using the A280 method employing the extinction coefficient for BSA. Based on these measurements, 7 μg of total protein containing 100 fmol of each synthetic peptide was injected into the Nano-LC-2D/LTQ.
For quantification of Cmo-FTL1 and Cmo-FTL2, monitoring of tandem MS and/or multiple reaction monitoring (MRM) of major fragments was used for the native and synthetic peptides. Doubly protonated peptide ions were selected for tandem MS monitoring: m/z 616.3 (TR*), 611.3 (TR), 609.3 (SR*), 604.3 (SR), 485.3 (LR*), and 480.3 (LR). Fragments monitored in MRM were as follows: m/z 1019.5 (y9), 847.4 (y7), and 748.4 (y6) from TR*; m/z 1009.5 (y9), 837.6 (y7), and 738.3 (y6) from TR; m/z 1005.4 (y9), 833.3 (y7), and 734.2 (y6) from SR*; m/z 995.4 (y9), 823.3 (y7), and 724.2 (y6) from SR; m/z 741.3 (y7), 628.3 (y6), and 571.3 (y5) from LR*; and m/z 731.3 (y7), 618.4 (y6), and 561.3 (y5) from LR. Absolute quantification was determined by comparing the abundance of the known internal standard peptide with its native peptide.
For analysis of phloem proteins collected from heterografted C. moschata scions, the following representative C. moschata and C. maxima peptides were monitored in MRM: m/z 616.3 (TR*), 611.3 (TR), 609.3 (SR*), 604.3 (SR), 597.3 (VIGDVVDSFSR and VIGDVIDSFTK for Cm-FTL1 and Cm-FTL2, respectively), 485.3 (LR*), and 480.3 (LR). Note VEIGGTDLR (LR) is common in all C. moschata and C. maxima FTL proteins.
Database Search
Database matching was performed through the use of the Mascot search algorithm (version 2.1; Matrix Science Limited). The sequence database consisted of 155,976 ESTs derived from vascular tissues of C. maxima, C. moschata PI441726, Cucumis sativus, and Sicyos angulatus assembled into ∼51,826 gene sets or consensi. Only statistically significant peptides were considered as a reliable match and were pursued for further analysis. SEQUEST search (Bioworks v2.1; Thermo Electron) was performed to calculate sequence coverage of Cm-FTL1 and Cm-FTL2. Parent mass tolerance of 2.5 D and fragment tolerance of 1 D were used. Only fully tryptic peptides were accepted, and Xcorr cutoff values of 1.5, 2.0, and 2.5 were used for singly, doubly, and triply charged ions. Manual validation was performed for every matched tandem mass spectrum.
Accession Numbers
Gene names and their nucleotide accessions can be found in Supplemental Table 1 online. Accession numbers for Populus trichocarpa genes are gene model identifiers found at the Joint Genome Institute (http://genome.jgipsf.org/Poptr1/Poptr1.home.html).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Methods.
Supplemental Table 1. Sequence Accession Numbers for Members of the FT-TFL Family Analyzed.
Supplemental Figure 1. Comparison of Arabidopsis FT, Cm-FTL1, Cm-FTL2, Cmo-FTL1, and Cmo-FTL2 Conceptual Amino Acid Sequences.
Supplemental Figure 2. Neighbor-Joining Phylogenetic Analysis of 40 FT-TFL Family Members in Angiosperms.
Supplemental Figure 3. Bayesian Phylogenetic Analysis of 40 FT-TFL Family Members in Angiosperms.
Supplemental Figure 4. Nucleotide Sequence Alignment of 40 FT-TFL Family Members in Angiosperms.
Supplemental Figure 5. Typical Sequence Coverage and Identified Peptides for Cm-FTL1 and Cm-FTL2 from LC-MS/MS Data Analysis.
Supplemental Figure 6. Phloem-Specific Expression of Cmo-FTL1/FTL2 in Plants Grown under Noninductive LD and Inductive SD Conditions.
Supplementary Material
Acknowledgments
We thank Shyi-Dong Yeh (National Chung Hsing University, Taiwan) for kindly providing the ZYMV viral expression vector. We also thank Prudence Grandison, Dieter Hermsmeier, and Justin Sweetman for assistance during the early phase of this project. This work was supported by grants from the National Science Foundation (IBN 03151174 to W.J.L. and DBI-0421679 to R.A.J.), the Department of Energy, Division of Energy Biosciences (DE-FG02-94ER20134 to W.J.L.), and the New Zealand Foundation for Research Science and Technology (to T.J.L.). B.X.-C. was supported by a University of California Institute for Mexico and the U.S.–Mexico National Council for Science and Technology Sabbatical Leave Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: William J. Lucas (wjlucas@ucdavis.edu).
Online version contains Web-only data.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615–3626. [DOI] [PubMed] [Google Scholar]
- Ayre, B.G., and Turgeon, R. (2004). Graft transmission of a floral stimulant derived from CONSTANS. Plant Physiol. 135 2271–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow, R., and Dean, C. (2003). Plant sciences. Deciding when to flower. Science 302 1695–1696. [DOI] [PubMed] [Google Scholar]
- Bernier, G. (1988). The control of floral evocation and morphogenesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 175–219. [Google Scholar]
- Bernier, G., and Périlleux, C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3 3–16. [DOI] [PubMed] [Google Scholar]
- Böhlenius, H., Eriksson, S., Parcy, F., and Nilsson, O. (2007). Retraction. Science 316 367. [DOI] [PubMed] [Google Scholar]
- Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A.M., Jansson, S., Strauss, S.H., and Nilsson, O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Haldeman, R., Dolja, V.V., and Restrepo-Hartwig, M.A. (1993). Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J. Virol. 67 6995–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailakhyan, M.K. (1936). New facts for hormonal theory of plant development. Dokl. Akad. Nauk SSSR 13 79–83. [Google Scholar]
- Chailakhyan, M.K. (1937). Concerning the hormonal nature of plant development processes. Dokl. Akad. Nauk SSSR 16 227–230. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., and Sundaresan, V. (2000). ‘Florigen’ enters the molecular age: Long-distance signals that cause plants to flower. Trends Biochem. Sci. 25 236–240. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., and Coupland, G. (2005). Photoperiodic flowering of Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 28 54–66. [Google Scholar]
- Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., and Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco, P., Kapitza, K., Kolasa, A., Buhtz, A., and Kehr, J. (2006). Towards the proteome of Brassica napus phloem sap. Proteomics 6 896–909. [DOI] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Golecki, B., Schulz, A., Carstens-Behrens, U., and Kollmann, R. (1998). Evidence for graft transmission of structural phloem proteins or their precursors in heterografts of Cucurbitaceae. Planta 206 630–640. [Google Scholar]
- Golecki, B., Schulz, A., and Thompson, G.A. (1999). Translocation of structural P proteins in the phloem. Plant Cell 11 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, G., Torres, H., and Pallas, V. (2005). Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long-distance RNA transport system. Plant J. 41 107–116. [DOI] [PubMed] [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Hsu, C.H., Lin, S.S., Liu, F.L., Su, W.C., and Yeh, S.D. (2004). Oral administration of a mite allergen expressed by zucchini yellow mosaic virus in cucurbit species downregulates allergen-induced airway inflammation and IgE synthesis. J. Allergy Clin. Immunol. 113 1079–1085. [DOI] [PubMed] [Google Scholar]
- Hsu, C.Y., Liu, Y., Luthe, D.S., and Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18 1846–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T., Böhlenius, H., Eriksson, S., Parcy, F., and Nilsson, O. (2005). The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309 1694–1698. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Imlau, A., Truernit, E., and Sauer, N. (1999). Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.L., Thomas, C.L., and Maule, A.J. (1998). De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 17 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, D.S., Gerber, S.A., and Gygi, S.P. (2005). The absolute quantification strategy: A general procedure for the quantification of proteins and post-translational modifications. Methods 35 265–273. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lang, A. (1965). Physiology of flower initiation. In Encyclopedia of Plant Physiology, Vol. XV, W. Ruhland, ed (Berlin: Springer-Verlag), pp. 1380–1536.
- Lang, A. (1977). Promotion and inhibition of flower formation in a day-neutral plant in grafts with a short-day plant and a long-day plant. Proc. Natl. Acad. Sci. USA 74 2412–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz, E., Eviatar, T., Rozman, A., Goldshmidt, A., Amsellem, Z., Alvarez, J.P., and Esched, Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.-S., Hou, R.F., and Yeh, S.-D. (2002). Construction of in vitro and in vivo infectious transcripts of a Taiwan strain of Zucchini yellow mosaic virus. Bot. Bull. Acad. Sin. 43 261–269. [Google Scholar]
- Lough, T.J., and Lucas, W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., and Lee, J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5 712–726. [DOI] [PubMed] [Google Scholar]
- Martens, H.J., Roberts, A.G., Oparka, K.J., and Schulz, A. (2006). Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiol. 142 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, R.E.F. (1991). Plant Virology. (San Diego, CA: Academic Press).
- Michaels, S.D., Himelblau, E., Kim, S.Y., Schomburg, F.M., and Amasino, R.M. (2005). Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 137 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97 743–754. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., and Santa Cruz, S. (2000). The great escape: Phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 323–347. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., and Turgeon, R. (1999). Sieve elements and companion cells: Traffic control centers of the phloem. Plant Cell 11 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Rosenfeld, J., Capdevielle, J., Guillemot, J.C., and Ferrara, P. (1992). In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203 173–179. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cázares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Searle, I., and Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., Wright, K.M., Lauterbach, C., Amon, G., Gahrtz, M., Feuerstein, A., Oparka, K.J., and Sauer, N. (2005). Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J. 41 319–331. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Goto, K. (2003). Terminal flower 2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki, S., Matsuo, S., Wong, H.L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316 1033–1036. [DOI] [PubMed] [Google Scholar]
- Truernit, E., Schmid, J., Epple, P., Illig, J., and Sauer, N. (1996). The sink-specific and stress-regulated Arabidopsis STP4 gene: Enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8 2169–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, A., Kobayashi, Y., Goto, K., Abe, M., and Araki, T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46 1175–1189. [DOI] [PubMed] [Google Scholar]
- Yoo, B.C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1962). Physiology of flowering. Science 137 723–731. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A.D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 27 321–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.